14-3-3γ mediates Cdc25A proteolysis to block premature mitotic entry after DNA damage
Kasahara et al offer new insights into the ATR-Chk1 kinase-dependent mechanisms of damage-induced cell cycle arrest, demonstrating that the underlying post-translational regulation is yet more complex than previously assumed
Keywords: Cdc25, Chk1, DNA damage checkpoint, phosphorylation, 14-3-3
Abstract
14-3-3 proteins control various cellular processes, including cell cycle progression and DNA damage checkpoint. At the DNA damage checkpoint, some subtypes of 14-3-3 (β and ζ isoforms in mammalian cells and Rad24 in fission yeast) bind to Ser345-phosphorylated Chk1 and promote its nuclear retention. Here, we report that 14-3-3γ forms a complex with Chk1 phosphorylated at Ser296, but not at ATR sites (Ser317 and Ser345). Ser296 phosphorylation is catalysed by Chk1 itself after Chk1 phosphorylation by ATR, and then ATR sites are rapidly dephosphorylated on Ser296-phosphorylated Chk1. Although Ser345 phosphorylation is observed at nuclear DNA damage foci, it occurs more diffusely in the nucleus. The replacement of endogenous Chk1 with Chk1 mutated at Ser296 to Ala induces premature mitotic entry after ultraviolet irradiation, suggesting the importance of Ser296 phosphorylation in the DNA damage response. Although Ser296 phosphorylation induces the only marginal change in Chk1 catalytic activity, 14-3-3γ mediates the interaction between Chk1 and Cdc25A. This ternary complex formation has an essential function in Cdc25A phosphorylation and degradation to block premature mitotic entry after DNA damage.
Introduction
The cell cycle checkpoint is a fundamental mechanism, not only for monitoring genomic stability, but also for coordinating repair and cell cycle progression. The protein kinase cascade from ATR to Chk1 has important functions in the DNA damage checkpoint (Zhou and Elledge, 2000; Bartek and Lukas, 2003; Kastan and Bartek, 2004). In response to damaged DNA or stalled replication, ATR phosphorylates Chk1 at Ser317 and Ser345; this phosphorylation is considered to elevate the catalytic activity of Chk1 (Zhao and Piwnica-Worms, 2001; Walker et al, 2009). Chk1 then phosphorylates and inhibits Cdc25 family phosphatases, which consist of Cdc25A, B and C in human cells (Boutros et al, 2007). For example, Chk1 induces Cdc25A-Ser76 phosphorylation, which results in βTrCP-dependent Cdc25A degradation (Busino et al, 2003, 2004; Jin et al, 2003; Neely and Piwnica-Worms, 2003; Melixetian et al, 2009). As Cdc25 dephosphorylates cyclin-dependent kinases (Cdks) at an inhibitory phosphorylation site (Cdk1 at Tyr15), Cdc25 inhibition results in Cdk inactivation and cell cycle arrest (Jackman and Pines, 1997; Zhou and Elledge, 2000; Bartek and Lukas, 2003; Kastan and Bartek, 2004).
The DNA damage checkpoint response is also modulated by phosphoserine/phosphothreonine-binding proteins/domains, such as 14-3-3 proteins, FHA domains and BRCT domains (Mohammad and Yaffe, 2009). Studies in fission yeast first suggested that there was a correlation between 14-3-3 proteins and checkpoint control: Rad24, one of 14-3-3 proteins in fission yeast, was identified in a search for irradiation-sensitive mutants (Ford et al, 1994). Further studies indicated that Chk1 and 14-3-3 proteins act through Cdc25 (Pines, 1999). In both human cells and fission yeast, Chk1 phosphorylates Cdc25 on a conserved serine residue (human Cdc25C on Ser216), creating a phosphoserine-binding site for 14-3-3 (Furnari et al, 1997; Peng et al, 1997; Sanchez et al, 1997). As Ser216 on human Cdc25C appears to be highly phosphorylated by C-TAK1 in the absence of DNA damage (Peng et al, 1998; Russell, 1998; Zhou and Elledge, 2000), it remains controversial how 14-3-3 modulates the signal from Chk1 to Cdc25 in mammals.
Some subtypes of 14-3-3 (β and ζ isoforms in mammalian cells and Rad24 in fission yeast) also bind Chk1 in a Ser345-phosphorylation-dependent manner (Jiang et al, 2003; Dunaway et al, 2005). This binding promotes the nuclear retention of Chk1 likely through the masking of NES on Chk1 (Jiang et al, 2003; Dunaway et al, 2005). As Chk1 is phosphorylated at several sites other than Ser317 and Ser345 (ATR sites) in DNA damage responses (Clarke and Clarke, 2005; Puc et al, 2005; Ikegami et al, 2008), we postulate that Chk1 phosphorylation at other site(s) may also modulate the checkpoint signalling through 14-3-3 binding. Here, we show that the γ subtype of 14-3-3 also forms a complex with Chk1 when DNA damage occurs. This binding depends on Chk1 autophosphorylation at Ser296, which occurs after ATR-induced phosphorylation of Chk1 (by implication, catalytic activation of Chk1) and then promotes dephosphorylation at the ATR sites. This phosphorylation shift from ATR sites to Ser296 not only has an important function in the spread of Chk1 signals, but also changes the Chk1-binding subtype of 14-3-3 (from β or ζ to γ). The 14-3-3γ serves as a platform between Cdc25A and Ser296-phosphorylated Chk1, promoting Chk1-induced Cdc25A phosphorylation at Ser76, a critical site for its degradation.
Results
Ser296 phosphorylation is catalysed by Chk1 itself after phosphorylation by ATR
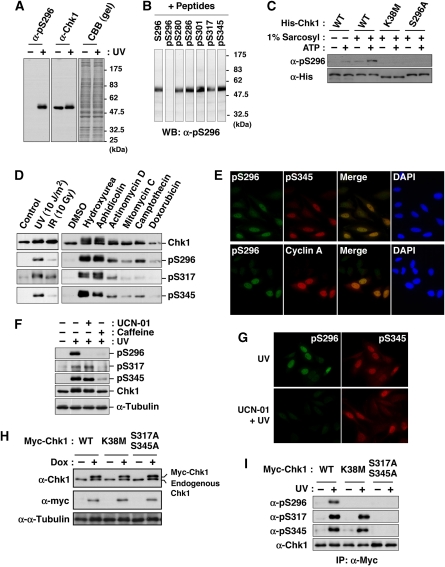
Chk1 was reported to be phosphorylated not only at Ser317 and Ser345, but also at Ser296 (Clarke and Clarke, 2005) in response to DNA damage, but only limited information has been available about Ser296 phosphorylation. We first produced an antibody that specifically recognizes Chk1 phosphorylated at Ser296 (Figure 1A and B; see also Supplementary Figure S1). Using Chk1 purified from baculovirus-infected Sf9 cells, we performed the in vitro autophosphorylation assay. After 30 min of the incubation with [γ-32P] ATP, radioactive phosphates (32P) were incorporated into Chk1 wild-type (WT) protein (Supplementary Figure S1E). The electrophoretic mobility of Chk1 was slower after the incubation with ATP; the anti-pS296 on Chk1 (α-pS296) reacted with WT specifically after the incubation (Supplementary Figure S1E). Chk1 mutation at Lys38 to Met (K38M), which lost the catalytic activity, almost completely abolished 32P incorporation, the mobility shift and α-pS296 immunoreactivity (Supplementary Figure S1E). Chk1 mutation at Ser296 to Ala (S296A) reduced 32P incorporation and abolished α-pS296 immunoreactivity. However, S296A did not completely abolish both 32P incorporation and the mobility shift (Supplementary Figure S1E). In the 2D phosphopeptide mapping analysis, S296A induced the disappearance of the radioactive spots 1 and 2, although other major spots (3–6) appeared to remain unchanged on the thin layer plate (Supplementary Figure S1E). To rule out the possibility that a contaminating kinase in insect cells may phosphorylate Chk1-Ser296, we used His-ProS2-Chk1 protein expressed in bacteria (Figure 1C; His-Chk1). In the extraction of protein without sarcosyl, α-pS296 immunoreactivity in WT was observed very weakly even after the incubation with ATP (Figure 1C; 1% sarcosyl: −). On the other hand, the extraction of WT protein with 1% sarcosyl elevated the α-pS296 immunoreactivity after the incubation with ATP much more than without ATP (Figure 1C) (Zhao and Piwnica-Worms, 2001). However, such phenomena were not observed in the case of K38M or S296A (Figure 1C). All these results suggested that Ser296 on Chk1 serves as one of the major autophosphorylation sites in vitro.
