Molecular steps of G-overhang generation at human telomeres and its function in chromosome end protection
Weihang Chai et al show that generation of G-strand overhangs, which are important for telomere protection and for promoting telomerase activity, is dynamically regulated during the cell cycle, involving both DNA polymerization and processing mechanisms.
Keywords: CDK1, cell cycle, G-overhang, polα, telomere
Abstract
Telomeric G-overhangs are required for the formation of the protective telomere structure and telomerase action. However, the mechanism controlling G-overhang generation at human telomeres is poorly understood. Here, we show that G-overhangs can undergo cell cycle-regulated changes independent of telomerase activity. G-overhangs at lagging telomeres are lengthened in S phase and then shortened in late S/G2 because of C-strand fill-in, whereas the sizes of G-overhangs at leading telomeres remain stable throughout S phase and are lengthened in G2/M. The final nucleotides at measurable C-strands are precisely defined throughout the cell cycle, indicating that C-strand resection is strictly regulated. We demonstrate that C-strand fill-in is mediated by DNA polymerase α (polα) and controlled by cyclin-dependent kinase 1 (CDK1). Inhibition of CDK1 leads to accumulation of lengthened G-overhangs and induces telomeric DNA damage response. Furthermore, depletion of hStn1 results in elongation of G-overhangs and an increase in telomeric DNA damage. Our results suggest that G-overhang generation at human telomeres is regulated by multiple tightly controlled processes and C-strand fill-in is under the control of polα and CDK1.
Introduction
Maintaining functional telomeres is essential for preserving genome stability. Human telomere DNA is composed of many kilobases of double-stranded (TTAGGG)n repetitive sequence and terminates with a single-stranded G-rich overhang (G-overhang) at the 3′ end. To protect telomere ends from being recognized as damaged DNA, telomeres form special nucleoprotein structures termed t-loops by inserting the ss G-overhangs into the ds telomere DNA repeats (Griffith et al, 1999; Stansel et al, 2001). Disruption of this protective structure activates DNA damage response pathways that initiate cell cycle arrest, senescence, or apoptosis (d'Adda di Fagagna et al, 2004).
Formation of the t-loop requires ds telomere DNA, ss G-overhang, and a number of proteins, which bind to ds and ss telomere DNA and promote chromosome end protection. G-overhangs have been observed in most organisms studied from yeast to humans (Wellinger et al, 1996; Makarov et al, 1997; Riha et al, 2000; Jacob et al, 2001; Chai et al, 2005; Raices et al, 2008). In addition to its essential function in t-loop formation, G-overhang generation seems to regulate telomerase activity in yeast, as inappropriate generation of G-overhang affects telomerase elongation of telomeres (Dionne and Wellinger, 1998; Bonetti et al, 2009). Moreover, the G-overhang is the binding site for ss telomere DNA-binding proteins that are central players in telomere end protection (Baumann and Cech, 2001; Lei et al, 2003; Loayza et al, 2004). Finally, in tumour cells using homologous recombination to maintain telomeres, the ss G-overhang DNA is presumed to initiate strand invasion (Lundblad, 2002; Muntoni and Reddel, 2005). Thus, deciphering the mechanism of G-overhang generation is critical for understanding the mechanisms for maintaining functional telomeres and is also essential for understanding telomere elongation through telomerase-dependent or -independent pathways.
Most studies on G-overhang generation come from lower eukaryotes that possess short overhangs. G-overhangs at budding yeast telomeres are short (∼10–14 nt) for the majority of the cell cycle, but their length increases transiently in the late S phase at the time of telomere replication (Wellinger et al, 1993). C-strands are then filled in, presumably by replication or repair synthesis, yielding very short G-overhangs (Wellinger et al, 1993; Larrivee et al, 2004). This cell cycle-dependent control of G-overhang formation is mediated by cyclin-dependent kinase 1 (CDK1) (Frank et al, 2006; Vodenicharov and Wellinger, 2006). Multiple nucleases and helicases have been identified in yeast that contribute to C-strand resection (Bonetti et al, 2009). The importance of G-overhang generation is underscored by the observation that telomerase extension of telomeres is regulated by G-overhang generation and C-strand fill-in in yeast (Nugent et al, 1996; Diede and Gottschling, 1999).
On the other hand, very little is known about the nature of G-overhang generation in mammalian cells. G-overhangs exist on most chromosome ends, with the sizes ranging from 12 nt to several hundred nt (Chai et al, 2006a; Zhao et al, 2008). A number of telomere-binding proteins including TRF2, the Mre11/Rad50/Nbs1 complex, Pot1, Stn1, and Ctc1 have been shown to influence G-overhang length (van Steensel et al, 1998; Chai et al, 2006b; Miyake et al, 2009; Surovtseva et al, 2009). However, the mechanism for G-overhang generation is unclear and the nucleases/helicases responsible for processing telomere ends remain unidentified. A recent report shows that G-overhang sizes transiently increase during the late S/G2 phase in telomerase-positive cells (Zhao et al, 2009), yet it is unknown whether this transient increase of G-overhang is important for telomere maintenance. As many proteins regulate telomerase in cis (Smogorzewska and de Lange, 2004; de Lange, 2005; Palm and de Lange, 2008), it is therefore imperative to determine the molecular machinery responsible for G-overhang generation and whether the activities of these proteins are subjected to regulation by proper generation of G-overhangs in human cells.
The ss G-overhang DNA is protected by telomeric ssDNA-binding proteins. These proteins are essential for protecting chromosome ends and have been identified in a wide range of organisms including vertebrates, plants, worms, ciliates, and yeasts. Their DNA-binding domains consist of structurally conserved oligonucleotide/oligosaccharide-binding folds. In yeast, the Cdc13/Stn1/Ten1 complex protects telomere ends in multiple ways by repressing telomerase activity, restricting extensive nuclease degradation of C-strand, and mediating C-strand fill-in (Nugent et al, 1996; Grandin et al, 1997, 2001,Grandin et al, 1997, 2001; Chandra et al, 2001; Lustig, 2001; Pennock et al, 2001; Puglisi et al, 2008). Dysfunction of Cdc13 leads to extensive C-strand degradation and G-overhang elongation (Garvik et al, 1995; Nugent et al, 1996), and partial loss of functional alleles of all three proteins cause telomere elongation (Chandra et al, 2001). The Stn1 homologs in Schizosaccharomyces pombe and Arabidopsis thaliana are essential for chromosome end protection (Martin et al, 2007; Song et al, 2008). A mammalian complex similar to yeast Cdc13/Stn1/Ten1 is formed by three RPA-like proteins Ctc1/Stn1/Ten1 (Miyake et al, 2009; Surovtseva et al, 2009). This complex binds to the ssDNA in a sequence-independent manner (Miyake et al, 2009). Although one report shows that hStn1 associates with another telomere capping protein TPP1 and that C-terminal deletion of hStn1 results in telomere elongation (Wan et al, 2009), another group shows that the CST complex is involved in telomere protection in a way redundant to the Pot1 pathway (Miyake et al, 2009).
