Abstract
The inability of proteins to potently penetrate mammalian cells limits their usefulness as tools and therapeutics. When fused to superpositively charged GFP, proteins rapidly (within minutes) entered five different types of mammalian cells with potency up to ∼100-fold greater than that of corresponding fusions with known protein transduction domains (PTDs) including Tat, oligoarginine, and penetratin. Ubiquitin-fused supercharged GFP when incubated with human cells was partially deubiquitinated, suggesting that proteins delivered with supercharged GFP can access the cytosol. Likewise, supercharged GFP delivered functional, nonendosomal recombinase enzyme with greater efficiencies than PTDs in vitro and also delivered functional recombinase enzyme to the retinae of mice when injected in vivo.
Proteins have demonstrated great value as research tools and as human therapeutics. Due to the inability of virtually all proteins to spontaneously enter cells, however, exogenous proteins are predominantly restricted to interaction with extracellular targets and targets accessible through the endocytic pathway. Over the past decade, a variety of reagents for the delivery of proteins into mammalian cells have been developed including lipid-linked compounds (1), nanoparticles (2), and fusions to receptor ligands (3,4). Perhaps the most commonly used method for protein delivery is genetic fusion to protein transduction domains (PTDs) including the HIV-1 transactivator of transcription (Tat) peptide, oligoarginine, and the Drosophila Antennapedia-derived penetratin peptide (5,6). Despite these advances, intracellular targets remain difficult to perturb using exogenous proteins; even modest success can require high concentrations of exogenous protein because of the limited potency of most current methods. Challenges for protein delivery are significantly increased in vivo, where cells in the context intact tissues and organs have proven especially difficult targets for functional protein delivery (7,8). The development of more potent protein transduction platforms would therefore significantly increase the scope of potential applications for protein reagents and therapeutics.
We recently described “supercharged” GFP variants that have been extensively mutated at their surface-exposed residues, resulting in extremely high theoretical net charge magnitudes ranging from −30 to +48 (9). We discovered that superpositively charged GFP variants can enter a variety of mammalian cells by binding to anionic cell-surface proteoglycans and undergoing endocytosis in an energy-dependent and clathrin-independent fashion (10). Further, we observed that superpositive GFPs are able to form stable noncovalent complexes with nucleic acids and that +36 GFP can deliver siRNA and plasmid DNA into a variety of mammalian cell lines without apparent cytotoxicity.
We hypothesized that +36 GFP may also serve as a potent and general platform for the delivery of proteins into mammalian cells. We began by generating a variety of fusion proteins with +36 GFP and observed that these fusions maintain the ability to rapidly (within 15 min) and potently (at low nanomolar concentrations) penetrate mammalian cells without toxicity (Supplementary Figures 1−3).
Next we directly compared the ability of +36 GFP, Tat, Arg10, and penetratin to deliver fused mCherry (11), a red fluorescent protein variant. We generated +36 GFP-mCherry, Tat-mCherry, Arg10-mCherry, and penetratin-mCherry with identical linkers and fusion orientations (Supplementary Figure 4), and incubated these four fusion proteins with HeLa cells, baby hamster kidney cells (BSR cells, a clone of BHK-21 cell line), NIH 3T3 cells, inner medullary collecting duct (IMCD) cells, and rat pheochromocytoma PC12 cells in serum-free media at various concentrations for 4 h at 37 °C.
