Abstract
The rugged energy landscape of biomolecules and associated large-scale conformational changes have triggered the development of many innovative enhanced sampling methods, either based or not based on molecular dynamics (MD) simulations. Surveyed here are methods in the latter class - including Monte Carlo methods, harmonic approximations, and coarse graining - many of which yield valuable conformational insights into biomolecular structure and flexibility, despite altered kinetics. MD-based methods are surveyed in an upcoming issue of F1000 Biology Reports.
Introduction and context
Computer modeling and simulation offer a modern ‘microscope’ by which to simulate a variety of conformational events in many molecular systems and subsequently extract related mechanistic, thermodynamic, and kinetic information. The governing force fields have been extensively developed on the basis of experimental data and fundamental physical laws. The force fields define complex ‘energy landscapes’ that relate motion and function, as described by Frauenfelder and Wolynes [1,2], and later Onuchic, Thirumalai, and others. These foundations for protein dynamics, folding, and function led to a hierarchical notion of energy landscapes with conformational substates separated by barriers that can be as high as of the order of 100 kJ/mol. Experimental studies, such as from fluorescence spectroscopy, nuclear magnetic resonance (NMR), single-molecule experiments, or four-dimensional electron microscopy provide detailed views on biomolecular motion and confirm a wide range of the timescales involved [3]. Sampling these rugged conformational landscapes to link dynamics to function and bridge the gap between experimental timescales and atomic-level behavior remains a grand challenge.
Methods not based on molecular dynamics (MD) include three broad classes: Monte Carlo (MC) approaches, harmonic approximations, and coarse graining. Although in their own right MC methods are not always satisfactory for large systems, they form essential components of more sophisticated methods (for example, transition path sampling or Markov chain MC sampling, surveyed in the MD-based sampling methods review in an upcoming issue of F1000 Biology Reports [4]). Harmonic approximation-based methods can provide valuable insights into structure/flexibility/function relationships of complex systems, and coarse-graining approaches allow studies of key features of complex systems not amenable to regular atomistic treatments. These methods will be surveyed, with promising directions highlighted.
Major recent advances
Monte Carlo approaches
MC approaches have long been used due to their simplicity and generality. For example, they can be applied to many types of potentials, even discontinuous ones, like the square-well potential for fluid or colloidal suspensions [5], or lattice and off-lattice protein models (for example, [6]). They also allow exploration of variable conditions not amenable to fixed potentials, for instance, the conformational dependencies on ionization states of proteins, which affect side-chain protonation states, as in the electrostatically driven MC (EDMC) method of Scheraga and colleagues [7]. EDMC in combination with different dihedral angle constraints was shown to successfully fold a villin headpiece in close agreement to the NMR structure [7]. For recent reviews on MC applications to biomolecules, see [8,9]; see [10] for a recent review of MC theory.
The general premise in canonical MC sampling is to generate a set of conformations under Boltzmann statistics. Based on the Metropolis acceptance criterion, states that decrease the energy are always accepted and those that increase the energy are accepted with a probability P = exp(-β∆U), where β = 1/kBT and ∆U is the energy difference between the internal energy of the new and old configurations. In practice, this probability is achieved by generating a uniform random variate ran on (0,1) and accepting the new state if P > ran in order to ensure detailed balance and the target thermal distribution. The result of this procedure is the acceptance probability:
(Note that, if P ≤ ran, the old state is re-counted and a new trial state is generated.) This approach allows the molecular system to overcome barriers in the vast conformational space and escape from local minima.
Because convergence of this protocol can be slow, simulated annealing (SA), a form of global optimization, has been developed so that the effective temperature is gradually lowered according to a specified cooling protocol to overcome barriers in the rugged landscape. SA can be used successfully as an extended form of MC, as well as molecular, Langevin, or Brownian dynamics simulations.
Still, selecting the appropriate trial move set and movement magnitudes for a biomolecule without high rejection rates can be challenging. Biased MC variants have been devised with trial moves and hence the conformational deformations designed to move the system to more probable states. Therefore, the Rosenbluth, instead of the Metropolis, criterion is used to factor in the probability (Boltzmann weights) of all trial positions that were skipped in favor of the biased moves:
Here, the Rosenbluth factor W is equal to the product of the sum of the Boltzmann weights of trial positions for each segment i insertion:
where N is the number of chain segments and is the potential energy of the kth trial of adding the ith segment. (One of these trial moves is selected for each segment i with a probability proportional to its Boltzmann weight, and this process is repeated for all segments until the entire chain is re-grown.) Thus, additional overhead is required in biased MC simulations to calculate that probability ratio.