To gain insights into the G-overhang generation and telomerase regulation in human cells, we analysed the cell cycle-regulated G-overhang dynamics. We found that the global G-overhang length gradually increased during S phase in both telomerase-positive and -negative cells. Further analysis of separated leading and lagging telomeres from synchronized HeLa cells revealed that G-overhangs at lagging telomeres were lengthened in S phase and then were shortened at late S/G2 because of delayed C-strand fill-in, whereas the sizes of G-overhangs at leading telomeres remained stable throughout S phase and were later lengthened in G2/M. No further shortening was detected at leading overhangs, suggesting that C-strand fill-in might be absent at leading telomeres. The final nucleotides at measurable C-strands remained precisely defined throughout the cell cycle, indicating that C-strand resection was tightly regulated. We further demonstrated that the delayed C-strand fill-in required lagging strand polymerases and was controlled by CDK1. Inhibition of CDK1 activity at late S/G2 phase led to accumulation of ss G-overhangs and triggered an ATM/ATR-dependent DNA damage response at telomeres, revealing a previously unidentified function of CDK1 in protecting chromosome ends. Furthermore, depletion of hStn1 resulted in elongation of G-overhangs and an increase in DNA damage at telomeres. Collectively, our results provided insights into the detailed molecular steps of G-overhang formation at leading and lagging telomeres, as well as the regulation of C-strand fill-in at human telomeres.
Results
The cell cycle-regulated G-overhang dynamics at human telomeres is independent of telomerase activity
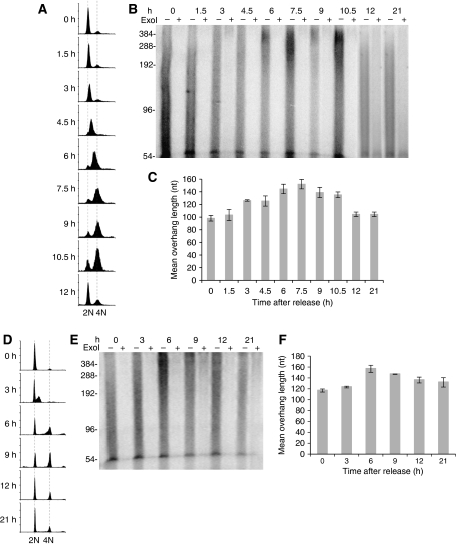
To determine whether G-overhangs at human telomeres undergo cell cycle-regulated changes, we synchronized HeLa cells at the G1/S phase boundary using the double-thymidine block. Cells were then released from the block and collected at 1.5 h intervals (Figure 1A). Genomic DNA was extracted from synchronized cells and the telomere overhang protection assay was carried out to measure the mean overhang length as previously described (Chai et al, 2005). We observed that the global G-overhang lengths gradually increased during S phase, peaked at late S and early G2 phase (6–7.5 h after release), then gradually decreased during the G2 phase (9–10.5 h), reaching normal size in the next cycle (12 and 21 h) (Figure 1B and C). Our results suggest that long G-overhangs are generated during the S phase, and C-strands are then filled in during late S/G2 phase, consistent with recently reported data (Zhao et al, 2009).
Figure 1.
Global telomeric G-overhangs undergo cell cycle-regulated dynamics regardless of telomerase expression. (A–C) HeLa cells. (D–F) IMR90 cells. (A, D) FACS analysis of DNA content in cells synchronized at the G1/S boundary and then released to S phase. Cells were collected at indicated time points for FACS analysis to determine the DNA content. A portion of IMR90 cells were permanently arrested at G0 and exited the cell cycle. (B, E) Measurement of G-overhang sizes using the overhang protection assay. The smears represent the heterogeneity of overhang sizes. ExoI digests 3′ overhangs and the ExoI plus (+) lanes show the background. The ∼54 nt band has been shown to be non-specific (Chai et al, 2005) and is not eliminated in the quantitations because its contribution to the overhang size is minimal. (C, F) Quantitation of the weighted mean lengths of G-overhangs at indicated time points during the cell cycle from (B, E). Results are representative of at least three independent experiments. Error bars represent s.d.
To determine whether telomerase is essential for the S phase-specific G-overhang lengthening, we examined the G-overhang dynamics in telomerase-negative cells. Human fetal lung fibroblasts IMR90 were subjected to synchronization using serum starvation followed by aphidicolin treatment to block the cells at the G1/S boundary (Figure 1D). Aphidicolin was then removed and cells were released into S phase and collected at 3 h intervals. Surprisingly, G-overhang length in IMR90 cells increased during S phase, peaked by late S/G2 phase, then decreased as cells proceeded to the subsequent stages of the cell cycle (Figure 1E and F). These cell cycle-regulated G-overhang dynamics were then confirmed by non-denaturing in-gel hybridization (Supplementary Figure S1). Thus, it appears that G-overhangs of human telomeres undergo dynamic changes during the cell cycle regardless of telomerase activity. However, our observation of G-overhang dynamics in telomerase-negative cells does not exclude the contribution of telomerase activity in G-overhang increase in telomerase-positive cells.
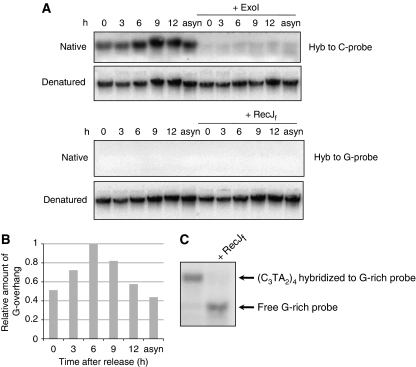
We next determined whether the observed increase of G-overhang sizes during S phase was caused by the collapse of replication forks before reaching chromosome ends. If so, long 3′ G-overhangs would be generated at lagging daughter telomeres whereas 5′ C-rich overhangs would be generated at leading daughter telomeres (Verdun and Karlseder, 2006). The 5′ C-rich overhangs could be detected by hybridizing DNA to oligonucleotides complementary to the C-strand, that is, the G-rich probe. To test this possibility, we hybridized genomic DNA isolated from synchronized cells collected at different time points during the cell cycle to G-rich probe under native conditions. No signal was detected when G-rich probe was hybridized to DNA collected throughout the cell cycle, whereas C-rich probe produced strong ExoI-sensitive signals (Figure 2A and B). These results indicate that C-rich overhangs were not generated during replication and that the increase of G-overhang was not due to the collapse of replication fork. The absence of hybridization signal with G-rich probe was not due to the failure of hybridization, as the same G-rich probe hybridized to a single-stranded C-rich oligo (Figure 2C). To conclude, the extended G-overhangs detected in S phase were not due to the collapse of replication fork, but rather caused by telomerase elongation and/or C-strand resection.
Figure 2.
The extended G-overhang is not due to the collapse of replication fork. (A) No 5′ C-rich overhang was detected during the cell cycle. Genomic DNA isolated from synchronized HeLa cells were hybridized to an 18-mer G-rich probe for detecting C-rich overhangs and hybridized to an 18-mer C-rich probe for detecting G-overhangs under native conditions. To determine the hybridization signals contributed by G- or C-rich overhangs, DNA was digested with 3′ → 5′ exonuclease ExoI, which specifically removes 3′ G-overhangs, or the 5′ → 3′ exonuclease RecJf, which specifically removes 5′ C-rich overhangs (Lovett and Kolodner, 1989) before hybridization. The same gels were then denatured to determine the total telomere signal. Asyn, asynchronized HeLa cells. (B) Quantitation of relative amount of G-overhangs from (A). (C) Hybridization of G-rich probe to (C3TA2)4 oligo. The ss (C3TA2)4 oligo was hybridized to G-rich probe under the native condition identical to (A) and separated in 20% native 0.5 × TBE polyacrylamide gel.