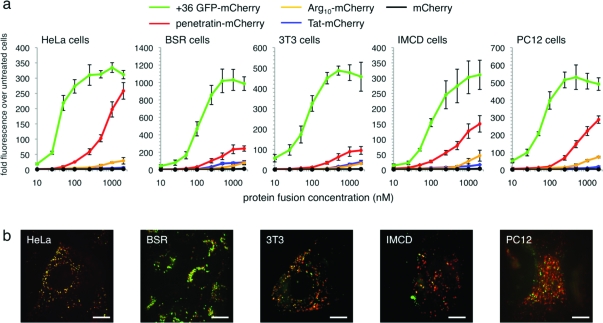
After incubation, cells were washed under conditions confirmed to remove surface-bound protein (Supplementary Figure 5), trypsinized, and assayed for internalized mCherry by flow cytometry (1, panel a). For all five cell lines and at all concentrations tested (10 nM to 2 μM), +36 GFP delivered ∼10- to 100-fold more mCherry than Tat or Arg10. At concentrations of ≤100 nM, +36 GFP also delivered ∼6- to 20-fold more mCherry than penetratin, which approached the potency of +36 GFP only in HeLa cells and only at the highest tested concentrations (1 and 2 μM). These results suggest that +36 GFP is a significantly more potent protein transduction agent than the widely used Tat, Arg10, and penetratin, especially at submicromolar concentrations. We also observed that the presence of serum in culture media modestly decreased (∼2-fold reduction in media containing 50% serum compared with media lacking serum) but did not abrogate the delivery of +36 GFP-mCherry into cells (Supplementary Figure 5). Delivery of mCherry into cells by +36 GFP was confirmed by live-cell confocal fluorescence microscopy (1, panel b) and by comparison with control experiments in which endocytosis is blocked at 4 °C and protein remains surface-bound (Supplementary Figure 6).
Figure 1.
Comparison of mCherry delivery by +36 GFP, Tat, Arg10, and penetratin. a) Flow cytometry of HeLa, BSR, 3T3, PC12, and IMCD cells incubated in the presence of the specified concentrations of +36 GFP-mCherry, Tat-mCherry, Arg10-mCherry, penetratin-mCherry, or wild-type mCherry alone for 4 h at 37 °C. Cells were washed three times with 20 U mL−1 heparin in PBS to remove membrane-bound protein before analysis. Error bars represent the standard error of three independent biological replicates. b) Confocal fluorescence microscopy of live cells incubated with 100 nM +36 GFP-mCherry for 4 h at 37 °C. Red color represents mCherry signal; green color represents +36 GFP signal. The scale bar is 15 μm.
To study the ability of proteins delivered with +36 GFP to access the cytosol, we generated a ubiquitin-+36 GFP fusion in which the C-terminus of ubiquitin was directly followed by +36 GFP. A direct fusion of this type is recognized and processed by cytosolic deubiquitinases (DUBs), and DUB-dependent deubiquitination has previously been used as an indicator of cytosolic exposure (12,13). A mutant form of ubiquitin (G76V) that is not a substrate for DUBs (12) was similarly fused to +36 GFP to distinguish the effect of cytosolic DUBs from nonspecific proteolysis.
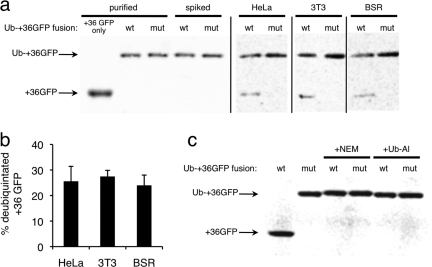
After a 1 h of incubation of HeLa, 3T3, or BSR cells with either 100 nM ubiquitin-+36 GFP or 100 nM ubiquitin G76V +36 GFP, a significant fraction (HeLa, 25 ± 5.8%; 3T3, 27 ± 2.4%; BSR: 24 ± 4.0%) of internalized +36 GFP was deubiquitinated, producing a protein equal in size to +36 GFP (2, panel a). In contrast, in all cases the G76V mutant-+36 GFP fusion was not appreciably cleaved, indicating that this reduction in size does not arise from nonspecific endosomal proteases but instead from the action of DUBs. Ubiquitin-+36 GFP spiked into the cell lysis buffer prior to harvesting untreated cells was not cleaved (2, panel a), indicating that the observed deubiquitination is a result of exposure to cytosolic DUBs and not due to contact with DUBs during the cell-harvesting procedure. Additionally, ubiquitin-+36 GFP was completely deubiquitinated when incubated in HeLa cytosolic extract for 1 h, while the DUB inhibitors N-ethylmaleimide(14) and ubiquitin-aldehyde(15) blocked deubiquitination (2, panel b), further suggesting that the cleavage of ubiquitin-+36 GFP is a result of DUB activity. Collectively, these results demonstrate that some of the ubiquitin-+36 GFP protein fusion can access cytosolic enzymes in three distinct mammalian cell lines.
Figure 2.