Figure 1.
Chk1 autophosphorylation at Ser296. (A, B) Characterization of an antibody specifically recognizing Chk1 phosphorylation at Ser296. The antibody (α-pS296) reacted specifically with a band corresponding to Chk1 in lysates of UV-irradiated (+) HeLa cells (Ikegami et al, 2008), but not of non-treated (−) cells (A). Immunoreactivity was impaired specifically by preincubation with pS296 corresponding to Ser296-phosphorylated Chk1, but not with non-phosphorylated peptide S296 and phosphopeptides for other sites within Chk1 (B). (C) The in vitro autophosphorylation assay using 6xHis-ProS2-Chk1 (His-Chk1) expressed in bacteria. E. coli strain BL21-CodonPlus®(DE3)-RP was transformed with pCold ProS2 carrying each Chk1. Each 6xHis-ProS2-tagged Chk1 was expressed in the presence of 1 mM IPTG at 15°C for 16 h. E. coli cells were lysed in the lysis buffer (40 mM Hepes-NaOH [pH 8.0], 300 mM NaCl, 1% Triton X-100) supplemented with (+) or without (−) 1% sarcosyl. After centrifugation (17 000 g) for 10 min at 4°C, the supernatant was rotated with TALON metal affinity resin at 4°C for 1 h. After washing with the lysis buffer and reaction buffer (25 mM Tris–HCl [pH 7.5], 10 mM MgCl2), the beads were incubated in the reaction buffer with (+) or without (−) 10 μM ATP at 30°C for 30 min. Chk1 phosphorylated at Ser296 or total Chk1 was detected through immunoblotting with an anti-pS296 (α-pS296) or anti-His (α-His) antibody, respectively. (D) HeLa cells were irradiated with UV, X-rays (IR; 10 Gy) or none (control) and then incubated for an additional 1 h. For treatment with drugs, cells were incubated with 2 mM hydroxyurea, 5 μg/ml aphidicolin, 5 μM actinomycin D, 5 μM mitomycin C, 5 μM camptothecin, 10 μM doxorubicin or 0.1% DMSO (as a solvent control) for 3 h. Extracts were subjected to immunoblotting with the indicated antibodies. (E) HeLa cells irradiated with UV were stained with the indicated antibodies and DAPI. (F, G) Immunoblots (F) or immunocytochemistry (G) shows effects of pre-treatment with UCN-01 or caffeine on Chk1 phosphorylation in UV-irradiated HeLa cells. (H, I) Establishment of HeLa cells in which each Myc-Chk1 (WT, K38 M or S317A/S345A) is expressed in a doxycycline (Dox)-dependent manner (H). Levels of Chk1 phosphorylation after UV-irradiation (I).
We next examined Ser296 phosphorylation in the checkpoint response. In response to various DNA damage stimuli and replication disorders, Chk1 phosphorylation at Ser296 was found to occur in a way similar to the phosphorylation at ATR sites (Figure 1D and E). Furthermore, treatment with UCN-01 (a Chk1 kinase inhibitor) attenuated Chk1 phosphorylation at Ser296, but not at ATR sites, although caffeine (an ATR and ATM inhibitor) showed non-specific reduction in phosphorylation rates (Figure 1F and G). Next, we established HeLa cell lines in which Myc-tagged Chk1 was expressed in a tetracycline- or doxycycline (Dox)-dependent manner; the protein level of each exogenous Chk1 was very similar to that of endogenous Chk1 under our experimental conditions (Figure 1H). Although Ser296 phosphorylation in response to ultraviolet (UV) irradiation was observed in WT, it rarely occurred on Chk1 mutated at ATR sites to Ala (S317A/S345A; Figure 1I). In a kinase dead Chk1 mutant (K38M), Ser296 phosphorylation was hardly detected, although Ser317 and Ser345 phosphorylation was observed (Figure 1I). The above observations suggested that Chk1 phosphorylation at Ser296 is catalysed by its own kinase activity, but not by ATR, although it requires pre-phosphorylation at ATR sites, which has implications for the catalytic activation of Chk1 (Zhao and Piwnica-Worms, 2001; Walker et al, 2009).
Ser296-phosphorylated Chk1 is rapidly dephosphorylated at ATR sites and distributed throughout the nucleoplasm
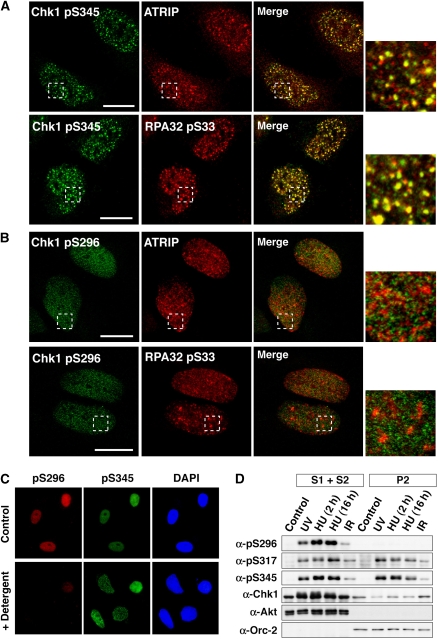
We then scrutinized the subcellular distribution of Ser296-phosphorylated Chk1. In UV-irradiated cells, Ser345-phosphorylated Chk1 was observed at nuclear foci of ATR-interacting protein (ATRIP) and the phosphorylated form of RPA32 (an ATR substrate; Figure 2A), in which ATR is considered to be activated (Zou and Elledge, 2003). In contrast, Ser296-phosphorylated Chk1 appeared to distribute diffusely in the nucleus (Figure 2B). In support of this observation, nuclear signals of anti-pS296, but not of anti-pS345, almost completely disappeared after brief extraction with Triton X-100 detergent (Figure 2C). Biochemical fractionation (Figure 2D) also showed that Ser296-phosphorylated Chk1 was clearly detectable in the soluble (S1 plus S2) but not the chromatin (P2) fractions, although Ser317- and Ser345-phosphorylated Chk1 existed in both (Jiang et al, 2003; Smits et al, 2006). These observations suggest that Ser296-phosphorylated Chk1 is distributed throughout the nucleoplasm, distinct from the localization of Chk1 phosphorylated by ATR.
Figure 2.
Spatial distributions of phosphorylated Chk1. (A, B) UV-irradiated HeLa cells were stained with the indicated antibodies. Scale bars are 10 μm. (C) UV-irradiated HeLa cells were briefly extracted with Triton X-100 detergent and then stained with the indicated antibodies. (D) HeLa cells exposed to UV light or ionizing radiation (IR) or cells treated with hydroxyurea (HU; 2 or 16 h) were fractioned into soluble (S1+S2) and chromatin (P2) fractions. Akt and Orc-2 were used as markers for soluble and chromatin fractions, respectively.