Distinct G-overhang dynamics exist at leading and lagging daughter telomeres during the cell cycle
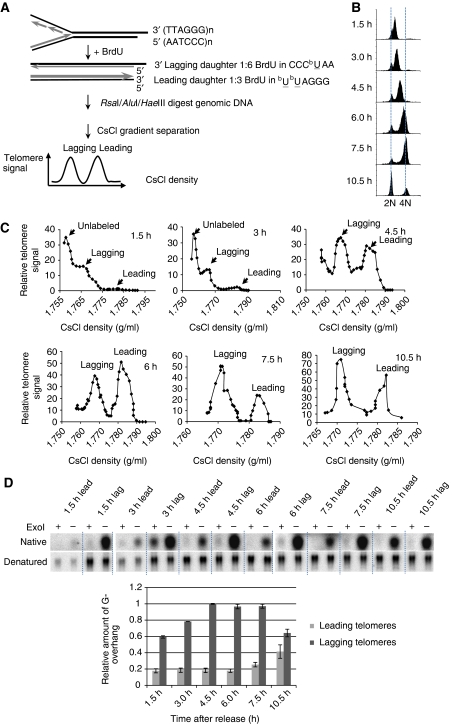
Replication of telomeric DNA by leading and lagging synthesis machinery generates two distinct termini at daughter telomeres. Although lagging telomeres possess G-overhangs because of the inability of lagging strand synthesis machinery to fill in the gap caused by the removal of the final RNA primer, the leading telomeres are initially blunt ended. A previous report suggested that leading telomeres are rapidly processed during S phase to allow telomerase elongation of most telomeres (Zhao et al, 2009). As telomerase elongation only requires a very short overhang (⩽6 nt), and such short overhangs are undetectable by the overhang protection assay, there is a possibility that the observed G-overhang dynamics (Figure 1) might have resulted from lagging telomeres only. To test this possibility, we purified leading and lagging telomeres during the cell cycle using CsCl ultracentrifugation (Chai et al, 2006a) (Figure 3A) and then applied non-denaturing in-gel hybridization (Wellinger et al, 1993) to measure the abundance of leading and lagging G-overhangs including short and long ones.
Figure 3.
G-overhangs at leading and lagging telomeres exhibit different dynamics during the cell cycle. (A) Scheme of separating leading and lagging telomeres. (B) FACS analysis of DNA content in synchronized HeLa released to S phase. (C) Separated leading and lagging telomeres from G1/S synchronized HeLa cells released into S phase for indicated time. (D) The abundance of G-overhangs was measured by in-gel hybridization assay. Results are from three experiments. Error bars represent s.d.
Synchronized HeLa cells were released into media containing 5′-bromo-2′-deoxyuridine (BrdU) and cells were collected at different time points during the cell cycle (Figure 3B). Leading daughter telomeres incorporate twice as much BrdU as lagging daughter telomeres, and thus are heavier than lagging telomeres, allowing separation of the two telomeres with CsCl density gradient ultracentrifugation (Figure 3C). Separated leading and lagging telomeres were then purified and analysed using the non-denaturing in-gel hybridization assay. Figure 3D shows that overhangs at lagging telomeres exhibited elongation during S phase and then shortened during late S/G2. Interestingly, much shorter overhangs (1/6–1/5 of lagging at 4.5 h) were present at leading telomeres during S phase, which remained constant throughout the S phase and then increased in G2/M (Figure 3D). No subsequent shortening of leading strand G-overhangs was observed (Figure 3D). These results suggest that different molecular mechanisms control the generation of mature G-overhangs at the ends of leading and lagging telomeres. Lagging telomeres possess long G-overhangs during S phase, which are then shortened during late S/G2. Meanwhile, a rapid initial processing followed by telomerase extension shortly after replication creates overhangs at leading telomeres. These overhangs are further elongated in G2/M. At present, the mechanism of the G2-specific lengthening of leading overhangs is unknown, necessitating further investigation into the detailed molecular steps for G-overhang generation at the two chromosome ends.
The terminal nucleotides at C-strands from measurable telomeres remain tightly regulated throughout the cell cycle
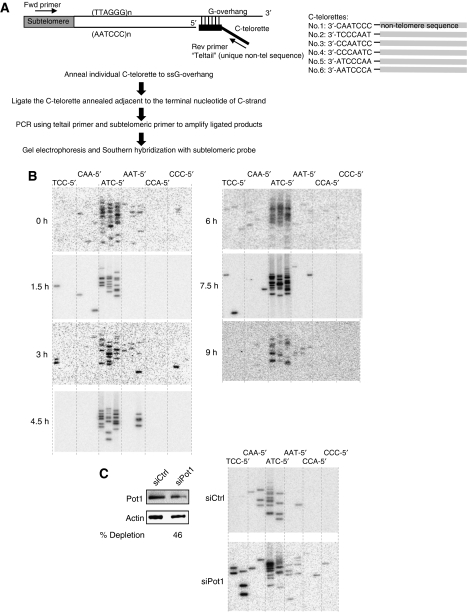
Analysis of asynchronous cells has shown that the last nucleotide at the 5′ end of C-strand is tightly regulated, with about 70–80% of telomeres preferentially ending with 5′-CTA (Sfeir et al, 2005). To determine whether this preference is regulated by the cell cycle, we used single telomere length analysis (STELA) (Baird et al, 2003; Sfeir et al, 2005) to determine the terminal nucleotide of the telomeric C-strand throughout the cell cycle. Six different C-telorettes comprising a unique sequence followed by telomeric repeat homology were individually annealed to the ss G-overhang of XpYp telomeres and then ligated to the 5′ end of the C-rich strands of the duplex telomeric DNA. The ligation products were amplified by PCR with primers specific for XpYp subtelomeres (Figure 4A). Using DNA from synchronized HeLa cells released into S phase, we found that most ligation products ended with 5′-CTA (Figure 4B), same as the terminal nucleotide detected in asynchronous cells (Sfeir et al, 2005). These results indicate that the terminal nucleotide at C-strand remains tightly controlled throughout the cell cycle. As a control, we performed STELA using DNA isolated from Pot1-depleted cells and showed that partial depletion of Pot1 randomized the last C (Figure 4C), consistent with previous report (Hockemeyer et al, 2005).
Figure 4.
The last nucleotide at C-strand is specifically defined throughout the cell cycle. (A) Scheme of STELA. (B) The specificity of the last nucleotide at C-strand determined by STELA with DNA isolated at different time points during the cell cycle from synchronized HeLa cells. The same set of DNA in Figure 1B was used. (C) Randomization of the last C in HeLa with partial depletion of Pot1. HeLa was transfected one time with Pot1 siRNA. Cells were collected 72 h after transfection for western blot and STELA. Percent of depletion is indicated below the blot.
As the XpYp telomere replicates between 2 and 4 h after release into S phase (Zhao et al, 2009), the last Cs at 1.5 h after release mainly arose from unreplicated XpYp telomere and should end precisely with 5′-CTA. The last Cs measured at 3 h resulted from a mixture of newly replicated and unreplicated XpYp, and the last Cs measured at 4.5 h and afterward represented fully replicated telomeres. As C-telorettes only anneal to G-overhangs of ⩾6 nt length, STELA excludes unprocessed blunt-ended leading telomeres, processed leading telomeres with <6 nt overhangs, and unprocessed RNA primer-containing lagging telomeres. At present, the population of these unmeasurable XpYp telomeres is unknown. Nonetheless, the specificity of last Cs observed here represent chromosome ends from processed leading telomeres containing overhangs ⩾6 nt plus processed lagging telomeres in which RNA primers were removed.