Configurational bias MC (CB-MC) is a biased MC variant that helps ‘grow’ a molecule toward particular states. Traditional CB-MC ‘re-grows’ a deleted position of a polymer at the same end in variable orientations (instead of trying out all neighboring sites randomly). This results in an exponential scaling time with polymer length to re-grow a self-avoiding lattice chain due to the high probability of segment overlaps. In certain applications, much more effective variants can be developed, as in the ‘end-transfer CB-MC’ for chromatin, where one end of the polymer is grown at the other end. Dramatic efficiency can be achieved - quadratic versus exponential scaling - in such applications [11].
Many hybrid MC methods [10] have also been developed to marry the advantages of MC (global sampling potential) with those of MD (continuous local sampling). The success of such methods has been highly application-dependent but can be very effective, especially for small systems [12].
Finally, J-walking or temperature jumps can be introduced to accelerate sampling (similar to SA) but here multiple simulations of non-interacting systems are involved. This parallel tempering approach [13,14] periodically exchanges replicas at different temperatures with a transition probability that maintains each temperature's equilibrium ensemble distribution:
where βi = 1/(kB Ti and Ui is the internal energy of state i (new and old). In this way, barriers over rough energy landscapes can be overcome. These parallel tempering methods have been particularly effective in their MD incarnation, termed replica exchange MD; see accompanying survey [4].
An MC method with advantages similar to parallel tempering in escaping from local barriers was introduced by Wang and Landau [15]. Their method performs multiple random walks in energy space, each to sample a different range of energy; the resulting information is combined to produce canonical averages for calculating thermodynamic quantities at any temperature. When performance of this energy-restricted multiple random walk protocol was compared with parallel tempering for protein conformational sampling, the two methods performed similarly and were faster by two orders of magnitude when compared with a canonical MC simulation at a low temperature; the Wang/Landau MC method was found to be easier to implement on single-processor systems, whereas parallel tempering is advantageous for multi-processor implementations [16].
Given recent successes [8,9,12], some advocate that recent improvements in MC methodology and increased computer memory and speed lend support for the increased application of MC algorithms for folding small biomolecules. Indeed, canonical, multi-canonical, and biased MC protocols that incorporate experimental information (knowledge-based dihedral angle distributions, hybrids involving global optimization techniques and MD, and so on) can significantly enhance the sampling of low energy configurations and reveal folding ensembles of small proteins. General and flexible MC modules have been built into standard programs like CHARMM (Chemistry at HARvard Macromolecular Mechanics) [12], with automatic optimization of step sizes and efficient combinations with minimization or MD modules. These optimized MC methods were found to outperform standard Langevin dynamics simulations in reaching folded states of small proteins.
In general, MC methods can become inefficient for large systems but can be effective for coarse-grained methods (for example, chromatin folding [17]) and as vital components of other methods (for example, transition path sampling; see accompanying survey [4]). These MC extensions and hybrids argue for further development of MC methods for biomolecular applications as a whole.
Harmonic approximations
Normal mode analysis (NMA) and principal component analysis (PCA) are based on harmonic theory. Thus, in their purest forms, spectral decompositions (diagonalization) of a mass-weighted Hessian at thermal equilibrium are performed [18]. This harmonic approximation is far from accurate at ambient temperatures when significant biomolecular fluctuations between minimum-energy regions, as well as occasional rearrangements, occur. Still, these techniques have provided valuable information on collective motions of biomolecules. Elastic networks [19-21] are modern extensions that forgo the computationally demanding diagonalization because the simplified bead/spring-type models are assumed by construction to reflect minimum states of the molecular system.
Besides elastic networks, a successful extension of these techniques that focuses on low-frequency high-amplitude vibrational modes is called ‘essential dynamics’ (ED), to which key contributions have been made by Berendsen, de Groot, Amadei, and others [22]. ED can be used to simulate the dynamics in the low-dimensional space spanned by the low-frequency modes. This is accomplished by constructing the variance/co-variance matrix of positional fluctuations, projecting the original configurations onto each of the principal components, and then following the principal motions in time. There is no explicit assumption of thermal equilibrium here.