We further confirmed that only faint signals for anti-pS317/pS345 and -pS296 were apparent in anti-pS296 and -pS345 immunoprecipitates, respectively (Supplementary Figure S2A). These findings raised the new question of why only a few Chk1 molecules are phosphorylated simultaneously at Ser296 and ATR sites despite the fact that Ser296 phosphorylation depends on ATR-induced phosphorylation. One possible explanation is rapid dephosphorylation at ATR sites on Ser296-phosphorylated Chk1. In support of this model, we found that dephosphorylation at ATR sites was remarkably delayed in Chk1 mutated at Ser296 to Ala (S296A), compared with the WT case (Supplementary Figure S2B). In addition, treatment with UCN-01 (Supplementary Figure S2C) or protein phosphatase 2A (PP2A)-specific small-interfering RNA (siRNA) (Supplementary Figure S2D) attenuated the dephosphorylation reaction of ATR sites after the release of hydroxyurea. These observations are consistent with a previous report that PP2A promptly dephosphorylates ATR sites in a Chk1 kinase activity-dependent manner (Leung-Pineda et al, 2006). Thus, Ser296 phosphorylation, which occurs only on Chk1 phosphorylated at ATR sites, is likely to promote rapid dephosphorylation at ATR sites by protein phosphatases such as PP2A.
14-3-3γ directly binds Ser296-phosphorylated Chk1
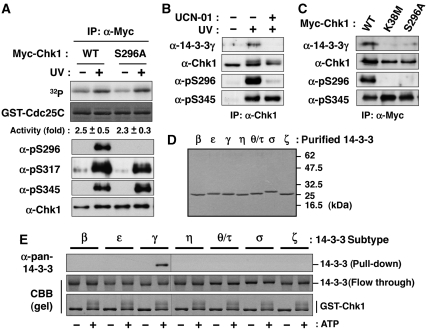
To elucidate the functional changes of Chk1 because of Ser296 phosphorylation, we first measured the in vitro kinase activity of each Myc-Chk1 purified from UV-irradiated or non-treated cells. Between WT and S296A, we observed only marginal differences in the elevation of catalytic activity after UV irradiation (Figure 3A). Together with the previous findings for purified Chk1 protein (Chen et al, 2000), our observation suggested that Chk1 autophosphorylation exerts limited effects on catalytic activity.
Figure 3.
Ser296-phosphorylated Chk1 binds 14-3-3γ. (A) In vitro kinase activity of individual immunoprecipitated Myc-Chk1 forms (WT or S296A) towards the GST-Cdc25C fragment (195-256 a.a.). Fold activation after UV irradiation is also indicated (mean±s.e.m. from three independent experiments). (B) Detection of endogenous 14-3-3γ in anti-Chk1 immunoprecipitates after UV irradiation. UCN-01 pre-treatment was performed as described in ‘Materials and methods'. (C) Effects of Chk1 mutation on the complex formation after UV irradiation. Myc-Chk1 WT, K38M or S296A was purified as an anti-Myc immunoprecipitate. (D, E) GST pull-down assays using GST-Chk1 purified from Sf9 cells and each subtype of 14-3-3 protein purified from bacteria. Each purified 14-3-3 protein was indicated in (D). A total of 0.4 μg of GST-Chk1-His was incubated in 20 μl of reaction mixture (25 mM Tris–HCl [pH 7.5], 10 mM MgCl2) with (+) or without (−) 100 μM ATP at 30°C for 60 min. Then, the mixture was rotated with glutathione beads at 4°C. Beads were washed with IP buffer and then rotated with 0.4 μg of each purified 14-3-3 protein in 100 μl of IP buffer at 4°C for 1 h. Beads were then washed and subjected to immunoblotting with anti-pan-14-3-3 (characterized in Supplementary Figure S3A).
We next searched for proteins binding to Chk1 in a Ser296 phosphorylation-dependent manner. As shown in Figure 3B, signals for anti-14-3-3γ (characterized in Supplementary Figure S3A) were detected in anti-Chk1 immunoprecipitates from UV-irradiated, but not non-treated cells. The signals were diminished by pre-treatment with UCN-01 (Figure 3B) or Chk1 mutations (S296A and K38M; Figure 3C). To further examine the relationship between Chk1 and 14-3-3, we performed the in vitro binding analyses using purified 14-3-3 proteins (Figure 3D) and GST-Chk1. As shown in Figure 3E, 14-3-3 bound to autophosphorylated Chk1 in a subtype-specific manner: γ had the highest affinity among all seven subtypes in vitro. GST pull-down assay from cell extracts revealed that K38M or S296A mutants lacked any ability to bind to 14-3-3γ (Supplementary Figure S3B). Although not only γ, but also β and ζ subtypes bound to Chk1 in an UV irradiation-dependent manner, Chk1 binding to β and ζ subtypes was not affected by the pre-treatment with UCN-01 (Supplementary Figure S3C). This observation was consistent with a previous report that β and ζ subtypes bind to Chk1 in a Ser345 phosphorylation-dependent manner (Jiang et al, 2003). Thus, all of these results together suggest that Chk1 selects specific subtypes of 14-3-3 in a phosphorylation site-dependent manner and that Ser296 phosphorylation generates binding sites specific for the γ subtype.
Ser296 phosphorylation on Chk1 is essential for DNA damage checkpoint
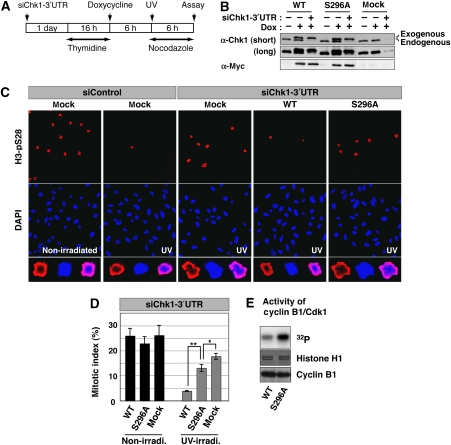
To examine the significance of Chk1-Ser296 phosphorylation for the DNA damage checkpoint, each Tet-On HeLa cell line was transfected with siRNA specific to Chk1 3′UTR sequence (Figure 4A). One day after transfection, thymidine was added to the growth medium to arrest cells at the G1/S boundary. After incubation with thymidine for 16 h, cells were washed and then incubated with Dox-containing medium to induce each Myc-Chk1 protein or EGFP (mock); we successfully replaced endogenous Chk1 with each exogenous Myc-Chk1 or EGFP (Figure 4B). Six hours after release, cells were irradiated with UV-C light and then incubated in fresh medium containing Dox and nocodazole (to block passage through mitosis) for an additional 6 h. Without UV irradiation, we observed only marginal change in the cumulative mitotic index in these cell lines (Figure 4D; non-irradi.). UV irradiation markedly reduced the cumulative mitotic index in control siRNA-treated cells in which EGFP (mock) was introduced (Figure 4C, compare ‘non-irradiated' and ‘UV' in ‘siControl'). As reported previously (Zhou and Elledge, 2000; Bartek and Lukas, 2003), this DNA damage checkpoint was impaired by reduction of the endogenous Chk1 protein level through the RNA interference mechanism (Figure 4C, compare ‘siControl, UV' and ‘siChk1-3′UTR UV' in ‘mock'). Induction of Myc-Chk1 WT almost nullifies this impairment (Figure 4C and D). However, in contrast, S296A induction hardly rescued checkpoint-deficient cells (Figure 4C and D): we obtained similar results on adding Dox 1 day before thymidine addition (Supplementary Figure S4). In support, we detected higher kinase activity of Cyclin B1/Cdk1 in cells replaced with S296A than with WT (Figure 4E). Our data suggest that Ser296 phosphorylation has critical functions in the DNA damage checkpoint.
Figure 4.