Lagging strand synthesis machinery is required for the late S/G2-specific C-strand fill-in
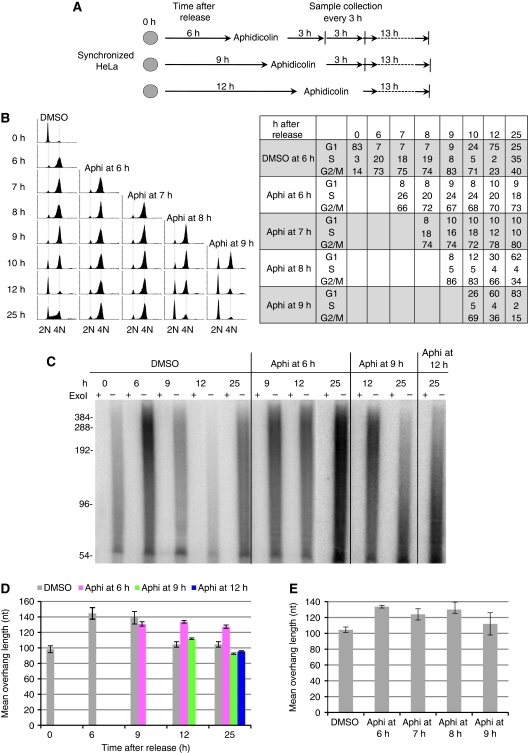
ChIP analysis has revealed that the lagging strand synthesis proteins reassociate with telomeres during the G2 phase after telomere DNA has completed replication (Verdun and Karlseder, 2006). We therefore reasoned that the lagging strand synthesis machinery may mediate the delayed C-strand fill-in at late S/G2 for generating proper G-overhangs. To test this hypothesis, we determined the effects of inhibiting lagging strand synthesis machinery on G-overhang dynamics. Aphidicolin, a specific inhibitor of DNA polymerases polα and δ, was used to block DNA synthesis at late S/G2 phase after the majority of telomere DNA had completed replication. FACS analysis showed that the majority of cells were in late S/G2 at 6 h after release from the G1/S arrest (Figure 5B; DMSO, 6 h). Aphidicolin was then added at this time point, and cells were cultured for an additional 3, 6, and 19 h in the presence of aphidicolin (Figure 5A). Remarkably, G-overhangs in aphidicolin-treated cells remained lengthened even 25 h after release, whereas G-overhangs in control cells already shortened by 12 h (Figure 5C and D). These results suggest that inhibition of polα/δ blocked the delayed telomeric C-strand fill-in. Therefore, we conclude that polα/δ is required for C-strand fill-in.
Figure 5.
The lagging strand synthesis machinery is required for C-strand fill-in. (A) Diagram of aphidicolin inhibition. (B) FACS analysis shows that inhibiting polα activity at late S or G2 phase arrested cells in G2 phase. HeLa was synchronized at G1/S boundary and then released into S phase. Aphidicolin (aphi) was added at different times during the cell cycle. Cells were arrested at G2 phase when polα was inhibited in late S/G2 phase (6 h) or G2 phase (7 and 8 h), whereas the cell cycle progression was delayed when aphidicolin was added at late G2/M phase (9 h). Percentage of cells at each phase of the cell cycle is shown next to the FACS profile. (C) G-overhangs remained elongated when polα activity was inhibited at G2 phase. Aphidicolin was added at late S/early G2 (6 h after release), late G2/M (9 h after release), or after G2/M (12 h after release) to inhibit polα activity. Cells were collected at 9, 12, and 25 h and G-overhang sizes were measured using overhang protection assay. ExoI digests 3′ overhangs and the ExoI plus (+) lanes show the background. (D) The weighted mean sizes of G-overhangs from (C). Results are representative of three independent experiments. Error bars represent s.d. (E) Excessive G-overhangs were produced upon polα inhibition at G2 phase. After synchronization of HeLa at G1/early S boundary, aphidicolin or DMSO was added at different times during the cell cycle and cells were collected at 12 h after release.
To determine the time window during which C-strand fill-in occurs, aphidicolin was added at 9 and 12 h after release (Figure 5A). FACS analysis showed that cells were at late G2/M and at the next G1 phase, respectively (Figure 5B; DMSO panel). Cells were cultured for additional 3 and 13 h in the presence of aphidicolin. We found that G-overhangs were reduced to normal sizes during the next G1 phase in both circumstances (Figure 5C and D), in contrast to the retention of lengthened G-overhangs when aphidicolin was added at late S/G2 (6 h). More thorough analysis revealed that G-overhangs remained lengthened when aphidicolin was added at 7 h (early G2) and 8 h (mid to late G2) after release (Figure 5E). Thus, C-strand fill-in at the majority of telomeres takes place in late S/G2 phase.
As a G2 arrest was induced by aphidicolin, it is possible that failure of C-strand fill-in was caused by the inability of cells to progress through the G2/M transition and not directly because of inhibition of polα/β. To address this, we determined G-overhang lengths in cells arrested at G2/M with either nocodazole or etoposide (Clifford et al, 2003) (Supplementary Figure S2A). Synchronized HeLa cells were released into media containing nocodazole or etoposide for 12 h, and G-overhang measurement showed no persistence of lengthened overhangs (Supplementary Figure S2B). In addition, we attempted to inhibit polα during late S/G2 while still allowing cells to progress through the cell cycle. When caffeine was added simultaneously with aphidicolin to HeLa cells 6.5 h after release to prevent the ATM/ATR-mediated DNA damage response, a significant portion of the cells (∼50%) bypassed G2/M arrest (Supplementary Figure S2A). Overhang determination revealed that lengthened G-overhangs still persisted (Supplementary Figure S2B). Taken together, we conclude that the aphidicolin-induced retention of lengthened G-overhangs was not due to cell cycle arrest.
The function of CDK1 in G-overhang generation
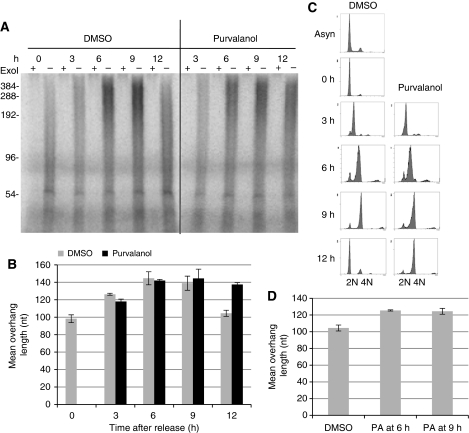
In budding yeast, G-overhang generation is regulated by CDK1, which is required for generating the S phase-specific long G-overhangs (Frank et al, 2006). To test whether the cell cycle-regulated G-overhang dynamics in human cells is regulated by CDK1, we treated synchronized HeLa cells with a highly selective CDK1 inhibitor, purvalanol A (PA) at 7.5 μM (Gray et al, 1998). The concentration of PA was carefully determined as lower concentration of PA (5 μM) did not completely block CDK1 activity in HeLa, whereas higher concentration (10 μM) induced apoptosis (data not shown).