The literature is vast with applications of PCA, NMA, and ED with both all-atom and coarse-grained models and in combination with various algorithms, including molecular, Langevin, and Brownian dynamics, to biomolecular conformational flexibility and dynamics. Clearly, these approaches have provided valuable insights into biomolecular flexibility and functional activity. However, the results depend strongly on the level of convergence of the sampling, which influences the results and hence the interpretations.
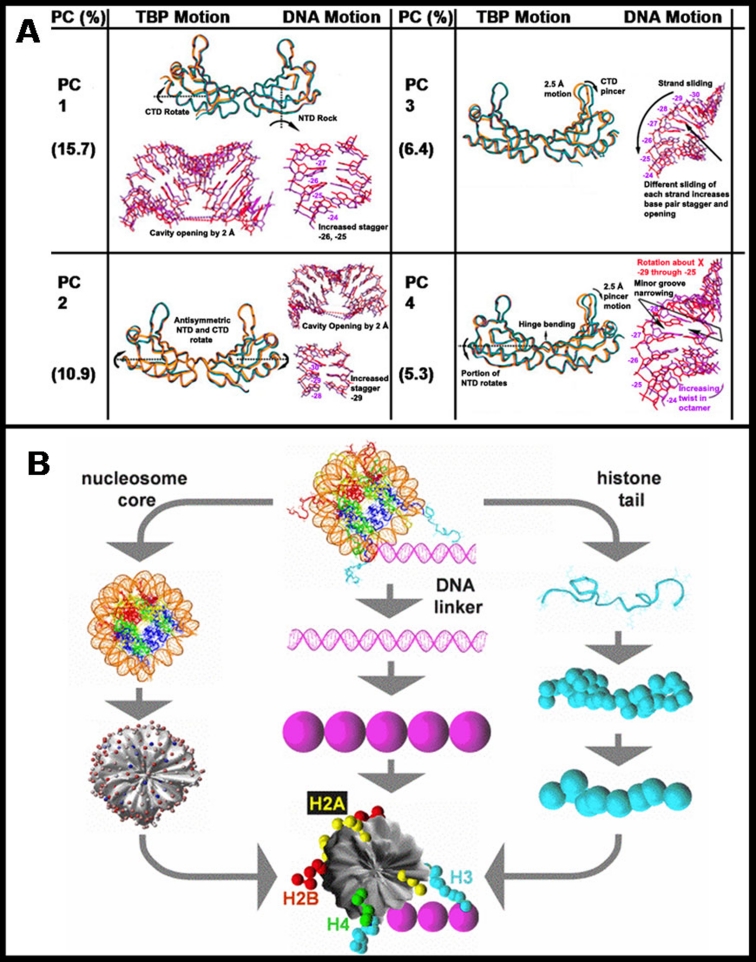
As one example, a PCA study of the closing conformational change of DNA polymerase β upon binding the nucleotide substrate revealed that the top three principal components involve correlations between the thumb subdomain and other regions of the protein (palm, 8-kDa) [23]. Another study, also using PCA, of 13 single-base variants of TATA-box DNA sequences bound to the TATA-binding protein [24], helped explain why these variants revealed a wide range of transcriptional efficiency despite remarkably similar structures: high-efficiency variants favored complexation motions while low-efficiency variants tended toward dissociation deformations. The dominant motions common to all complexes are shown in Figure 1A and are dissected for the protein and bound TATA-box DNA separately.
Figure 1. Examples of principal component analysis and coarse-grained models.
(A) Principal component analysis (PCA) for the group of TATA-binding protein (TBP) structures bound to several TATA-DNA sequences showing top principal components as dissected separately for the protein and the DNA. Adapted from [24]. (B) Mesoscale chromatin model. Beads represent the linker DNAs following the wormlike chain model for DNA, and coarse-grained beads denote the histone tails. An electrostatic surface charge treatment for the nucleosome core (discrete surface charge optimization [DiSCO] to approximate the Poisson-Boltzmann potential as a function of salt) is shown [39,40]. CTD, C-terminal domain; NTD, N-terminal domain; PC, principal component.
Network models have been particularly effective for applications to molecular machines like GroEL and the ribosome modeled by coarse-grained formulations. For example, in an application to the ribosome [25], collective ratchet-like motions were identified that are key in the translocation of the mRNA-tRNA complex.