Deubiquitination suggests cytosolic exposure of a ubiquitin-+36 GFP fusion protein. a) Western blots using anti-GFP antibodies. Lanes 1−3: purified protein samples of +36 GFP, wild-type ubiquitin-+36 GFP fusion (wt), or G76V mutant ubiquitin-+36 GFP fusion (mut). Lanes 4 and 5: purified protein spiked into HeLa cell lysate to confirm that lysis conditions do not affect fusion protein integrity. Lanes 6−11: the indicated cells were treated with 100 nM of either the wt or mutant ubiquitin-+36 GFP for 1 h and then lysed. b) Mean extent of deubiquitination of wt ubiquitin-+36 GFP fusion protein in HeLa, 3T3, and BSR cells. Error bars reflect the standard deviation of three independent biological replicates. c) In vitro deubiquitination control experiment. Ubiquitin-+36 GFP fusion proteins were incubated in either HeLa cytosolic extract or in HeLa cytosolic extract containing one of two DUB inhibitors, 10 mM N-ethylmaleimide (NEM) or 20 μg mL−1 ubiquitin-aldehyde (Ub-Al), for 1 h at 37 °C.
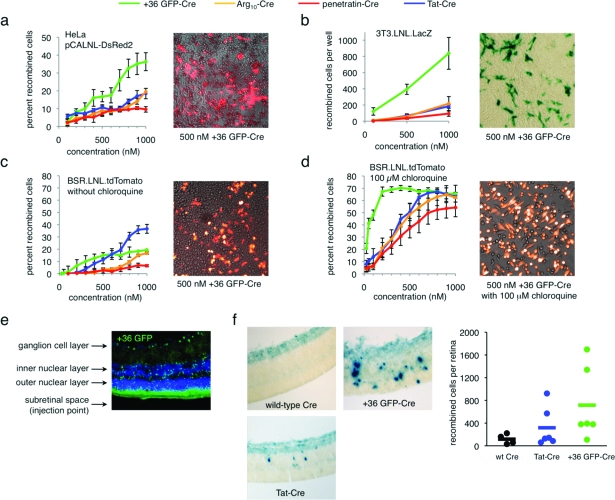
Next we compared the ability of +36 GFP, Tat, Arg10, and penetratin to deliver a functional enzyme, Cre recombinase, into a variety of mammalian cells. Exogenously delivered Cre must escape the endosome, localize to the nucleus, and tetramerize to mediate DNA recombination (16). We generated +36 GFP-Cre, Tat-Cre, Arg10-Cre, and penetratin-Cre fusion proteins (Supplementary Figure 7) and tested their ability to effect recombination in HeLa cells transiently transfected with pCALNL-DsRed2 (17), a DsRed2-based Cre activity reporter plasmid. After incubation with 100−1000 nM of each Cre fusion protein for 4 h in serum-free media, cells were washed to remove surface bound protein and incubated in full media for 48 h. Delivery of Cre was assayed by following DsRed2 expression using flow cytometry and fluorescence microscopy (3, panel a). We observed that +36 GFP-Cre generated ∼2- to 5-fold more recombinants than the corresponding fusions with Tat, Arg10, or penetratin.
Figure 3.
Delivery of active Cre recombinase into mammalian cells in vitro and in vivo. a) Cre-mediated recombination in HeLa cells transiently transfected with pCALNL-DsRed2 and treated with +36 GFP-Cre, Tat-Cre, Arg10-Cre, or penetratin-Cre for 4 h at 37 °C. The image is an overlay of DsRed2 signal and brightfield images of HeLa cells transfected with pCALNL-DsRed2 and treated with 100 nM +36 GFP-Cre. b) Cre-mediated recombination in 3T3.LNL.LacZ cells treated with +36 GFP-Cre, Tat-Cre, Arg10-Cre, or penetratin-Cre for 4 h at 37 °C. The image is of 3T3.loxP.lacZ cells treated with 500 nM +36 GFP-Cre and stained with X-Gal. c) Cre-mediated recombination in BSR.LNL.tdTomato cells treated with +36 GFP-Cre, Tat-Cre, Arg10-Cre, or penetratin-Cre for 4 h at 37 °C. The image is an overlay of tdTomato signal and brightfield images of BSR.LNL.tdTomato cells treated with 100 nM +36 GFP-Cre. d) Identical to panel c but with the addition of 100 μM chloroquine during and after protein treatment. In panels a−d, error bars reflect the standard error of three independent biological replicates. e) Fluorescence microscopy of a retinal section of a CD1 adult mouse injected with 0.5 μL of 100 μM +36 GFP. The retina was harvested and analyzed 6 h after injection. GFP fluorescence is shown in green, and DAPI nuclear stain is shown in blue. f) Retinal sections of neonatal RC::PFwe mouse pups harboring a nuclear LacZ reporter of Cre activity. Three days after injection of 0.5 μL of 40 μM wild-type Cre, Tat-Cre, or +36 GFP-Cre, retinae were harvested, fixed, and stained with X-gal. Dots on the graph represent the total number of recombined cells counted in each retina. The horizontal bar represents the average number of recombined cells per retina for each protein injected (n = 4 for wild-type Cre, n = 6 for Tat-Cre, n = 6 for +36 GFP-Cre).