Requirement of Ser296 phosphorylation for Chk1-induced DNA damage checkpoint. (A) Scheme of experimental procedures for evaluation of the DNA damage checkpoint. (B) Replacement of endogenous Chk1 with each Myc-Chk1 or EGFP (mock) was assessed by immunoblotting with anti-Myc and anti-Chk1 antibodies. (C, D) Evaluation of mitotic entry after UV irradiation. For calculation of mitotic indices, cells were stained with anti-H3-pSer28 (as a mitotic marker; C). Higher magnification image of a mitotic cell in each group is also indicated (C, in last row). The graph (D) shows the cumulative mitotic index (%) in UV-irradiated or non-irradiated cells in which endogenous Chk1 was replaced with Myc-Chk1 (WT or S296A) or EGFP (mock). Data are mean±s.e.m. values from three independent experiments (**P<0.01, 0.01<*P<0.05). (E) H1 kinase activity of Cyclin B1/Cdk1 complex immunoprecipitated from UV-irradiated cells in which endogenous Chk1 was replaced with different Myc-Chk1 forms.
14-3-3γ mediates interaction between Chk1 and Cdc25A
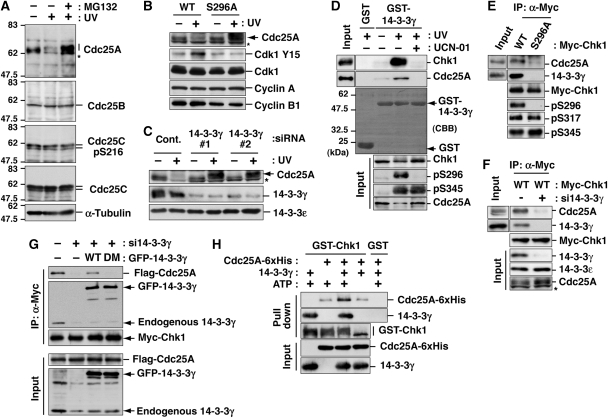
How does Ser296 phosphorylation participate in signalling for the DNA damage checkpoint? Higher Cdk1 activity in S296A-replaced cells (Figure 4E) provides some clues. Cdk1 is activated through dephosphorylation of Cdk1-Tyr15 (an inhibitory phosphorylation site) by Cdc25 family phosphatases (Jackman and Pines, 1997), which Chk1 phosphorylates to inhibit their contribution to the DNA damage checkpoint (Sanchez et al, 1997; Mailand et al, 2000; Zhou and Elledge, 2000; Bartek and Lukas, 2003; Jin et al, 2003; Busino et al, 2004). Among the phosphatases, we focused on Cdc25A because it appeared to be most affected by UV irradiation in HeLa cells; UV irradiation-induced Cdc25A degradation in a proteasome-dependent manner (Figure 5A) as reported previously (Mailand et al, 2000; Busino et al, 2004). As shown in Figure 5B, replacement with S296A, but not with WT, caused inhibition of UV damage-dependent Cdc25A degradation, which promoted Cdk1-Tyr15 dephosphorylation. These results suggest that Ser296 phosphorylation is required for Cdc25A degradation.
Figure 5.
14-3-3γ mediates the interaction between Chk1 and Cdc25A. (A) Changes in protein level of each Cdc25 phosphatase or Cdc25C-Ser216 phosphorylation level after UV irradiation. In the last lane, cells were also treated with MG132 from 30 min before UV irradiation to inhibit the proteasome-dependent degradation. (B, C) Effects of Chk1 mutation (B) or 14-3-3γ depletion (C) on Cdc25A degradation after UV irradiation. Replacement of endogenous Chk1 with exogenous Myc-tagged Chk1 (B; also see Figure 4B) or siRNA treatment of HeLa cells (C) was performed as described in ‘Materials and methods'. Each cell extract was proved with the indicated antibodies. Asterisks indicate non-specific bands of anti-Cdc25A. (D) GST pull-down assays using GST-14-3-3γ. Cells were irradiated with UV and further cultured for 10 min. In the last lane, cells were also treated with UCN-01 from 30 min before UV irradiation. (E, F) Effects of Chk1 mutation (E) or 14-3-3γ knock down (F) on ternary complex formation. After Myc-Chk1 induction, cells were irradiated with UV light in the presence of MG132. In panel F, cells were transfected with control (−) or 14-3-3γ (#1; +) siRNA at 48 h before UV irradiation. Each anti-Myc immunoprecipitate or cell extract (input) was subjected to the immunoblotting. (G) Effect of 14-3-3γ dimerization on the complex formation between Chk1 and Cdc25A. Two days before UV irradiation, cells were transfected with control (−) or 14-3-3γ (#1; +) siRNA. At 1 day before UV irradiation, cells were also transfected with or without (−) GFP-14-3-3γ WT or a dimerization-defective mutant (DM) in addition to Myc-Chk1 WT and Flag-Cdc25A. Cells were irradiated with UV light in the presence of MG132, and then subjected to the immunoprecipitation with an anti-Myc antibody. (H) The ternary complex formation among autophosphorylated Chk1, 14-3-3γ and Cdc25A in a purified system. A total of 10 μg of GST-Chk1-His (purified from Sf9 cells) in the presence of 25 μg of 14-3-3γ and/or 25 μg of Cdc25A-6xHis-Myc were incubated in 250 μl of the buffer (25 mM Tris–HCl [pH 7.5], 10 mM MgCl2) with or without 100 μM ATP at 30°C for 1 h. As a negative control, we used 10 μg of GST with the same amounts of 14-3-3γ and Cdc25A-6xHis-Myc. After the incubation, the mixture was rotated with glutathione beads. After washing with IP buffer three times, beads were subjected to immunoblotting.
As Ser296-phosphorylated Chk1 bound to 14-3-3γ (Figure 3; Supplementary Figure S3), we examined whether 14-3-3γ participates in the Cdc25A degradation pathway. As shown in Figure 5C, the transfection with 14-3-3γ-specific siRNA (#1 or #2), but not with control siRNA (cont.) inhibited Cdc25A degradation after UV irradiation, suggesting that 14-3-3γ mediated Cdc25A degradation. GST pull-down assays revealed that 14-3-3γ bound not only Chk1, but also Cdc25A, in UV-irradiated cells (Figure 5D). The pre-treatment with UCN-01 impaired not only Cdc25A degradation, but also 14-3-3γ binding to Chk1 or Cdc25A (Figure 5D). Immunoprecipitation analyses also showed that not only 14-3-3γ but also Cdc25A bound to Myc-Chk1 WT in UV-irradiated cells (Figure 5E and F). This Cdc25A binding to Chk1 was diminished by the S296A mutation (Figure 5E) or 14-3-3γ depletion (Figure 5F). As 14-3-3 proteins form a dimer (Liu et al, 1995; Mohammad and Yaffe, 2009), we examined whether the dimerization of 14-3-3γ is required for the complex formation between Chk1 and Cdc25A. For this assay, we constructed the dimerization-defective GFP-14-3-3γ mutant (GFP-14-3-3γ DM; Supplementary Figure S5) (Liu et al, 1995), which did not lose the ability to bind target phosphoprotein(s) because it bound to Chk1 after UV irradiation (Figure 5G). Although the expression of GFP-14-3-3γ WT recovered the complex formation between Chk1 and Cdc25A in endogenous 14-3-3γ-depleted cells, the induction of GFP-14-3-3γ DM did not rescue it (Figure 5G). To show direct interaction among three proteins, we performed the in vitro binding assay using each purified protein. As shown in Figure 5H, GST-Chk1 formed a complex with Cdc25A in a 14-3-3γ-dependent and autophosphorylation-dependent manner. All these results suggest that 14-3-3γ serves as a platform for complex formation between Chk1 and Cdc25A.