A tour de force computational comparison between coarse-grained NMA and atomistic ED studies on many proteins [26] showed that both techniques are valid for describing the spectrum of the low-frequency modes and tracing protein flexibility in water, despite the fact that individual eigenvectors from NMA have small values.
An extensive PCA study of a beta-protein WW domain using the coarse-grained protein model UNRES (united residue) [27] showed that dynamics of fast, slow, and non-folding MD trajectories can be well characterized by PCA and that the top few principal components describe the dynamics processes well.
Note that, besides normal-mode-based methods, another class of harmonic approximation methods based on MD includes internal (for example, torsion-angle) MD propagation and variable transformation of classical statistical mechanical configuration partition functions. The latter will be mentioned in the forthcoming MD-based survey [4]. As for the former, internal coordinate dynamics approaches have long been attempted with the rationale that the fewer degrees of freedom (compared to Cartesian coordinates) allow for longer integration timesteps, and hence greater sampling. Indeed, peptide folding and refinement with dihedral angle MD demonstrated a computational advantage of several orders of magnitude compared to Cartesian analogues [28], as well as the capturing of folding pathways of helical peptides and local side-chain and domain dynamics [29]. Another recent study combined dihedral space MD with PCA (dPCA) in a clever way to systematically construct the low-dimensional free energy landscape from a classical MD simulation [30]. Although this analysis is interpretive, it shows that major conformational states, barriers, and reaction pathways for solvated peptides can be visualized from the constructed energy landscape.
In general, such dihedral angle MD approaches for propagating biomolecular motion have not yet caught on at large, perhaps due to both the added cost of the transformation involved in the Newtonian laws of motion and the fact that biomolecular vibrational modes are intricately coupled and hence dynamics can be critically altered by neglecting the high-frequency bond-length and bond-angle modes. However, the increase of coarse-graining models argues for their resurgence.
Coarse graining
System-specific coarse-grained methods are attractive because they drastically reduce the number of degrees of freedom. However, their formulations are highly system-dependent and require as much art as science in constructing, testing/validating, and applying them to appropriately formulated questions. Coarse graining can involve bead models, implicit solvent approximations, discrete lattice models, and general multiscale formulations.
The simplest type of coarse graining involves bead models, long used for proteins (for example, Warshel and Levitt's united residue model [31]) and supercoiled DNA (wormlike chain model of Allison and McCammon [32]), and more recently developed for RNA (for example, [33]). Such methods can lead to meaningful insights into larger-scale rearrangements, including folding, not typically amenable to all-atom simulations. However, the neglect of many details (for example, solvent/solute interactions, which at best can only be accounted for indirectly, as in Langevin or Brownian dynamics) should be considered in the biological interpretations.
In addition to bead models, coarse graining can involve implicit solvent approaches (developed by McCammon, Case, Karplus, Roux, Honig, Truhlar, and many others, and reviewed recently [34]) that reduce the number of degrees of freedom drastically, accounting for them in an average sense in the form of solvation free-energy estimates. Such treatments can be effective, especially when combined with coarse-grained models of molecular systems. However, Chen and Brooks [34] caution that current surface-area-based non-polar models have significant limitations and thus could benefit from incorporating several non-polar solvation aspects.
Lattice models also reduce the conformational degrees of freedom to a discrete set, therefore allowing (in theory) exhaustive sampling of the conformational space. Lattice models of proteins, such as those developed by Gō and Taketomi [21] and by Miyazawa and Jernigan [35], are associated with ideal funnel energy landscapes: a protein chain is modeled by attractive interactions between pairs of residues that interact in the native structures and repulsive interactions of the other pairs, based on statistical data. Recently, Coluzza and Frenkel [36] also applied such lattice models to study the effect of substrates on the folding of their partner proteins. Such lattice models of polymers are typically sampled by MC methods, with tailored moves like corner-flip, crankshaft (rotation by 90° of two consecutive particles), branch rotation, and center-of-mass translation. Protein lattice models have also been extended to off-lattice protein versions.