Cre delivery was further evaluated in a NIH-3T3 cell line harboring an integrated lacZ-based Cre-reporter(5). After incubation, treatment, and washing as described above, these cells were stained with X-Gal to identify recombinants. Consistent with the HeLa cell results, +36 GFP-Cre resulted in more efficient generation of recombinants than Tat, Arg10, or penetratin. The efficacy of +36 GFP-Cre was 10- to 100-fold higher than that of the other Cre fusions at 100 nM, 10-fold higher at 500 nM, and 5-fold higher at 1 μM (3, panel b). These findings together indicate that +36 GFP can deliver substantially more functional Cre than Tat, Arg10, or penetratin in these cell lines.
Next, we used BSR cells to generate a Cre reporter cell line conditionally expressing the tdTomato fluorescent protein after Cre-mediated DNA recombination. Following treatment as described above, Cre-mediated recombination was quantified by flow cytometry. In this cell line, +36 GFP was 2- to 15-fold more effective than Arg10 or penetratin. At low concentrations, +36 GFP delivered modestly higher levels of functional Cre than Tat, while higher concentrations of Tat-Cre generated ∼2-fold more recombinants than +36 GFP-Cre. This cell line exhibits unusual features, including a high metabolic rate (doubling time = ∼12 h(18)), which led us to speculate that endosomal trafficking to lysosomes may be unusually efficient in these BSR cells compared with HeLa- and 3T3-based reporter cells. Indeed, when BSR cells were incubated with the Cre fusions in the presence of 100 μM chloroquine, an inhibitor of lysosomal protein degradation (19), we observed a dramatic increase in the number of recombinants arising from +36 GFP-Cre treatment and modest improvements for Tat-Cre, Arg10-Cre, and penetratin-Cre, such that at all concentrations tested +36 GFP-Cre delivered more recombinase activity than any of the other proteins (3, panel c). These results suggest that the unique cell-penetration potency of +36 GFP can be more fully exploited by extending the time available for internalized protein to escape endosomes, resulting in even higher levels of delivered functional protein. In support of this hypothesis, in vitro assays reveal that while +36 GFP-Cre is not active as the intact fusion (Supplementary Figure 7), at pH 5.5−6.5 cathepsin B, a ubiquitous mammalian endosomal protease, will cleave +36 GFP-Cre to generate +36 GFP and active Cre (Supplementary Figures 7 and 8). At pH 5.0, the exopeptidase activity of cathepsin B is maximized(20), and we observed complete degradation of Cre and the loss of recombinase activity in vitro (Supplementary Figure 8). These findings are consistent with a model in which choloroquine’s ability to prevent complete acidification of lysosomes(21) extends the endosomal lifetime of active Cre and thereby enables a larger amount of the internalized enzyme to escape endosomes.