Ternary complex formation facilitates Cdc25A-Ser76 phosphorylation by Chk1, which promotes Cdc25A degradation in a βTrCP-dependent manner
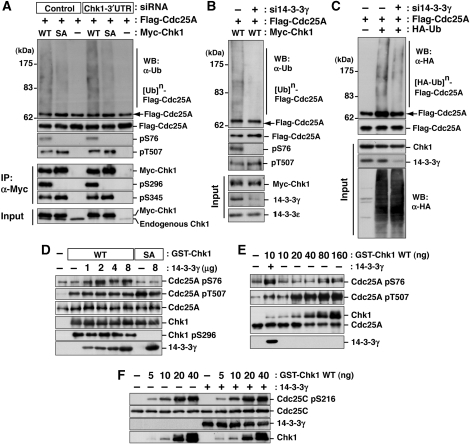
Chk1 is known to phosphorylate Cdc25A at multiple sites after DNA damage (Chen et al, 2003; Jin et al, 2003; Busino et al, 2004). Cdc25A-Ser76 is a rate-limiting phosphorylation site for the generation of a βTrCP recognition motif (phosphodegron) by NEK11 (Jin et al, 2003; Busino et al, 2004; Melixetian et al, 2009), although Thr507 phosphorylation creates a docking site for 14-3-3 (Chen et al, 2003). We used three strategies to elucidate the effect of Chk1-Ser296 phosphorylation or 14-3-3γ on UV-irradiation-induced Cdc25A phosphorylation/polyubiquitylation (also see Supplementary Figure S6). First, cells were transfected with control or Chk1-3′UTR-specific siRNA and then with Flag-Cdc25A in addition to Myc-Chk1 WT or S296A before UV irradiation. As we observed only marginal changes between mock- and Chk1-depleted cells after UV irradiation (Figure 6A), endogenous Chk1 was unlikely to affect both Flag-Cdc25A phosphorylation and polyubiquitylation in this experimental system. Immunoprecipitation analyses revealed that Flag-Cdc25A was phosphorylated both at Ser76 and Thr507 in cells in which Myc-Chk1 WT was introduced (Figure 6A). However, S296A induction impaired Flag-Cdc25A phosphorylation at Ser76, but not at Thr507 (Figure 6A). S296A expression also reduced Cdc25A polyubiquitylation, compared with the WT case (Figure 6A). Second, cells were transfected with control (−) or 14-3-3γ-specific (+) siRNA and then with Flag-Cdc25A and Myc-Chk1 WT before UV irradiation. As shown in Figure 6B, 14-3-3γ depletion abolished Cdc25A-Ser76 phosphorylation and Cdc25A polyubiquitylation in WT-expressing cells. Finally, cells were transfected with control (−) or 14-3-3γ-specific (+) siRNA and then with Flag-Cdc25A and HA-tagged ubiquitin before UV irradiation. The 14-3-3γ-depletion decreased Flag-Cdc25A polyubiquitylation even in the absence of Myc-Chk1 expression (Figure 6C). Thus, these results indicated that the complex formation between Chk1 and Cdc25A on 14-3-3γ accelerates Cdc25A-Ser76 phosphorylation, which was critical for βTrCP-dependent Cdc25A polyubiquitylation/degradation (Jin et al, 2003; Busino et al, 2004; Melixetian et al, 2009).
Figure 6.
The 14-3-3γ facilitates Chk1 access to Cdc25A-Ser76. (A–C) Effects of Chk1 mutation (A) or 14-3-3γ knock down (B, C) on Cdc25A phosphorylation and polyubiquitylation after UV irradiation. Experiments were performed as described in Supplementary Figure S6. Some aliquots of cell extracts were also subjected to the immunoprecipitation with anti-Myc (IP, α-Myc; A) or the immunoblotting with the indicated antibodies (Input; A–C). (D) A total of 1 μg of Cdc25A was incubated with 10 ng of GST-Chk1 (WT or S296A) with or without 14-3-3γ (0–8 μg) in 20 μl of reaction mixture (25 mM Tris–HCl [pH 7.5], 10 mM MgCl2, 100 μM ATP) at 30°C for 5 min. (E) In total, 1 μg of Cdc25A was incubated with 10–160 ng of GST-Chk1 (WT) with or without 14-3-3γ (2 μg) in 20 μl of the above mixture at 30°C for 5 min. (F) In total, 1 μg of Cdc25C was incubated with 0–40 ng of GST-Chk1 (WT) with or without 14-3-3γ (2 μg) in 20 μl of the above mixture at 30°C for 5 min.
To show the importance of 14-3-3γ in Cdc25A-Ser76 phosphorylation more clearly, we performed an in vitro assay. In the absence of 14-3-3γ, Chk1 appeared to phosphorylate Cdc25A-Thr507 more effectively than Cdc25A-Ser76 (Figure 6D). Cdc25A-Thr507 phosphorylation depended on the amount of Chk1 protein (Figure 6E) rather than the presence of 14-3-3γ (Figure 6D). On the other hand, Cdc25A-Ser76 appeared to be well correlated with the amount of 14-3-3γ (Figure 6D) rather than that of Chk1 protein (Figure 6E). The Ser296A mutation abolished Cdc25A-Ser76 phosphorylation by Chk1 even in the presence of 14-3-3γ (Figure 6D, see S296A lanes). Similar to Cdc25A-Thr507 phosphorylation, Cdc25C-Ser216 phosphorylation depended on the amount of Chk1 protein rather than that of 14-3-3γ (Figure 6F), suggesting that the 14-3-3γ requirement was relatively specific to Cdc25A-Ser76 phosphorylation. All these observations suggest that 14-3-3γ has a critical function in Cdc25A degradation through the complex formation of Chk1 and Cdc25A on 14-3-3γ during DNA damage.
Discussion
A long-standing concept in the functions of mammalian 14-3-3 during DNA damage has been that 14-3-3 binds Cdc25C phosphorylated at Ser216 by Chk1 and negatively regulates Cdc25C activity. The 14-3-3-binding sequence including a phosphorylation site (Ser216 on human Cdc25C) is conserved from yeast to mammals, and Chk1 phosphorylates Cdc25C at Ser216 in mammalian cells (Furnari et al, 1997; Peng et al, 1997; Sanchez et al, 1997; Pines, 1999). Thus, mammalian 14-3-3 proteins have long been believed to modulate the checkpoint signal from Chk1 to Cdc25C. However, Cdc25C-Ser216 is highly phosphorylated (probably by C-TAK1) throughout interphase in the absence of DNA damage (Peng et al, 1998; Russell, 1998; Zhou and Elledge, 2000): we obtained a similar result (Figure 5A). Cdc25B is also reported to be phosphorylated in the absence of DNA damage (Schmitt et al, 2006). Genetic studies raised similar question of whether Chk1 and 14-3-3 proteins act through Cdc25C (or Cdc25B) in the checkpoint response, because normal cell cycle progression and checkpoint responses were observed in mice and cells lacking Cdc25B and Cdc25C (Chen et al, 2001; Lincoln et al, 2002; Ferguson et al, 2005). In sharp contrast, Cdc25A null (−/−) mice die on embryonic day 5–7 during the peri-implantation period (Ray and Kiyokawa, 2007; Ray et al, 2007), suggesting the possibility that Cdc25A may be the most effective Cdk activator during cell cycle progression. Here, we report that 14-3-3 binds Chk1 autophosphorylated at Ser296 in a γ-subtype-specific manner. The complex formation of Chk1 and Cdc25A on 14-3-3γ facilitates Cdc25A phosphorylation at Ser76, a critical phosphorylation site for degradation (Jin et al, 2003; Busino et al, 2004; Melixetian et al, 2009).