General coarse-grained or multiscale models are most challenging to formulate and validate because the various components need to be resolved by different approaches and combined effectively. For example, simplified models of the chromatin fiber developed by the groups of Langowski [37], Schiessel [38], Schlick [39], and others, necessarily select the molecular parts to resolve in more detail and those that can be effectively approximated. For example, in studies aimed at deducing the architecture of the 30-nm chromatin fiber, the nucleosome core, histone tails, linker DNA, and linker histones are each modeled differently in a mesoscale model sampled by MC (Figure 1B). The nucleosome core - DNA wrapped around a histone octamer - is represented as an irregular surface with Debye-Hückel point charges that approximate the electrostatic field, as evaluated by the non-linear Poisson-Boltzmann equation; the linker DNA, histone tails, and linker histone protein are described by coarse-grained bead models. Such a chromatin model, when carefully parameterized, can reveal the dynamics/structure of each component as a function of internal and external factors [40].
An impressive example of general coarse graining is the membrane system as modeled by Arkhipov et al. [41] and highlighted in [42]. Six coarse-grained amphiphysin BAR-domain proteins placed on top of a coarse-grained planar membrane patch of lipids triggered a re-shaping of the electrostatics-dominated surface by inducing global curvature within several microseconds, in agreement with curvature dimensions observed experimentally. Other successful and insightful coarse-grained membrane systems were reported recently, revealing pore formation [43], membrane architecture [44], and protein/membrane-binding interactions [45]. Dynamics simulations of virus capsids were also pursued by a coarse-grained model to study the factors affecting capsid stability [46]. An ambitious coarse-grained model of the GroES chaperone showed that the equatorial region of the GroEL/GroES chaperonin complex creates a channel that blocks the passage of folded proteins while at the same time welcomes the passage of secondary segments of diameter up to that of an alpha helix [47].
A minimalist coarse-graining model for proteins based on a ‘switching Gō model’ was also developed and applied to derive a rotational mechanism of a biomolecular machine, an ATP-driven molecular motor, F1-ATPase [48].
A key question that was recently addressed by the Voth group [49] was how, in general, should the coarse graining be chosen. Their work proposed a systematic elastic network coarse-graining approach that essentially selects beads to represent groups of atoms so that atoms in the same domain reflect the collective motions as computed by PCA [49]. These beads are determined by minimizing a residual of displacement differences. As shown for models of the HIV-1 capsid protein dimer, six- and eight-site models both approximate the system's ‘essential dynamics’ well, as determined by subdomain dynamics. They also showed that such coarse-grained models for peptides can visit and re-visit the folded state, unlike atomistic MD simulations, which reveal limited sampling [50].
A systematic parameterization of protein side chains for a coarse-grained peptide model in coarse-grained solvent was also reported by Han et al. [51], who demonstrated comparable solvation free energies with respect to atomistic models and a factor of 1,000 speedup.
Another rigorous approach to multiscale formulations was described recently by Noid et al. [52], who developed a formal statistical mechanical framework for multiscale coarse-grained models by constructing a many-body potential of mean force that generates equilibrium probability distributions for the coarse-grained sites using information from atomistic simulations. Thus, the work rigorously connects equilibrium ensembles of all-atom and multiscale models. Many interesting applications of multiscale models in various scientific fields are collected in a special volume [53].
Future directions
With advances in computer memory and speed, MC methods are enjoying increased applications in biomolecular simulations, both for atomistic and coarse-grained models. They are vital components of various enhanced sampling methods like transition path sampling (see [4]) and thus deserve further consideration and development as our molecular models and force-field potentials evolve and become more complex and hence more amenable to MC methods.
While harmonic approximation methods like PCA, NMA, ED, and elastic networks continue to add valuable insights into biomolecular flexibility and function, they are also participating in more applications with the growth of network models for molecular machines that help dissect and distill complex functional motions.
Coarse-grained models are clearly emerging as a favored approach to study either long-time behavior of small systems like peptides, as in folding trajectories, or large supramolecular systems that are too complex to study at atomic resolution, such as the chromatin fiber, membrane systems, and viruses. Exciting new rigorous frameworks for coarse-graining general systems are also under development and will likely increase.
Significantly, all of these approaches for enhanced sampling can be combined for cumulative and significant computational advantages. For example, a coarse-grained energy function with parallel tempering MC was used to study protein-protein binding through creating equilibrium ensembles of various complexes to help interpret paramagnetic relaxation enhancement experimental data [54]. The combined populations of the specific complexes and the relatively small number of distinct but non-specific complexes helped explain the existence of observed transient encounter complexes.