Finally, we tested +36 GFP as a protein delivery agent in vivo. First we examined the tissue penetration of +36 GFP in the adult mouse retina. We injected 0.5 μL of 100 μM +36 GFP into the subretinal space of CD1 adult mice. After 6 h, the retinae were harvested and analyzed by fluorescence microscopy (2, panel d). Most of +36 GFP signal was observed near the photoreceptor outer segments, but significant signal was observed throughout the retina, including all three nuclear layers (the outer, inner, and ganglion cell layers) as well as in the cell processes. To test the ability of +36 GFP to deliver functional protein in vivo, we injected +36 GFP-Cre into the subretinal space of RC::PFwe mouse p0 pups containing a LoxP-flanked transcriptional terminator upstream of a nuclear lacZ reporter gene (22). Three days after injection of 0.5 μL of 40 μM wild-type Cre, Tat-Cre, or +36 GFP-Cre, retinae were harvested, fixed, and stained with X-gal (2, panel e). Injection of +36 GFP-Cre generated an average of 715 recombined cells per injected retina (n = 6), Tat-Cre generated an average of 318 recombined cells (n = 6), and wild-type Cre generated an average of 117 recombined cells per retina (n = 4). To our knowledge, this is the first report of functional delivery of an enzyme into retinal cells in vivo.
We also performed additional complete sets of mCherry and Cre delivery experiments in vitro using +36 GFP, Tat, and oligoarginine fusion proteins constructed with linkers and fusion orientations distinct from the examples presented above. For both sets of additional experiments we observed results that are qualitatively similar to those in 3 (Supplementary Figures 9 and 10). The delivery of mCherry by +36 GFP in these alternative fusion proteins was ∼10- to 100-fold more potent than mCherry delivery by either Tat or oligoarginine, and the delivery of functional Cre recombinase by +36 GFP in these alternative fusion proteins was ∼3- to 5-fold more potent than Cre delivery by Tat or oligoarginine. These results collectively indicate that the findings described in this work are not unique to a specific protein construct but instead are more generally applicable.
In conclusion, side-by-side comparisons of +36 GFP, Tat, Arg10 and penetratin fused to mCherry or Cre recombinase revealed that fusions with supercharged GFP result in dramatically higher levels of internalized protein (up to ∼100-fold) and in significantly greater efficiencies of Cre-induced recombination (up to ∼10-fold) than three currently used protein transduction domains in a variety of mammalian cell lines. Functional Cre recombinase can also be delivered to cells upon injection in vivo using +36 GFP. These results collectively demonstrate the potential of supercharged proteins as an unusually potent in vitro and in vivo protein delivery platform.
Methods
For complete experimental methods including details of cell lines, cell culture, protein purification, deubiquitination assay, and all sequences of all proteins, see Supporting Information.
Live-Cell Imaging
Cells were plated onto 35-mm glass-bottom microwell dishes with a No. 1.5 cover glass (MatTek) at a density of 106 cells per plate. After 18 h, cells were washed once with cold PBS and incubated with protein in serum-free DMEM. After incubation, cells were washed three times with cold 20 U mL−1 heparin in PBS to remove membrane-bound protein, imaged in prewarmed HEPES imaging solution (1 mM MgCl2, 5 mM KCl, 5 mM CaCl2, 150 mM NaCl, 1.9 g L−1 glucose, 1.9 g L−1 albumin, 20 mM HEPES, pH 7.4, 37 °C). Cells were imaged on an Olympus IX71 spinning disk confocal microscope on a heated stage with a 100X objective lens. GFP and mCherry were visualized with a 491 and 561 nm excitation laser, respectively. Images were prepared using OpenLab software.
For live-cell images of Cre reporter cells, cells were treated with 500 nM +36 GFP-Cre as described below for analysis of Cre recombination and imaged on an Olympus IX51 fluorescent microscope with a DP30BW black and white camera. Images are false-color overlays of the fluorescent signal on a bright-field image. LacZ-positive cells were imaged on an IX70 microscope under bright-field illumination with a DP70 camera.
Flow Cytometry and mCherry Delivery Assays
Cells were plated onto a 48-well plate at a density of 5 × 105 cells per well. After 18 h, cells were washed once with cold PBS and incubated with protein in serum-free DMEM. After incubation, cells were washed three times with cold 20 U mL−1 heparin in PBS to remove membrane-bound protein, trypsinized, resuspended in 500 μL of full media and placed on ice. Cells were analyzed on either a LSRII or Fortessa flow cytometer (BD Biosciences) for GFP internalization (ex, 488 nm) or mCherry internalization (ex, 561 nm). Cells were gated for live cells and at least 5 × 103 live cells were analyzed for each treatment. Data was analyzed with FlowJo software (Tree Star, Inc.)