HeLa cells were first arrested at the G1/S boundary, released into media containing PA, and collected at 3 h intervals. In the presence of PA, the length of G-overhangs continued to increase throughout the S phase and peaked at late S/G2 phase, identical to DMSO-treated cells (Figure 6A and B). However, G-overhangs remained markedly lengthened 12 h after release in the presence of PA (Figure 6A and B), whereas G-overhangs had shortened at this time in DMSO-treated cells, suggesting that CDK1 inhibition prevented C-strand fill-in. The failure of C-strand fill-in was unlikely to be caused by the delay of cell cycle progression, as the movement from G1/S to late S/G2 was only slightly delayed by PA treatment (Figure 6C), and it has been shown that PA does not block the completion of DNA synthesis in S-phase cells (Gray et al, 1998; Goga et al, 2007). Consistent with the previous finding that CDK1 inhibition causes a G2 arrest (Gray et al, 1998; Goga et al, 2007), cells were arrested at G2/M upon PA treatment (Figure 6C; 12 h). Furthermore, when PA was added after the majority of DNA synthesis was completed (6 and 9 h after release from G1/S arrest), G-overhangs remained lengthened at 12 h after release (Figure 6D), strongly arguing against the notion that inhibition of C-strand fill-in by PA arose from the delay of DNA synthesis. Thus, we conclude that the kinase activity of CDK1 is essential for completing C-strand fill-in.
Figure 6.
The function of CDK1 in proper generation of G-overhangs. (A) Measurement of G-overhang lengths from cells treated with PA during the cell cycle. HeLa cells were synchronized at G1/S boundary and then released into media containing 7.5 μM PA or DMSO. Cells were collected at 3 h intervals to measure G-overhang lengths. ExoI digests 3′ overhangs and the ExoI plus (+) lanes show the background. (B) Quantitation of the weighted mean length of G-overhangs calculated from (A). Results are representative of three independent experiments. Error bars represent s.d. (C) FACS analysis showing that cells were arrested at G2 phase when CDK1 was inhibited with PA, whereas progression through S phase was largely unaffected. (D) Inhibition of CDK1 at late S/G2 blocked C-strand fill-in. HeLa was synchronized at G1/S boundary and released into S phase. Purvalanol (PA) was added at 6 or 9 h after release and cells were collected at 12 h for measuring mean overhang length. Results are from three experiments. Error bars represent s.d.
CDK1 inhibition leads to DNA damage response at telomeres
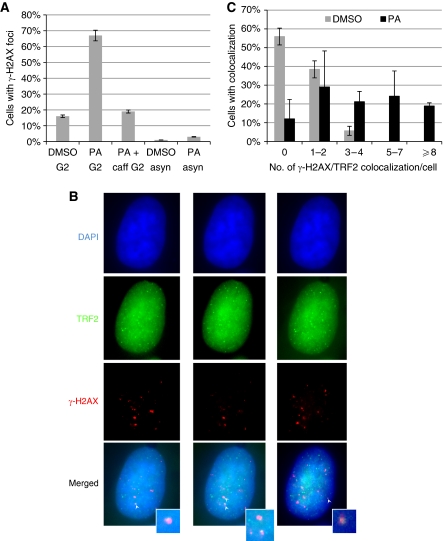
We next determined the importance of CDK1 in telomere stability. Synchronized HeLa cells were released into S phase in the presence of PA and then collected 11 h after release, which corresponded to the G2/M phase (Figure 6C). As shown in Figure 7A, PA treatment resulted in a large increase in cells containing γ-H2AX foci, which were completely abolished upon caffeine treatment. The increase was not caused by PA per se, as asynchronized cells treated with PA for 9 h did not significantly increase γ-H2AX-stained cells (Figure 7A). We also observed that a marked number of γ-H2AX foci colocalized with TRF2 (Figure 7B and C), suggesting that at least a portion of telomeres became dysfunctional and contributed to the induction of DNA damage response.
Figure 7.
Inhibition of CDK1 at late S/G2 leads to DNA damage response at telomeres. (A) Increased ATM/ATR-dependent DNA damage response upon CDK1 inhibition at G2 phase. PA was added at 2 h after the G1/S arrested HeLa was released into S phase. Cells were fixed at 9 h (G2 phase) for anti-γ-H2AX immunostaining (PA G2). Caffeine was added at 6 h after release (PA+caff G2). A total of over 100 cells in random fields were counted for each sample and cells containing >4 γ-H2AX foci were considered as positive. Treatment of asynchronized HeLa PA for 9 h (PA asyn) did not significantly increase γ-H2AX staining. Results are from two experiments. (B) Representative images showing colocalization of γ-H2AX with TRF2. Cells were stained with antibodies specific for TRF2 (green) and γ-H2AX (red), and nuclei were counterstained with DAPI (blue). Cells were optically sectioned at 0.275 μM intervals, and colocalizations (yellow) were observed in multiple sections. Here, sections from the same nucleus containing colocalization (indicated by white arrow) are shown, and inserts show enlarged images of colocalization. (C) Percent of cells containing TRF2/γ-H2AX colocalization. A total of over 50 nuclei were analysed for each sample. Results are average from three experiments. Error bars represent s.d. A full-colour version of this figure is available at The EMBO Journal Online.
The elevated telomeric DNA damage upon CDK1 inhibition might have resulted from the accumulation of ss G-rich DNA. However, a significant portion of normal human telomeres contain long overhangs that are protected from activating DNA damage response. Given that the observed G-overhang increase is <50% when CDK1 is inhibited, there still might be an adequate amount of ssDNA-binding protein such as Pot1 or CST to protect most elongated overhangs. The residual elongated overhangs may be insufficient to trigger massive telomeric DNA damage response. Indeed, the number of telomeric γ-H2AX foci is fairly modest compared with that induced by deficiencies in shelterin proteins (Takai et al, 2003; Denchi and de Lange, 2007; Konishi and de Lange, 2008; Sfeir et al, 2009), indicating that only a portion of telomeres become dysfunctional. Alternatively, CDK1 activity may be needed for resolving other replication defects such as replication fork stalling at telomeres by an unidentified mechanism, which might, for example, involve post-translational modification of the shelterin complex.
The human homolog of Stn1 is involved in regulating G-overhang length
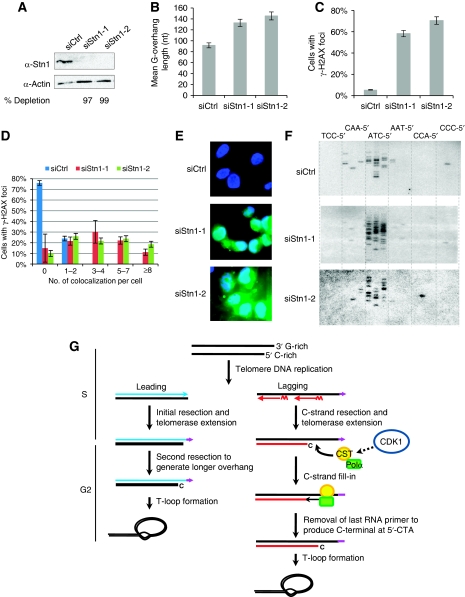
It has been proposed that the Cdc13/Stn1/Ten1 complex in budding yeast binds to telomeres and recruits polα for C-strand fill-in after replication (Lustig, 2001; Grossi et al, 2004). The human homolog of Stn1 stimulates the activity of polα (Casteel et al, 2009), localizes at telomeres, and forms a complex with Ctc1 and Ten1 that contains a high affinity for ssDNA (Miyake et al, 2009). We reasoned that the hStn1 might be involved in regulating G-overhang length, probably by recruiting polα to telomeres to complete C-strand fill-in. If so, dysfunction of hStn1 would abolish C-strand fill-in and G-overhangs would become elongated. Indeed, when hStn1 was depleted by RNAi (Figure 8A), G-overhang length increased by ∼45–55% (Figure 8B), consistent with the recent report by Miyake et al (2009). Therefore, hStn1, like its counterparts in yeasts and plants, regulates telomeric G-overhang length.