Of course, careful testing, parameterization, and cautious interpretations are especially warranted in these creative coarse-grained approaches. Still, all of these advances, including elastic networks, coarse-grained approaches, implicit solvation, internal coordinate PCA, and low-frequency vibrational mode propagation, are collectively opening the way to exciting applications of a rich variety of biomolecular systems regarding large-scale conformational changes and functional dynamics on millisecond and longer timescales that are helping to close the gap between experimental and theoretical time frames.
Acknowledgments
Support from the National Science Foundation, the National Institutes of Health, the American Chemical Society, and Philip Morris International is gratefully acknowledged. The author thanks Meredith Foley, Shereef Elmetwaly, Hin Hark Gan, Christian Laing, and Ravi Radhakrishnan for valuable assistance and comments on the manuscript.
Abbreviations
- CB-MC
configurational bias Monte Carlo
- CHARMM
Chemistry at Harvard Macromolecular Mechanics
- DiSCO
discrete surface charge optimization
- dPCA
dihedral angle principal component analysis
- ED
essential dynamics
- EDMC
electrostatically driven Monte Carlo
- MC
Monte Carlo
- MD
molecular dynamics
- NMA
normal mode analysis
- NMR
nuclear magnetic resonance
- PCA
principal component analysis
- SA
simulated annealing
Competing interests
The author declares that she has no competing interests.
The electronic version of this article is the complete one and can be found at: http://F1000.com/Reports/Biology/content/1/48
References
- 1.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 2.Wolynes PG. Recent successes of the energy landscape theory of protein folding and function. Quart Rev Biophys. 2005;38:405–10. doi: 10.1017/S0033583505004075. [DOI] [PubMed] [Google Scholar]
- 3.Zewail AH. Physical Biology: From Atoms to Medicine. London: Imperial College Press; 2008. Physical biology: 4D visualization of complexity; pp. 23–50. [Google Scholar]
- 4.Schlick T. F1000 Biology Reports. 2009. Molecular-dynamics based approaches for enhanced sampling of long-time, large-scale conformational changes in biomolecules. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulougouris GC, Frenkel D. Novel Monte Carlo scheme for systems with short-ranged interactions. J Chem Phys. 2005;122:244106. doi: 10.1063/1.1931652. [DOI] [PubMed] [Google Scholar]
- 6.Chen WW, Yand JS, Shakhnovich EI. A knowledge-based move set for protein folding. Proteins. 2007;66:682–8. doi: 10.1002/prot.21237. [DOI] [PubMed] [Google Scholar]
- 7.Ripoll DR, Vila JA, Scheraga HA. Folding of the villin headpiece subdomain from random structures. Analysis of the charge distribution as a function of the pH. J Mol Biol. 2004;339:915–25. doi: 10.1016/j.jmb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Liwo A, Czaplewski C, Oldziej S, Scheraga HA. Computational techniques for efficient conformational sampling of proteins. Curr Opin Struct Biol. 2008;18:134–9. doi: 10.1016/j.sbi.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmermann O, Hansmann UH. Understanding protein folding: small proteins in silico. Biochim Biophys Acta. 2008;1784:252–8. doi: 10.1016/j.bbapap.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earl DJ, Deem MW. Monte Carlo simulations. Methods Mol Biol. 2008;443:25–36. doi: 10.1007/978-1-59745-177-2_2. [DOI] [PubMed] [Google Scholar]
- 11.Arya G, Schlick T. Efficient global biopolymer sampling with end-transfer configurational bias Monte Carlo. J Chem Phys. 2007;126:044107. doi: 10.1063/1.2428305. [DOI] [PubMed] [Google Scholar]
- 12.Hu J, Ma A, Dinner R. Monte Carlo simulations of biomolecules: the MC module in CHARMM. J Comput Chem. 2006;27:203–16. doi: 10.1002/jcc.20327. [DOI] [PubMed] [Google Scholar]
- 13.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics methods for protein folding. Chem Phys Lett. 1999;314:141–51. doi: 10.1016/S0009-2614(99)01123-9. [DOI] [Google Scholar]
- 14.Hansmann UEH. Parallel-tempering algorithm for conformational studies of biological molecules. Chem Phys Lett. 1997;281:140–50. doi: 10.1016/S0009-2614(97)01198-6. [DOI] [Google Scholar]