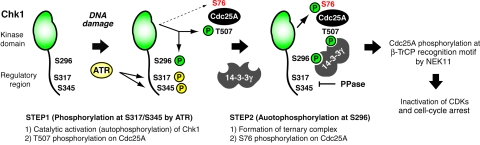
Our model is summarized in Figure 7. At sites (foci) where DNA damage occurs, ATR is first activated and then phosphorylates Ser317 and Ser345 on Chk1. This ATR-induced Chk1 phosphorylation stimulates the catalytic activity of Chk1, resulting in autophosphorylation at Ser296, which generates a docking site for 14-3-3γ. Chk1 activation is also likely to elevate the level of Cdc25A phosphorylation at Thr507, but not at Ser76. Dimer formation by 14-3-3 proteins facilitates their function as platforms for generation of complexes between Chk1 and Cdc25A. Such ternary complex formation enables Chk1 to phosphorylate Cdc25A at Ser76. This phosphorylation induces the generation of a βTrCP recognition motif (phosphodegron) by NEK11 (Jin et al, 2003; Busino et al, 2004; Melixetian et al, 2009), which results in βTrCP-dependent Cdc25A degradation. This proteasome-dependent Cdc25A degradation prevents premature activation of Cdks and induces cell cycle arrest after DNA damage (Zhou and Elledge, 2000; Bartek and Lukas, 2003).
Figure 7.
Model for sequential actions of Chk1 after DNA damage.
During the checkpoint response, Chk1 is accumulated at the nucleus (Jiang et al, 2003), where Cdc25A is enriched. This nuclear accumulation reportedly depends on Chk1-Ser345 phosphorylation (Jiang et al, 2003). As Chk1-Ser296 phosphorylation depends on ATR-induced phosphorylation, our observation also raised another possibility that Chk1-Ser296 autophosphorylation and/or 14-3-3γ might have an important function in Chk1 translocation from cytoplasm to nucleus. We examined the subcellular localization of Myc-Chk1 WT or S296A before or after UV irradiation, but found only marginal changes in the nucleus/cytoplasm ratio between the two proteins (data not shown). In addition, 14-3-3γ depletion had little impact on Chk1 localization before or after UV irradiation (data not shown). Hence, nuclear accumulation of Chk1 is likely to be regulated by Chk1-Ser345 phosphorylation rather than Chk1-Ser296 autophosphorylation and 14-3-3γ association.
Our present data thus provide strong evidence that 14-3-3γ mediates Chk1-induced Cdc25A proteolysis through complex formation between Chk1 and Cdc25A. We propose that Chk1 and 14-3-3 act through Cdc25A in the DNA damage checkpoint response. However, checkpoint regulation by 14-3-3 proteins appears to be more complex in mammalian cells. β and ζ subtypes of 14-3-3 also bind Chk1 in a Ser345 phosphorylation-dependent manner (Jiang et al, 2003) (see also Supplementary Figure S3C). This binding appears to have different functions in checkpoint signalling, as it promotes the nuclear retention of Chk1 likely through the masking of NES on Chk1 (Jiang et al, 2003). Once Ser296 phosphorylation occurs, Ser345 rapidly undergoes dephosphorylation (Leung-Pineda et al, 2006). In contrast, Ser296 phosphorylation induces binding to 14-3-3γ, which promotes Cdc25A degradation. Such ordered binding of Chk1 to specific 14-3-3 subtype(s) may indeed have a critical function in checkpoint signalling.
In our in vitro assays, Chk1-Ser296 phosphorylation occurred (Figure 1C; Supplementary Figure S1E), although we observed little or no phosphorylation of Chk1 by ATR (Supplementary Figure S1E). Thus, these in vitro phenomena appear to contrast with what happens in vivo. As the Chk1 C-terminal regulatory domain was reported to inhibit the catalytic activation of Chk1 (Katsuragi and Sagata, 2004; Walker et al, 2009), this contrast may be due to the difference in the way inhibition of the Chk1 C-terminal regulatory domain is cancelled. Under the in vitro condition, the detergent treatment was reported to induce the structural change of Chk1 C-terminal regulatory domain, which cancelled C-terminal inhibition and then stimulated the catalytic activity in the absence of ATR-induced phosphorylation (Walker et al, 2009). Similar observations were obtained by using Chk1 protein expressed in bacteria. The catalytic activity towards Chk1-Ser296 was elevated by the usage of 1% sarcosyl for bacteria lysis (Figure 1C) (Zhao and Piwnica-Worms, 2001). Therefore, we consider that the C-terminal inhibition is also cancelled during Chk1 purification from insect cells. However, in cells, such cancellation was performed by ATR-induced Chk1 phosphorylation (Zhao and Piwnica-Worms, 2001; Walker et al, 2009). Thus, Ser296 autophosphorylation is likely to require pre-phosphorylation at Ser317 and Ser345 in cells.
In this study, we showed the Chk1 autophosphorylation at Ser296 after ATR-induced phosphorylation. Ser296 phosphorylation makes a docking site for 14-3-3 in a γ-subtype-specific manner. Chk1 binding to 14-3-3γ promotes the complex formation between Chk1 and Cdc25A, which induces Cdc25A-Ser76 phosphorylation and then Cdc25A polyubiquitylation/degradation. Our in vitro analyses also revealed that Chk1 had several autophosphorylation sites other than Ser296 (Supplementary Figure S1E). In this way, Chk1 autophosphorylation at other site(s) may also have a critical function in checkpoint signalling. Further studies in this regard will be undertaken in future.
Materials and methods
Cell culture
We established HeLa cells in which each type of Myc-tagged Chk1 was expressed in a tetracycline/Dox-dependent manner, as described previously (Ikegami et al, 2008). HeLa or Tet-ON HeLa cells were grown in DME medium supplemented with 10% FBS. For Myc-Chk1 induction, Tet-ON HeLa cells were treated with 0.1–1.0 μg/ml of Dox (Sigma, St Louis, MO) for 1 day, except in the case illustrated in Figure 4.
For inhibitor experiments, cells were pre-treated with or without 300 nM UCN-01 (a Chk1 inhibitor; Merck, Whitehouse Station, NJ) or 20 mM caffeine (an ATR and ATM inhibitor; Wako Pure Chemical, Osaka, Japan) in the presence or absence of 10 μM MG132 (Merck) for 30 min. After culture medium was removed, cells in uncovered tissue culture dishes were treated with or without 254-nm UV light at a dose of 5 or 10 J/m2 (FUNA-UV-LINKER, Funakoshi, Tokyo, Japan). The same medium was re-added back and incubated for an additional 10 (Figure 5D), 30 (Figures 5A–C, E–G, and 6A–C) or 60 min (Ikegami et al, 2008). Biochemical fractionation was performed as reported (Jiang et al, 2003; Smits et al, 2006).
For immunocytochemistry, cells grown on coverslips were fixed with 2% formaldehyde for 20 min and then permeabilized with 0.1% Triton X-100 in phosphate-buffered saline at room temperature.
Transfection
siRNA duplexes were purchased from Qiagen (Valencia, CA). Target sequences were as follows: a negative control, AATTCTCCGAACGTGTCACGT; Chk1, AAGGTGAATATAGTGCTGCTA (3′UTR); 14-3-3γ, AAGAGCTATATCCTTAACCAT (#1) and CACTGTCGAATGAGGAACGAA (#2). Transfection was performed with mixtures of each siRNA (final concentration, 20 nM) and LipofectamineTM RNAiMAX reagent, according to the reverse transfection procedures (Invitrogen, Carlsbad, CA).
Human Cdc25A or myc-Chk1 DNA was inserted into pCMV-Tag2B (Stratagene, La Jolla, CA) or pIRES-Puro3 (Clontech, Mountain View, CA), respectively. pEGFP-C1-14-3-3 or pCGN-HA-ubiquitin was kindly provided by Dr Kaibuchi (Nagoya University) or Dr Kikuchi (Osaka University). We constructed the dimerization-defective 14-3-3γ mutant (GFP-14-3-3γ DM) using site-directed mutagenesis: 14-3-3γ was mutated at Glu5 to Lys (E5K), at Leu12 to Gln (L12Q), at Ala13 to Gln (A13Q), at Glu14 to Arg (E14R), at Tyr85 to Gln (Y85Q), at Lys88 to Asn (K88N) and at Glu89 to Gln (E89Q) as described previously (Liu et al, 1995). Plasmid transfection was performed with TransIT-LT1® according to the manufacturers' protocol (Mirus, Madison, WI).