Figure 8.
hStn1 is involved in regulating G-overhang length. (A) Western blot analysis of HeLa cells treated with hStn1 RNAi. Cells were collected 72 h after siRNA transfection. Numbers below the image indicate percent knockdown of hStn1 protein. (B) Measurement of G-overhang sizes using overhang protection assay from cells collected 72 h after siRNA transfection. Results are representative of three independent experiments. Error bars represent s.d. (C) Depletion of hStn1 increased overall γ-H2AX staining. A total of over 125 cells in random fields from each RNAi sample were counted for each sample and cells containing >4 γ-H2AX foci were considered positive. (D) Percent of cells containing γ-H2AX/TRF2 colocalization among total number of cells was plotted. Results are representative of two independent experiments. Error bars represent s.d. (E) Apoptosis induced by hStn1 depletion. Cells were collected 72 h after siRNA transfection for Annexin V-FITC staining. Results are representative of two independent experiments. (F) STELA analysis for determination of the specificity of the last nucleotide at C-strand upon hStn1 depletion. (G) Model of G-overhang generation in telomerase-positive cells. See text for explanation. A full-colour version of this figure is available at The EMBO Journal Online.
In addition, we found that depletion of hStn1 dramatically increased the number of cells containing γ-H2AX foci (Figure 8C). Elevated colocalization of γ-H2AX and TRF2 was also observed (Figure 8D; Supplementary Figure S3), indicating that disruption of hStn1 induced telomeric DNA damage response. Apoptosis was noticeable 3 days after transfection (Figure 8E), indicating that hStn1 is essential for cell viability. The above results are reminiscent of the effects of Ctc1 depletion (Surovtseva et al, 2009). We were unable to determine telomere abnormalities in hStn1 knockdown cells using metaphase FISH, as the frequency of metaphase cells was too low. No chromatin bridges were observed in interphase cells when hStn1 was depleted (data not shown).
Yeast studies suggest that Stn1 may modulate telomere length by restricting extensive nuclease processing of the C-strand. To determine whether hStn1 controls end resection of the C-strand, we performed STELA on DNA isolated from hStn1-depleted cells. Surprisingly, upon hStn1 depletion, the C-strand still preferred to end with 5′-CTA (Figure 8F), indicating that hStn1 is not required for the tight regulation of nuclease processing of the C-strand. Together with our results that the last nucleotide at C-strand remained precisely defined during the cell cycle, we conclude that hStn1 is unlikely to be involved in regulating nuclease resection of the C-strand.
Discussion
Despite the critical function of G-overhangs in telomere maintenance, the molecular mechanism for G-overhang generation in human cells is poorly understood. Results presented here provide evidence that G-overhang generation at human telomeres is a dynamic process consisting of multiple tightly regulated steps. We propose that different mechanisms are used at leading and lagging daughter telomeres for generating mature overhangs in telomerase-positive cells (Figure 8G). At leading telomeres, an initial C-strand resection and telomerase extension of G-strand take place shortly after replication. During the G2/M phase, leading overhangs are further lengthened, most likely by a second C-strand resection. No subsequent C-strand fill-in seems to occur at leading telomeres. In contrast, replication of lagging telomeres produces excessively long G-overhangs in S phase. This may be caused by the inability of lagging strand synthesis machinery to place the final RNA primers at the very end of chromosomes, C-strand resection, and/or telomerase extension of G-strand. The resection of C-strand is tightly controlled, ending with a defined nucleotide at C-strand terminus. Then, at late S/G2 phase, C-strands at lagging telomeres are filled in by polα, which is probably recruited to telomere ends by the CST complex (or individual components of the CST complex), yielding mature G-overhangs. CDK1 may control C-strand fill-in by phosphorylating a yet unidentified target. Following C-strand fill-in, a subsequent tightly controlled processing step rapidly removes the terminal RNA primer, producing a 5′-CTA end on C-strand.
We have observed that the telomerase-negative IMR90 cells exhibit transient G-overhang lengthening in S phase similar to telomerase-positive cells (Figure 1), suggesting that telomerase action is not essential for generating elongated overhangs. This observation is reminiscent of the telomerase-independent G-overhang increase at late S phase in budding yeast (Wellinger et al, 1993, 1996,Wellinger et al, 1993, 1996). The telomerase-independent G-overhang lengthening may be caused by two mechanisms. If the replication machinery is unable to position the final RNA primer at the very end of chromosome DNA, a long G-overhang may be generated at lagging telomeres. Alternatively, extensive resection at C-strand of lagging telomeres following replication may also create excessive ss G-overhangs. However, our observation of G-overhang dynamics in telomerase-negative cells does not exclude the contribution of telomerase in G-overhang increase in telomerase-positive cells. In telomerase-positive cells, the elongated G-overhangs may result from telomerase extension of G-strand in addition to C-strand resection.
Mature G-overhangs are generated differently at leading and lagging daughter telomeres
Figure 3D shows that leading telomeres possess overhangs throughout S phase, in agreement with the previous report that the blunt-ended leading telomeres are processed immediately following replication to yield overhangs in order for telomerase to extend leading telomeres (Zhao et al, 2009). Interestingly, we found that whereas lagging overhangs were elongated during S phase and then shortened in late S/G2 by C-strand fill-in, leading overhangs remained stable during S phase and were further extended in G2/M phase (Figure 3D), indicating that the late S/G2-specific C-strand fill-in may be absent at leading telomeres. Although it seems surprising, Zhao et al (2009) also reports that the lengths of leading overhangs remain largely unchanged from 30 min after replication to late S/G2 phase. The authors' measurement of leading overhangs 30 min after replication was ∼70 nt, whereas in the late S/G2 phase, the length of leading overhangs was barely diminished to ∼63 nt. During the same time period, lagging overhangs shortened from 100 to 60 nt (Zhao et al, 2009), indicating that C-strand fill-in had occurred at lagging telomeres. If similar C-strand fill-in also occurred at leading telomeres, the leading overhangs should have been decreased to ∼33 nt (=73–40). Taken together, these observations support the notion that following telomerase elongation of leading telomeres, the sizes of leading overhangs remain largely constant during S phase and C-strand fill-in is likely to be absent at leading telomeres. Although there is a great discrepancy in the amount of G-overhangs measured from the two studies, this is probably caused by differences in methods used for measuring G-overhangs. This study uses non-denaturing in-gel hybridization assay, which measures the relative abundance of ss overhangs from all of the leading telomeres including the ones with blunt ends and with extremely short overhangs. The drawback of this assay is that it does not measure the absolute length of overhangs. In contrast, the DSN assay used in the previous study is able to measure the length of overhangs but excludes telomeres with blunt ends and those with extremely short overhangs. Therefore, it is likely that the length of leading overhangs measured by Zhao et al was an overestimation.