In Vitro Cre Delivery Assays
HeLa cells were plated at 3 × 104 cells per well in 48-well plates. After 16 h, cells were transfected with pCALNL-DsRed2(17) using Effectene transfection reagent (Qiagen). After incubation with 100−1000 nM of each Cre fusion protein for 4 h in serum-free DMEM, cells were washed three times with 20 U mL−1 heparin in PBS and incubated in full media for 48 h. Delivery of Cre was assayed by following DsRed2 expression using flow cytometry and fluorescence microscopy.
Cre reporter 3T3 cells were plated at 1 × 105 cells per well in 48-well plates. After 16 h, cells were incubated with various concentrations of protein for 4 h in serum-free media. Cells were washed with three times with 20 U mL−1 heparin in PBS and incubated in full media for 48 h. Recombined cells were quantified by X-gal staining and manual counting.
BSR cells were obtained from Matthias Schnell (Thomas Jefferson University). A pQCXIX MMLV retrovirus (Clontech) containing the tdTomato cre reporter construct was generated by subcloning the tdTomato gene (Clontech) into a pCALNL backbone(17) and packaged using Plat-E cells(23). BSR cells were infected with retrovirus and integrants were selected for 1 week in the presence of 1 mg mL−1 G418 (Sigma). BSR.LNL.tdTomato cells were plated at 1 × 105 cells per well in 48-well plates. After 16 h, cells were incubated with various concentrations of protein for 4 h in serum-free media. Cells were washed with three times with 20 U mL−1 heparin in PBS and incubated in full media for 48 h.
For chloroquine treatment of BSR cells, cells were incubated with Cre fusion proteins for 4 h in serum-free media containing 100 μM chloroquine, washed three times with 20 U mL−1 heparin in PBS, and incubated 12 h in full media containing 100 μM chloroquine. Following this incubation, cells were washed once with PBS and incubated a further 36 h in full media without chloroquine. Delivery of Cre was assayed by following tdTomato expression using flow cytometry and fluorescence microscopy.
For fluorescent Cre reporters, recombinants were identified by flow cytometry as those cells of the live-cell population that exhibited fluorescence significantly higher than that of nontreated reporter cells. Typically, the recombined population exhibited fluorescence at least 10-fold higher than the nonrecombined cells and were readily detected as a distinct subpopulation. Fluorescence gates were drawn accordingly to quantitate recombined and nonrecombined cells.
In Vivo Cre Delivery
RC::PFwe mice were obtained from Susan Dymecki (Harvard University). All of the experiments in this study were approved by the Institutional Animal Care and Use Committee at Harvard University. Adult CD1 mice were subretinally injected with 0.5 μL of 100 μM +36 GFP. After 6 h, the retinae were harvested and analyzed by fluorescence microscopy. RC:PFwe p0 pups were subretinally injected with 0.5 μL of 40 μM wtCre, Tat-Cre, or +36 GFP-Cre. After 72 h, retinae were harvested and fixed with 0.5% glutaraldehyde. Fixed retinae were stained with X-gal overnight and embedded in 50% OCT, 50% of 30% sucrose and stored at −80 °C. Retinae were cut into 30 μm sections and imaged for X-gal staining on a Zeiss Axiophot brightfield microscope with a Nikon CXM-1200F camera. Delivery of Cre was assayed by manually counting LacZ+ cells.
Acknowledgments
We thank Steven Dowdy (UCSD) for the 3T3 Cre reporter cell line. This work was supported by the National Institutes of Health/NIGMS (R01 GM 065400) and by the Howard Hughes Medical Institute. J.J.C. is supported by a National Science Foundation Graduate Student Fellowship. D.B.T. is supported by a National Defense Science and Engineering Fellowship.