- 15.Wang F, Landau DP. Efficient, multiple-range random walk algorithm to calculate the density of states. Phys Rev Lett. 2001;86:2050–3. doi: 10.1103/PhysRevLett.86.2050. [DOI] [PubMed] [Google Scholar]
- 16.Junghans D, Hansmann UHE. Numerical comparison of Wang-Landau sampling and aprallel tempering for met-enkephlin. Intl J Mod Phys C. 2006;17:817–24. doi: 10.1142/S012918310600931X. [DOI] [Google Scholar]
- 17.Sun J, Zhang Q, Schlick T. Electrostatic mechanism of nucleosomal array folding revealed by computer simulation. Proc Natl Acad Sci U S A. 2005;102:8180–5. doi: 10.1073/pnas.0408867102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayward S, de Groot BL. Normal modes and essential dynamics. Methods Mol Biol. 2008;443:89–106. doi: 10.1007/978-1-59745-177-2_5. [DOI] [PubMed] [Google Scholar]
- 19.Tirion MM. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys Rev Lett. 1996;77:1905–8. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 20.Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–81. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 21.Gō N, Taketomi H. Respective roles of short- and long-range interactions in protein folding. Proc Natl Acad Sci U S A. 1978;75:559–63. doi: 10.1073/pnas.75.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Aalten DMF, de Groot BL, Findlay JBC, Berendsen HJC, Amadei A. A comparison of techniques for calculating protein essential dynamics. J Comput Chem. 1997;18:169–81. [Google Scholar]
- 23.Arora K, Schlick T. In silico evidence for DNA polymerase-β's substrate-induced conformational change. Biophys J. 2004;87:3088–99. doi: 10.1529/biophysj.104.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strahs D, Barash D, Qian X, Schlick T. Sequence-dependent solution structure and motions of 13 TATA/TBP complexes. Biopolymers. 2003;69:216–43. doi: 10.1002/bip.10409. [DOI] [PubMed] [Google Scholar]
- 25.Tama F, Valle M, Frank J, Brooks CL., 3rd Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Natl Acad Sci U S A. 2003;100:9319–23. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rueda M, Chacon P, Orozco M. Thorough validation of protein normal mode analysis: a comparative study with essential dynamics. Structure. 2007;15:565–75. doi: 10.1016/j.str.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Maisuradze G, Liwo A, Scheraga H. Principal component analysis for protein folding dynamics. J Mol Biol. 2009;385:312–29. doi: 10.1016/j.jmb.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Im W, Brooks CL., 3rd Application of torsion angle molecular dynamics for efficient sampling of protein conformations. J Comput Chem. 2005;26:1565–78. doi: 10.1002/jcc.20293. [DOI] [PubMed] [Google Scholar]
- 29.Yun M-R, Lavery R, Mousseau N, Zakrzewska K, Derreumaux P. ARTIST: an activated method in internal coordinate space for sampling protein energy landscapes. Proteins. 2006;63:967–75. doi: 10.1002/prot.20938. [DOI] [PubMed] [Google Scholar]
- 30.Altis A, Otten M, Nguyen PH, Hegger R, Stock G. Construction of the free energy landscape of biomolecules via dihedral angle principal component analysis. J Chem Phys. 2008;128:245102. doi: 10.1063/1.2945165. [DOI] [PubMed] [Google Scholar]
- 31.Warshel A, Levitt M. Theoretical studies of enzymic reactions: dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme. J Mol Biol. 1976;103:227–49. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 32.Allison SA, McCammon JA. Multistep Brownian dynamics: application to short wormlike chains. Biopolymers. 1984;23:363–75. doi: 10.1002/bip.360230214. [DOI] [PubMed] [Google Scholar]
- 33.Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA. 2008;14:1164–73. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Brooks CL., 3rd Implicit modeling of nonpolar solvation for simulating protein folding and conformational transitions. Phys Chem Chem Phys. 2008;10:471–81. doi: 10.1039/b714141f. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by B Montgomery Pettitt 11 Feb 2008
- 35.Miyazawa S, Jernigan RL. A new substitution matrix for protein sequence searches based on contact frequencies in protein structures. Prot Eng. 1993;6:267–78. doi: 10.1093/protein/6.3.267. [DOI] [PubMed] [Google Scholar]