Antibodies
We produced site- and phosphorylation state-specific antibodies for Ser28 on histone H3 and Ser296, Ser317 and Ser345 (Ikegami et al, 2008) on Chk1 (rat monoclonal antibodies), as described previously (Goto and Inagaki, 2007). Competition experiments using peptides (Figure 1B) were also performed as detailed earlier (Sekimata et al, 1996). Antibodies from commercial sources were as follows: mouse anti-Chk1 (G4), anti-14-3-3ɛ (8C3), anti-pan-14-3-3 (H-8), anti-Cyclin B1 (GNS-1), anti-GST (B-14), anti-His (H-3) and rabbit anti-14-3-3γ (C-16) from Santa Cruz Biotechnology (Santa Cruz, CA); rabbit anti-Chk1-pSer317 and -pSer345 (133D3), anti-Akt, anti-Cdc25C (5H9) and anti-Cdc25C pSer216 (63F9) from Cell Signaling Technology (Beverly, MA); mouse anti-Cdc25A (DCS-120+DCS-121), rabbit anti-Cdc25A pSer75 and anti-ATRIP from Abcam (Cambridge, UK); mouse anti-Cyclin A, -Cdc25B, -Cdk1, -Cdk1 pTyr15 and -Orc-2 from BD Biosciences (San Jose, CA); mouse anti-Chk1 (DCS-310) and -α-tubulin (B-5-12) from Sigma; rabbit anti-RPA32 pSer33 (BL744) from Benthyl Laboratories (Montgomery, TX); rabbit anti-Cdc25A pThr506 from ABGENT (San Diego, CA); mouse anti-Myc (4A6) from Millipore (Bedford, MA), mouse anti-HA (12CA5) and anti-GFP (clone 7.1 and 13.1) from Roche Diagnostics (Mannheim, Germany); and mouse anti-Ubiquitin (P4G7) from COVANCE (Berkeley, CA).
Evaluation of mitotic entry
Each Tet-On HeLa cell line was transfected with siRNA for Chk1-3′UTR sequence according to the reverse transfection protocol (Invitrogen). One day after the transfection, we added 2 mM thymidine to the growth medium. After incubation with thymidine for 16 h, cells were extensively washed with pre-warmed PBS and then incubated with Dox-containing medium. Six hours after release, cells were irradiated with UV (5 J/m2) and then incubated in fresh medium containing Dox and 0.1 μg/ml nocodazole (to block passage through mitosis) for an additional 6 h. For the calculation of mitotic indices, cells were fixed and then stained with anti-H3-pSer28 as a mitotic marker (Goto et al, 1999). We judged mitotic cells not only by morphological features of nuclei or chromosomes (DAPI-staining patterns), but also by the detection of mitotic H3-Ser28 phosphorylation in chromosomes.
Protein purification
For bacterial expression of GST-tagged 14-3-3 and Cdc25C fragment (residues 195–256), Escherichia coli strain DH5α (Invitrogen) was transformed with pGEX-6P-1 (GE Healthcare, Little Chalfont, Buckinghamshire, UK) carrying each protein. Each GST-fusion protein was expressed in the presence of 0.2 mM IPTG at 30°C for 4 h. Each protein was purified through glutathione chromatography (GE Healthcare).
GST-Chk1-His (WT, S296A or K38M), Cdc25A-6xHis-Myc or Cdc25C-6xHis-HA was expressed in Sf9 cells for 3 days. After the manufacturers' protocol (GE Healthcare or Qiagen), GST-tagged or His-tagged protein was purified through glutathione- or nickel-affinity chromatography, with slight modifications. As proteins purified from Sf9 cells were already phosphorylated at several sites (data not shown), these proteins attached on the beads were treated with λ protein phosphatase (λPPase; Cell Signaling Technology) at 30°C for 1 h. After washing of the beads, recombinant proteins were eluted according to each protocol.
In some experiments, GST was removed from GST-14-3-3 (Figures 3D, E, 5H, and 6D–F) and GST-Chk1-His (Supplementary Figure 1E) through cleavage at the linker region by PreScission Protease (GE Healthcare) and then absorbed onto the glutathione beads.
Immunoprecipitation and GST pull-down assays from cell extracts
Cells were lysed in IP buffer (50 mM Tris–HCl [pH 7.5], 0.1 M NaCl, 50 mM β-glycerophosphate, 50 mM NaF, 1 mM Na3VO4, 50 mM sodium pyrophosphate, 2 mM EDTA, 1 mM EGTA, 1% NP-40 and 1 mM PMSF) at 4°C. After centrifugation (17 000 g) for 15 min at 4°C, the supernatant was subjected to immunoprecipitation (Ikegami et al, 2008) or GST pull-down assays. For the latter, supernatant was incubated with GST-fusion protein at 4°C for 2 h. After centrifugation (17 0000 g) for 15 min at 4°C, the mixture was incubated with glutathione beads for 30 min at 4°C with rotation. The beads were then washed with IP buffer twice and subjected to immunoblotting. We used appropriate HRP-conjugated, light chain-specific antibodies (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) for the detection of immunoprecipitated Chk1.
In vitro kinase assays
For measurement of Myc-Chk1 kinase activity (Figure 3A), each Myc-Chk1 protein was purified as an anti-Myc immunoprecipitate (Ikegami et al, 2008) from 5 × 106 cells and incubated with 5 μg of GST-Cdc25C fragment (195–256 a.a.) in 20 μl of buffer (25 mM Tris–HCl [pH 7.5], 10 mM MgCl2 and 10 mM [γ-32P] ATP [10 μCi]) at 30°C for 30 min. For measurement of cyclin B1/Cdk1 kinase activity (Figure 4E), cyclin B1/Cdk1 complexes were purified with anti-Cyclin B1 (Ikegami et al, 2008) from 2 × 105 cells and incubated with 4 μg of histone H1 (Roche Diagnostics) in 20 μl of reaction buffer (25 mM Tris–HCl [pH 7.5], 10 mM MgCl2 and 100 μM [γ-32P] ATP [1 μCi]) at 30°C for 5 min.
In vivo ubiquitylation assays
Cells were lysed in hot buffer (95°C) containing 25 mM Tris–HCl (pH 8.0), 1.5% SDS, 0.15% sodium deoxycholate, 0.15% NP-40, 1 mM EDTA, 1 μM okadaic acid and 5 mM N-ethylmaleimide. The lysates were diluted (1:10) with NP-40 lysis buffer (50 mM Hepes at pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA and 0.15% NP-40) and used for immunoprecipitation of Flag-Cdc25A with Anti-FLAG M2 affinity gel (Sigma). Immunoprecipitates were washed three times with NP-40 lysis buffer supplemented with 1 M NaCl and then analysed by SDS–PAGE.