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Zelphati O.; Wang Y.; Kitada S.; Reed J. C.; Felgner P. L.; Corbeil J. (2001) Intracellular delivery of proteins with a new lipid-mediated delivery system. J. Biol. Chem. 276, 3735103–35110. [DOI] [PubMed] [Google Scholar]
- Hasadsri L.; Kreuter J.; Hattori H.; Iwasaki T.; George J. M. (2009) Functional protein delivery into neurons using polymeric nanoparticles. J. Biol. Chem. 284, 6972–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel C. A.; Foster S. A. (1986) Mannose 6-phosphate receptor-mediated endocytosis of acid hydrolases: internalization of beta-glucuronidase is accompanied by a limited dephosphorylation. J. Cell Biol. 103, 51817–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk S. S.; Luchniak A.; Uysal S.; Brawley C. M.; Rock R. S.; Kossiakoff A. A. (2009) An engineered substance P variant for receptor-mediated delivery of synthetic antibodies into tumor cells. Proc. Natl. Acad. Sci. U.S.A. 106, 2711011–11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadia J. S.; Dowdy S. F. (2003) Modulation of cellular function by TAT mediated transduction of full length proteins. Curr. Protein Pept. Sci. 4, 297–104. [DOI] [PubMed] [Google Scholar]
- Heitz F.; Morris M. C.; Divita G. (2009) Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br. J. Pharmacol. 157, 2195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S. R.; Xu G.; Becker-Hapak M.; Ma M.; Dowdy S. F.; McLeod H. L. (2006) The kinetics and tissue distribution of protein transduction in mice. Eur. J. Pharm. Sci. 27, 4311–319. [DOI] [PubMed] [Google Scholar]
- Caron N. J.; Torrente Y.; Camirand G.; Bujold M.; Chapdelaine P.; Leriche K.; Bresolin N.; Tremblay J. P. (2001) Intracellular delivery of a Tat-eGFP fusion protein into muscle cells. Mol. Ther. 3, 3310–318. [DOI] [PubMed] [Google Scholar]
- Lawrence M. S.; Phillips K. J.; Liu D. R. (2007) Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129, 3310110–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaughton B. R.; Cronican J. J.; Thompson D. B.; Liu D. R. (2009) Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 156111–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C.; Campbell R. E.; Steinbach P. A.; Giepmans B. N.; Palmer A. E.; Tsien R. Y. (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 121567–1572. [DOI] [PubMed] [Google Scholar]
- Loison F.; Nizard P.; Sourisseau T.; Le Goff P.; Debure L.; Le Drean Y.; Michel D. (2005) A ubiquitin-based assay for the cytosolic uptake of protein transduction domains. Mol. Ther. 11, 2205–214. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. (2005) Ubiquitin fusion technique and related methods. Methods Enzymol. 399, 777–799. [DOI] [PubMed] [Google Scholar]
- Borodovsky A.; Kessler B. M.; Casagrande R.; Overkleeft H. S.; Wilkinson K. D.; Ploegh H. L. (2001) A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20, 185187–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A.; Rose I. A. (1987) Ubiquitin-aldehyde: a general inhibitor of ubiquitin-recycling processes. Proc. Natl. Acad. Sci. U.S.A. 84, 71829–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F.; Gopaul D. N.; van Duyne G. D. (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389, 664640–46. [DOI] [PubMed] [Google Scholar]
- Matsuda T.; Cepko C. L. (2007) Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. U.S.A. 104, 31027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L. S.; Pater M. M.; Hutchinson N. I.; di Mayorca G. (1984) Transformation by purified early genes of simian virus 40. Virology 133, 2341–353. [DOI] [PubMed] [Google Scholar]
- Seglen P. O.; Grinde B.; Solheim A. E. (1979) Inhibition of the lysosomal pathway of protein degradation in isolated rat hepatocytes by ammonia, methylamine, chloroquine and leupeptin. Eur. J. Biochem. 95, 2215–225. [DOI] [PubMed] [Google Scholar]
- Almeida P. C.; Nantes I. L.; Chagas J. R.; Rizzi C. C.; Faljoni-Alario A.; Carmona E.; Juliano L.; Nader H. B.; Tersariol I. L. (2001) Cathepsin B activity regulation. Heparin-like glycosaminogylcans protect human cathepsin B from alkaline pH-induced inactivation. J. Biol. Chem. 276, 2944–951. [DOI] [PubMed] [Google Scholar]
- Anderson R. G.; Falck J. R.; Goldstein J. L.; Brown M. S. (1984) Visualization of acidic organelles in intact cells by electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 81, 154838–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farago A. F.; Awatramani R. B.; Dymecki S. M. (2006) Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron 50, 2205–218. [DOI] [PubMed] [Google Scholar]
- Morita S.; Kojima T.; Kitamura T. (2000) Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 121063–1066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.