- 36.Coluzza I, Frenkel D. Monte Carlo study of substrate-induced folding and refolding of lattice proteins. Biophys J. 2007;92:1150–6. doi: 10.1529/biophysj.106.084236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voltz K, Trylska J, Tozzini V, Kurkal-Siebert V, Langowski J, Smith J. Coarse-grained force field for the nucleosome from self-consistent multiscaling. J Comput Chem. 2008;29:1429–39. doi: 10.1002/jcc.20902. [DOI] [PubMed] [Google Scholar]
- 38.Mühlbacher F, Schiessel H, Holm C. Tail-induced attraction between nucleosome core particles. Phys Rev E. 2006;74:031919. doi: 10.1103/PhysRevE.74.031919. [DOI] [PubMed] [Google Scholar]
- 39.Arya G, Schlick T. Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc Natl Acad Sci U S A. 2006;103:16236–41. doi: 10.1073/pnas.0604817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arya G, Schlick T. A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J Phys Chem A. 2009;113:4045–59. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arkhipov A, Yin Y, Schulten K. Four-scale description of membrane sculpting by BAR domains. Biophys J. 2008;95:2806–21. doi: 10.1529/biophysj.108.132563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein ML, Shinoda W. Large-scale molecular dynamics simulations of self-assembling systems. Science. 2008;321:798. doi: 10.1126/science.1157834. [DOI] [PubMed] [Google Scholar]
- 43.Gkeka P, Sarkisov L. Spontaneous formation of a barrel-stave pore in a coarse-grained model of the synthetic LS3 peptide and a DPPC lipid bilayer. J Phys Chem B. 2009;113:6–8. doi: 10.1021/jp808417a. [DOI] [PubMed] [Google Scholar]
- 44.Scott KA, Bond PJ, Ivetac A, Chetwynd AP, Khalid S, Sansom MSP. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure. 2008;16:621–30. doi: 10.1016/j.str.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 45.Kim YC, Hummer G. Coarse-grained models for simulations of multiprotein complexes: application to ubiquitin binding. J Mol Biol. 2008;375:1416–33. doi: 10.1016/j.jmb.2007.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arkhipov A, Freddolino PL, Schulten K. Stability and dynamics of virus capsids described by Coarse-grained modeling. Structure. 2006;14:1767–77. doi: 10.1016/j.str.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Coluzza I, De Simon A, Fraternali F, Frenkel D. Multi-scale simulations provide supporting evidence for the hypothesis of intramolecular protein translocation in GroEL/GroES complexes. PLoS Comp Biol. 2008;4:e1000006. doi: 10.1371/journal.pcbi.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koga N, Takada S. Folding-based molecular simulations reveal mechanisms of the rotary motor F1-ATPase. Proc Natl Acad Sci U S A. 2008;103:5367–72. doi: 10.1073/pnas.0509642103. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Wolfgang Junge 20 Apr 2006
- 49.Lyman E, Pfaendtner J, Voth GA. Systematic multiscale parameterization of heterogeneous elastic network models of proteins. Biophys J. 2008;95:4183–92. doi: 10.1529/biophysj.108.139733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorpe IF, Zhou J, Voth GA. Peptide folding using multiscale coarse-grained models. J Phys Chem B. 2008;112:13079–90. doi: 10.1021/jp8015968. [DOI] [PubMed] [Google Scholar]
- 51.Han W, Wan C-K, Wu Y-D. Toward a coarse-grained protein model coupled with a coarse-grained solvent model: solvation free energies of amino acid side chains. J Chem Theo Comp. 2008;4:1891–901. doi: 10.1021/ct800184c. [DOI] [PubMed] [Google Scholar]
- 52.Noid WG, Chu J-W, Ayton GS, Krishna V, Izvekov S, Voth GA, Das A, Anderson HC. The multiscale coarse-grained method: I. a rigorous bridge between atomistic and coarse-grained models. J Chem Phys. 2008;128:244114. doi: 10.1063/1.2938860. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Nathan Baker 05 Jan 2009
- 53.Schlick T. From Macroscopic to Mesoscopic Models of Chromatin Folding. In: Fish J, editor. Multiscale Methods: Bridging the Scales in Science and Engineering. Oxford: Oxford University Press; 2009. [Google Scholar]
- 54.Kim YC, Tang C, Clore GM, Hummer G. Replica exchange simulations of transient encounter complexes in protein-protein association. Proc Natl Acad Sci U S A. 2008;105:12855–60. doi: 10.1073/pnas.0802460105. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Joan Shea 27 Oct 2008