Analysis of the dynamics of leading overhangs in the previous study was limited to 8 h after release (Zhao et al, 2009), leaving the question of what happens to leading telomeres at later time points after cells enter G2 phase. In this study, we observed an unexpected increase of leading overhangs in G2/M (Figure 3D), suggesting that additional enzymatic events might have acted on leading telomeres after replication. The G2/M-specific elongation of leading overhangs could be due to a second processing event. In agreement with this view, it has been shown that nucleases involved in DSB processing are recruited to telomeres during the G2 phase of the cell cycle (Verdun et al, 2005). Our findings further stress the need to identify nucleases and helicases responsible for the two processing steps at leading telomeres. Multiple nucleases and helicases have been identified in budding yeast, including ExoI, Dna2, Sgs1, Sae2, and the Mre11/Rad50/Xrs2 complex, that contribute to C-strand resection (Bonetti et al, 2009). It remains to be determined whether similar nucleases/helicases are involved in processing telomere ends in human cells. An alternative explanation for the G2/M-specific lengthening of leading overhangs is that telomerase might extend leading G-strands at G2. This is considered less likely, as telomerase is recruited to telomeres during S phase (Tomlinson et al, 2006) and extends the majority of telomeres during S phase (Zhao et al, 2009).
Protection of elongated ss G-rich DNA
The S/G2 phase-specific excessive ss G-overhangs ought to be protected by proteins other than RPA, as RPA binding may trigger ATR-dependent DNA damage response. It would be interesting to know what proteins protect excessive ssDNA during the cell cycle. The ss telomeric DNA-binding protein Pot1 may fulfill this protective function during S phase, as it associates with telomeres throughout S phase (Verdun et al, 2005) and represses the ATR signalling pathway (Denchi and de Lange, 2007). Interestingly, the association of Pot1 to telomeres decreases at G2, whereas the association of DNA repair proteins, including ATM and ATR, to telomeres increases at G2 (Verdun et al, 2005), coinciding with the appearance of the longest G-overhangs. The reason for the plunge of Pot1 association with telomeres during G2 remains to be elucidated. One possibility could be that the continuing telomeric association of Pot1 at G2 may block C-strand fill-in. As discussed below, if the CST complex recruits polα to telomere ends for C-strand fill-in during late S/G2, the CST complex may bind to ss G-overhang DNA specifically at late S/G2. Binding of the CST complex may temporarily displace Pot1 from telomeres, allowing polα to finish C-strand synthesis. It will be interesting to know whether the CST complex can repress DNA damage signalling at late S/G2.
The molecular machinery involved in completing C-strand fill-in
G-overhangs at budding yeast telomeres also experience transient lengthening in late S phase (Wellinger et al, 1993; Larrivee et al, 2004; Takata et al, 2005). The delayed C-strand fill-in is mediated by DNA polα and regulates telomerase extension of telomeres. The yeast CST complex interacts with the two subunits of DNA polα: Pol1 and Pol12 (Qi and Zakian, 2000; Grossi et al, 2004; Puglisi et al, 2008). It has been proposed that the yeast CST complex binds to the telomere and recruits polα for completion of C-strand fill-in after replication (Chandra et al, 2001; Lustig, 2001; Grossi et al, 2004). The mammalian CST complex binds to ssDNA (Miyake et al, 2009) and Ctc1/Stn1 stimulates polα activity in vitro (Casteel et al, 2009). Depletion of hStn1 leads to elongated G-overhangs (Figure 8) (Miyake et al, 2009). Therefore, the human CST complex may be the telomere-specific polymerase recruiting factor for completing C-strand fill-in (Figure 8G) (Surovtseva et al, 2009). Alternatively, the CST complex may restrict nuclease processing of C-strand. However, as hStn1 depletion has no effect on the specificity of the last nucleotide at C-strand (Figure 8F), we favour the explanation that hStn1 is involved in C-strand fill-in. It will be important to determine the function of human CST complex in telomere maintenance.
In summary, the results presented here have dissected the molecular steps in G-overhang generation at human telomeres and suggested that C-strand fill-in may be critical for cell cycle progression. Targeting C-strand fill-in may be an effective approach to induce growth suppression in the cancer cells. Furthermore, as many telomere-binding proteins regulate telomerase in cis, it will be interesting to determine their functions in regulating C-strand fill-in. Understanding the mechanisms of G-overhang generation will be important for our knowledge of telomere maintenance and may assist in developing novel cancer therapy.
Materials and methods
Cell culture and synchronization
HeLa and IMR90 cells were purchased from American Type Culture Collection. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% cosmic calf serum (Hyclone). For synchronizing HeLa cells at G1/S boundary, exponentially growing cells were arrested at G1/S boundary with thymidine (2 mM) for 14 h, followed by a cell wash with prewarmed DMEM (three times). Cells were then trypsinized and released into fresh DMEM-10% FBS for 16 h. Thymine (2 mM) or aphidicolin (1 μg/ml) was added to medium for 12–16 h, followed by a cell wash with prewarmed DMEM (three times) before cells were released into fresh medium containing serum. For synchronizing IMR90 cells at G1/S boundary, cells were serum starved for 48 h and then released into DMEM-20% serum supplemented with aphidicolin (1 μg/ml) for 24 h. Aphidicolin was then removed by washing cells with prewarmed DMEM (three times) and cells were released to DMEM-20% serum. Cells collected at different time points were fixed in 70% ethanol, digested with RNase A (0.02 μg/μl), stained with propidium iodide (50 μg/ml), and DNA contents were analysed using a Becton-Dickinson FACSCalibur or Beckman Coulter EPICS® XL™ flow cytometer.
Antibodies
The following primary antibodies were used: mouse anti-hStn1 (Santa Cruz), rabbit anti-γ-H2AX (Active Motif), mouse anti-TRF2 (Imgenex), and mouse anti-actin (BD). Secondary antibodies were: horseradish peroxidase-conjugated anti-mouse IgG (BD), Dylight 488-conjugated anti-mouse IgG (ThermoFisher), and Dylight 549-conjugated anti-rabbit IgG (ThermoFisher).
Separation of leading and lagging daughter telomeres
Separation of leading and lagging telomeres was performed as described (Chai et al, 2006a) with minor modifications. All procedures were performed in dark. Briefly, HeLa cells synchronized at G1/S boundary by thymidine-aphidicolin block were released into media containing 100 μM BrdU. Genomic DNA isolated from BrdU-labelled cells was digested with three blunt-end restriction enzymes AluI, RsaI, and HaeIII, mixed with CsCl solution, and subjected to ultracentrifugation at 44 000 r.p.m. for 13 h. Fractions were then collected, followed by measurement of CsCl density in each fraction with a refractometer. The amount of telomere DNA in each fraction was determined by hybridization to telomeric probe with slot blotting. Fractions containing leading or lagging telomeres were pooled. DNA was then desalted, concentrated, and used in G-overhang analysing assays.
Telomeric G-overhang measurement
The length of the telomeric G-overhang was measured by telomere overhang protection assay as described (Chai et al, 2005).
Non-denaturing in-gel hybridization assay
Non-denaturing in-gel hybridization was performed as described (Chai et al, 2005).
Enzymatic treatments
To remove the 3′ G-rich overhang, total DNA was treated with the 3′ → 5′ exonuclease Escherichia coli ExoI (0.3 U/μg DNA, NEB) in 15 μl buffer (10 mM HEPES pH 7.5, 100 mM LiCl, 2.5 mM MgCl2, 5 mM CaCl2, 20 mM β-mercaptoethanol, 0.067 μg/μl DNase-free RNaseA) at 37°C for 1 h to overnight. To remove 5′ C-rich overhang, total DNA was treated with the 5′ → 3′ exonuclease RecJf (15 U/μg DNA, NEB) in 10 μl buffer (10 mM Tris–HCl pH 7.9, 50 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol) at 25°C for 1 h to overnight and then heat inactivated at 65°C for 20 min.