Supplementary Material
Acknowledgments
We thank K Kaibuchi (Nagoya University) for providing human 14-3-3 cDNAs, A Kikuchi (Osaka University) for pCGN-HA-ubiquitin, M Nakanishi (Nagoya City University) for a baculovirus carrying Chk1 and a DNA construct for GST-Cdc25C fragment expression and K Yamashita (Kanazawa University) for baculoviruses carrying Cdc25A and Cdc25C. We are indebted to M Matsuyama for performing the experiments in Figure 1C and Y Ikegami (Nagoya City University), Y Hayashi, C Yuhara and K Kobori for technical assistance. We are grateful to Y Takada for secretarial expertise, and M Moore and J Shields for critical comments on the paper. This work was supported in part by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and from the Ministry of Education, Science, Technology, Sports and Culture of Japan; by a Grant-in-Aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control from the Ministry of Health and Welfare, Japan; by the Uehara Memorial Foundation; the Naito Foundation; the Takeda Science Foundation and a Research Grant from the Princess Takamatsu Cancer Research Fund.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bartek J, Lukas J (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3: 421–429 [DOI] [PubMed] [Google Scholar]
- Boutros R, Lobjois V, Ducommun B (2007) CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer 7: 495–507 [DOI] [PubMed] [Google Scholar]
- Busino L, Chiesa M, Draetta GF, Donzelli M (2004) Cdc25A phosphatase: combinatorial phosphorylation, ubiquitylation and proteolysis. Oncogene 23: 2050–2056 [DOI] [PubMed] [Google Scholar]
- Busino L, Donzelli M, Chiesa M, Guardavaccaro D, Ganoth D, Dorrello NV, Hershko A, Pagano M, Draetta GF (2003) Degradation of Cdc25A by beta-TrCP during S phase and in response to DNA damage. Nature 426: 87–91 [DOI] [PubMed] [Google Scholar]
- Chen MS, Hurov J, White LS, Woodford-Thomas T, Piwnica-Worms H (2001) Absence of apparent phenotype in mice lacking Cdc25C protein phosphatase. Mol Cell Biol 21: 3853–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Ryan CE, Piwnica-Worms H (2003) Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol Cell Biol 23: 7488–7497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Luo C, Deng Y, Ryan K, Register J, Margosiak S, Tempczyk-Russell A, Nguyen B, Myers P, Lundgren K, Kan CC, O′Connor PM (2000) The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell 100: 681–692 [DOI] [PubMed] [Google Scholar]
- Clarke CA, Clarke PR (2005) DNA-dependent phosphorylation of Chk1 and Claspin in a human cell-free system. Biochem J 388: 705–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaway S, Liu HY, Walworth NC (2005) Interaction of 14-3-3 protein with Chk1 affects localization and checkpoint function. J Cell Sci 118: 39–50 [DOI] [PubMed] [Google Scholar]
- Ferguson AM, White LS, Donovan PJ, Piwnica-Worms H (2005) Normal cell cycle and checkpoint responses in mice and cells lacking Cdc25B and Cdc25C protein phosphatases. Mol Cell Biol 25: 2853–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JC, al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Carr AM (1994) 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science 265: 533–535 [DOI] [PubMed] [Google Scholar]
- Furnari B, Rhind N, Russell P (1997) Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science 277: 1495–1497 [DOI] [PubMed] [Google Scholar]
- Goto H, Inagaki M (2007) Production of a site- and phosphorylation state-specific antibody. Nat Protoc 2: 2574–2581 [DOI] [PubMed] [Google Scholar]
- Goto H, Tomono Y, Ajiro K, Kosako H, Fujita M, Sakurai M, Okawa K, Iwamatsu A, Okigaki T, Takahashi T, Inagaki M (1999) Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem 274: 25543–25549 [DOI] [PubMed] [Google Scholar]
- Ikegami Y, Goto H, Kiyono T, Enomoto M, Kasahara K, Tomono Y, Tozawa K, Morita A, Kohri K, Inagaki M (2008) Chk1 phosphorylation at Ser286 and Ser301 occurs with both stalled DNA replication and damage checkpoint stimulation. Biochem Biophys Res Commun 377: 1227–1231 [DOI] [PubMed] [Google Scholar]
- Jackman MR, Pines JN (1997) Cyclins and the G2/M transition. Cancer Surv 29: 47–73 [PubMed] [Google Scholar]
- Jiang K, Pereira E, Maxfield M, Russell B, Goudelock DM, Sanchez Y (2003) Regulation of Chk1 includes chromatin association and 14-3-3 binding following phosphorylation on Ser-345. J Biol Chem 278: 25207–25217 [DOI] [PubMed] [Google Scholar]
- Jin J, Shirogane T, Xu L, Nalepa G, Qin J, Elledge SJ, Harper JW (2003) SCFbeta-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev 17: 3062–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Katsuragi Y, Sagata N (2004) Regulation of Chk1 kinase by autoinhibition and ATR-mediated phosphorylation. Mol Biol Cell 15: 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Pineda V, Ryan CE, Piwnica-Worms H (2006) Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol Cell Biol 26: 7529–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln AJ, Wickramasinghe D, Stein P, Schultz RM, Palko ME, De Miguel MP, Tessarollo L, Donovan PJ (2002) Cdc25b phosphatase is required for resumption of meiosis during oocyte maturation. Nat Genet 30: 446–449 [DOI] [PubMed] [Google Scholar]
- Liu D, Bienkowska J, Petosa C, Collier RJ, Fu H, Liddington R (1995) Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376: 191–194 [DOI] [PubMed] [Google Scholar]
- Mailand N, Falck J, Lukas C, Syljuasen RG, Welcker M, Bartek J, Lukas J (2000) Rapid destruction of human Cdc25A in response to DNA damage. Science 288: 1425–1429 [DOI] [PubMed] [Google Scholar]
- Melixetian M, Klein DK, Sorensen CS, Helin K (2009) NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat Cell Biol 11: 1247–1253 [DOI] [PubMed] [Google Scholar]
- Mohammad DH, Yaffe MB (2009) 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 8: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely KE, Piwnica-Worms H (2003) Cdc25A regulation: to destroy or not to destroy—is that the only question? Cell Cycle 2: 455–457 [PubMed] [Google Scholar]
- Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ III, Wu Z, Stephenson MT, Piwnica-Worms H (1998) C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ 9: 197–208 [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277: 1501–1505 [DOI] [PubMed] [Google Scholar]
- Pines J (1999) Cell cycle. Checkpoint on the nuclear frontier. Nature 397: 104–105 [DOI] [PubMed] [Google Scholar]
- Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, Mansukhani M, Murty VV, Gaciong Z, Meek SE, Piwnica-Worms H, Hibshoosh H, Parsons R (2005) Lack of PTEN sequesters CHK1 and initiates genetic instability. Cancer Cell 7: 193–204 [DOI] [PubMed] [Google Scholar]
- Ray D, Kiyokawa H (2007) CDC25A levels determine the balance of proliferation and checkpoint response. Cell Cycle 6: 3039–3042 [DOI] [PubMed] [Google Scholar]
- Ray D, Terao Y, Nimbalkar D, Hirai H, Osmundson EC, Zou X, Franks R, Christov K, Kiyokawa H (2007) Hemizygous disruption of Cdc25A inhibits cellular transformation and mammary tumorigenesis in mice. Cancer Res 67: 6605–6611 [DOI] [PubMed] [Google Scholar]
- Russell P (1998) Checkpoints on the road to mitosis. Trends Biochem Sci 23: 399–402 [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ (1997) Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277: 1497–1501 [DOI] [PubMed] [Google Scholar]
- Schmitt E, Boutros R, Froment C, Monsarrat B, Ducommun B, Dozier C (2006) CHK1 phosphorylates CDC25B during the cell cycle in the absence of DNA damage. J Cell Sci 119: 4269–4275 [DOI] [PubMed] [Google Scholar]
- Sekimata M, Tsujimura K, Tanaka J, Takeuchi Y, Inagaki N, Inagaki M (1996) Detection of protein kinase activity specifically activated at metaphase-anaphase transition. J Cell Biol 132: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits VA, Reaper PM, Jackson SP (2006) Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol 16: 150–159 [DOI] [PubMed] [Google Scholar]
- Walker M, Black EJ, Oehler V, Gillespie DA, Scott MT (2009) Chk1 C-terminal regulatory phosphorylation mediates checkpoint activation by de-repression of Chk1 catalytic activity. Oncogene 28: 2314–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 21: 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ (2000) The DNA damage response: putting checkpoints in perspective. Nature 408: 433–439 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.