STELA
The terminal nucleotide at C-rich strand was determined by STELA as described (Sfeir et al, 2005) with minor modifications. Briefly, 20 ng EcoRI digested genomic DNA was ligated to each telorette (10−4 μM) at 35°C for 16 h in 10 μl ligation reaction containing 1 × ligase buffer and 200 U T4 ligase (NEB). The ligated DNA was diluted to 250 pg/μl for subsequent PCRs. Multiple PCR reactions (28 cycles of 95°C for 15 s, 62°C for 20 s, and 68°C for 10 min) were carried out in 25 μl containing 250 pg ligated DNA, 0.2 μM primers (XpYpE2 forward primer and C-Teltail reverse primer), and 2 U FailSafe enzyme mix (Epicentre). Each telorette was used for three independent assays and PCR products were resolved on 0.7% agarose gels in separate lanes. After electrophoresis, DNA was denatured and transferred onto positively charged nylon membranes (Hybond N+, GE healthcare), followed by UV cross-linking and hybridization with a subtelomeric probe. Signals were detected by PhosphorImager screen (GE healthcare).
RNA interference
Small interfering RNAs (siRNAs) were purchased from Qiagen for the target sequences of hStn1: TGGATCCTGTGTTTCTAGCCT (siRNA-1) and CAGCTTAACCTCACAACTTAA (siRNA-2). Control siRNA targeted luciferase and the sequence was CGUACGCGGAAUACUUCGA. Pot1 siRNA targeted the sequence at exon 7: AAGGGTGGTACAATTGTCAAT. HeLa cells were transfected with 20 nM siRNA with X-tremeGENE transfection reagent (Roche) according to the manufacturer's instructions. Cells were harvested 72 h after transfection for isolating DNA for telomere overhang measurement, STELA, Annexin V-FITC staining, and immunoblotting analysis.
TRF2/γ-H2AX colocalization analysis
Cells were washed with PBS, fixed on glass chamber slides with 4% paraformaldehyde, permealized with 0.15% Triton X-100, and then blocked with 10% bovine serum albumin at 37°C for 1 h. Cells were incubated with mouse anti-TRF2 and rabbit anti-γ-H2AX at 37°C for 1 h, washed three times with PBS, and incubated with Dylight 488-conjugated anti-mouse IgG and Dylight 549-conjugated anti-rabbit IgG. After the slides were washed with PBS (three times), DNA was counterstained with DAPI, and Z-stack images were taken at a 0.275-μM thickness per slice under Zeiss AxioImager M2 epifluorescence microscope.
Supplementary Material
Acknowledgments
We thank Chengtao Her and Ken Roberts for comments on the paper, and Amruta Mahadik and Jennifer Eldridge for technical assistance. This work was supported in part by NIH grant R15CA132090 and American Cancer Society to WC.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D (2003) Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet 33: 203–207 [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP (2009) Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB (2009) A DNA polymerase-{alpha}{middle dot}primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem 284: 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Du Q, Shay JW, Wright WE (2006a) Human telomeres have different overhang sizes at leading versus lagging strands. Mol Cell 21: 427–435 [DOI] [PubMed] [Google Scholar]
- Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE (2006b) The involvement of the MRN complex in the generation of G-overhangs at human telomeres. EMBO Rep 7: 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Shay JW, Wright WE (2005) Human telomeres maintain their overhang length at senescence. Mol Cell Biol 25: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A, Hughes TR, Nugent CI, Lundblad V (2001) Cdc13 both positively and negatively regulates telomere replication. Genes Dev 15: 404–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford B, Beljin M, Stark GR, Taylor WR (2003) G2 arrest in response to topoisomerase II inhibitors: the role of p53. Cancer Res 63: 4074–4081 [PubMed] [Google Scholar]
- d'Adda di Fagagna F, Teo SH, Jackson SP (2004) Functional links between telomeres and proteins of the DNA-damage response. Genes Dev 18: 1781–1799 [DOI] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE (1999) Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99: 723–733 [DOI] [PubMed] [Google Scholar]
- Dionne I, Wellinger RJ (1998) Processing of telomeric DNA ends requires the passage of a replication fork. Nucleic Acids Res 26: 5365–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CJ, Hyde M, Greider CW (2006) Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol Cell 24: 423–432 [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goga A, Yang D, Tward AD, Morgan DO, Bishop JM (2007) Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med 13: 820–827 [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M (2001) Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J 20: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N, Reed SI, Charbonneau M (1997) Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev 11: 512–527 [DOI] [PubMed] [Google Scholar]
- Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, Kim SH, Lockhart DJ, Schultz PG (1998) Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science 281: 533–538 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D (2004) Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev 18: 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T (2005) POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 24: 2667–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob NK, Skopp R, Price CM (2001) G-overhang dynamics at Tetrahymena telomeres. EMBO J 20: 4299–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi A, de Lange T (2008) Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev 22: 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee M, LeBel C, Wellinger RJ (2004) The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 18: 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Baumann P, Cech TR (2003) DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature 426: 198–203 [DOI] [PubMed] [Google Scholar]
- Loayza D, Parsons H, Donigian J, Hoke K, de Lange T (2004) DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem 279: 13241–13248 [DOI] [PubMed] [Google Scholar]
- Lovett ST, Kolodner RD (1989) Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci USA 86: 2627–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V (2002) Telomere maintenance without telomerase. Oncogene 21: 522–531 [DOI] [PubMed] [Google Scholar]
- Lustig AJ (2001) Cdc13 subcomplexes regulate multiple telomere functions. Nat Struct Biol 8: 297–299 [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88: 657–666 [DOI] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P (2007) Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA 104: 14038–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell 36: 193–206 [DOI] [PubMed] [Google Scholar]
- Muntoni A, Reddel RR (2005) The first molecular details of ALT in human tumor cells. Hum Mol Genet 14 (Spec No. 2): R191–R196 [DOI] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V (1996) Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D (2008) Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J 27: 2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA (2000) The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev 14: 1777–1788 [PMC free article] [PubMed] [Google Scholar]
- Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J (2008) C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132: 745–757 [DOI] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Fajkus J, Vyskot B, Shippen DE (2000) Analysis of the G-overhang structures on plant telomeres: evidence for two distinct telomere architectures. Plant J 23: 633–641 [DOI] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir AJ, Chai W, Shay JW, Wright WE (2005) Telomere-end processing: the terminal nucleotides of human chromosomes. Mol Cell 18: 131–138 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE (2008) STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansel RM, de Lange T, Griffith JD (2001) T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J 20: 5532–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE (2009) Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T (2003) DNA damage foci at dysfunctional telomeres. Curr Biol 13: 1549–1556 [DOI] [PubMed] [Google Scholar]
- Takata H, Tanaka Y, Matsuura A (2005) Late S phase-specific recruitment of Mre11 complex triggers hierarchical assembly of telomere replication proteins in Saccharomyces cerevisiae. Mol Cell 17: 573–583 [DOI] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP (2006) Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J (2005) Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell 20: 551–561 [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J (2006) The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 127: 709–720 [DOI] [PubMed] [Google Scholar]
- Vodenicharov MD, Wellinger RJ (2006) DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell 24: 127–137 [DOI] [PubMed] [Google Scholar]
- Wan M, Qin J, Songyang Z, Liu D (2009) OB-fold containing protein 1 (OBFC1), a human homologue of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem 284: 26725–26731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Ethier K, Labrecque P, Zakian VA (1996) Evidence for a new step in telomere maintenance. Cell 85: 423–433 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA (1993) Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72: 51–60 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hoshiyama H, Shay JW, Wright WE (2008) Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res 36: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE (2009) Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.