Abstract
Compared to the mechanisms of axon guidance, relatively little is known about the transcriptional control of dendrite guidance. The Drosophila olfactory system with its stereotyped organization provides an excellent model to study the transcriptional control of dendrite wiring specificity. Each projection neuron (PN) targets its dendrites to a specific glomerulus in the antennal lobe and its axon stereotypically to higher brain centers. Using a forward genetic screen, we identified a mutation in Rpd3 that disrupts PN targeting specificity. Rpd3 encodes a class I histone deacetylase (HDAC) homologous to mammalian HDAC1 and HDAC2. Rpd3−/− PN dendrites that normally target to a dorsolateral glomerulus mistarget to medial glomeruli in the antennal lobe, and axons exhibit a severe overbranching phenotype. These phenotypes can be rescued by postmitotic expression of Rpd3 but not HDAC3, the only other class I HDAC in Drosophila. Furthermore, disruption of the atypical homeodomain transcription factor Prospero (Pros) yields similar phenotypes, which can be rescued by Pros expression in postmitotic neurons. Strikingly, overexpression of Pros can suppress Rpd3−/− phenotypes. Our study suggests a specific function for the general chromatin remodeling factor Rpd3 in regulating dendrite targeting in neurons, largely through the postmitotic action of the Pros transcription factor.
Introduction
The Drosophila olfactory system with its stereotyped organization provides an excellent model to study wiring specificity. Each olfactory projection neuron (PN) targets its dendrites to one of ∼50 glomeruli in the antennal lobe to make synaptic connections with a specific class of olfactory receptor neurons, and sends its axon stereotypically to higher brain centers (Jefferis et al., 2001; Marin et al., 2002; Wong et al., 2002). Previous studies have identified several transcription factors that act cell-autonomously to regulate PN dendrite targeting specificity (Komiyama et al., 2003; Zhu et al., 2006; Komiyama and Luo, 2007; Spletter et al., 2007). We report here a genetic screen for additional factors that regulate PN dendrite wiring specificity, and identify Rpd3 [homolog of mammalian histone deacetylases 1 and 2 (HDAC1/2)] and the transcription factor Prospero (Pros) as additional regulators of PN dendrite wiring specificity.
HDACs are best studied for their role in deacetylating lysines on histones, resulting in a more compact chromatin structure correlated with gene repression (Thiagalingam et al., 2003). In the nervous system, HDACs have been most intensely studied in neurodegeneration. For instance, inhibition of HDACs can improve memory in mouse models of neurodegenerative diseases (Fischer et al., 2007) and can reduce polyglutamine-dependent neurodegeneration in Drosophila (Steffan et al., 2001). There are three major classes of HDACs, grouped based on their homology to yeast histone deacetylases: class I HDACs bear high homology to the yeast RPD3 protein, class II to Hda1, and class III to Sir2 (Thiagalingam et al., 2003). In Drosophila, Rpd3 and HDAC3 are the only two class I HDACs (De Rubertis et al., 1996; Johnson et al., 1998). They are present in the Drosophila nucleus and function in chromatin remodeling (Foglietti et al., 2006). In addition, RNAi knockdown of Rpd3 in embryos causes increased dendrite arborization in class I da neurons (Parrish et al., 2006). The role of HDACs in wiring specificity has not been characterized.
Prospero is an evolutionarily conserved atypical homeodomain transcription factor originally identified in Drosophila for its role in regulating homeobox gene expression in the developing CNS (Doe et al., 1991). Pros has been shown to regulate axon pathfinding (Doe et al., 1991), axon outgrowth (Vaessin et al., 1991) and dendrite morphogenesis (Gao et al., 1999). During asymmetric cell division of embryonic neuroblasts (Fig. 1A), pros is only transcribed in neuroblasts (Broadus et al., 1998), and Pros protein is asymmetrically segregated into the nucleus of ganglion mother cells (Knoblich et al., 1995; Spana and Doe, 1995). Pros is thought to act in ganglion mother cells to repress neuroblast genes required for self renewal while activating genes for terminal differentiation (Choksi et al., 2006). Although several studies show that Pros is downregulated in embryonic postmitotic neurons (Doe et al., 1991; Vaessin et al., 1991; Choksi et al., 2006), recent studies show that Pros is present in some larval neurons (Guenin et al., 2007, 2010). Our study reveals a function for Rpd3 in regulating dendrite targeting in neurons, largely through the postmitotic action of the Pros transcription factor.
Figure 1.
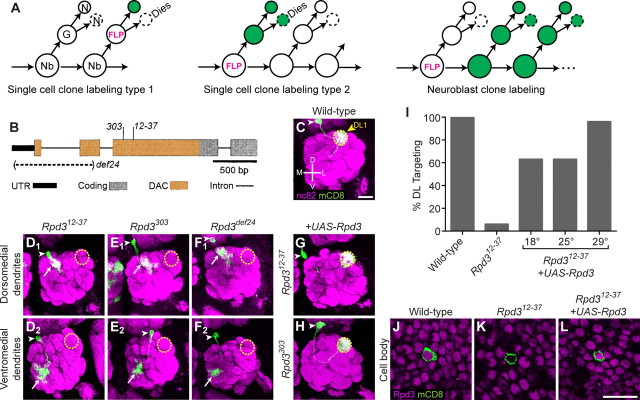
Rpd3 acts in postmitotic neurons to regulate projection neuron dendrite targeting. A, Schematic of MARCM single-cell or neuroblast clone generation. Using MARCM, single-cell or neuroblast clones can be positively labeled (shown in green) after FLP-mediated mitotic recombination in the ganglion mother cell or neuroblast, as indicated by the three models. Recent studies show that the sibling of the DL1 neuron dies during development (Lin et al., 2010; Potter et al., 2010). Nb, Neuroblast; G, ganglion mother cell; N, postmitotic neuron; FLP, Flippase. B, Schematic of the Rpd3 gene locus, with indicated mutations (modified after Mottus et al., 2000). Allele Rpd312–37 was generated in this study; Rpd3303 and Rpd3def24 have been previously described. Allele Rpd3303 is a cysteine to tyrosine missense mutation which retains anti-Rpd3 immunoreactivity (data not shown). Allele Rpd3def24 is a large deletion encompassing the transcriptional start and much of the N terminus which results in a loss of anti-Rpd3 immunoreactivity (data not shown). UTR, Untranslated region; DAC, deacetylase domain. C, WT DL1 PN dendrites target specifically to the glomerulus DL1 (yellow dashed circle). White arrowhead denotes cell body. D, Representative images of Rpd312–37 mutant single-cell clones. Mistargeting to the dorsomedial (DM) quadrant of the antennal lobe occurs in 47% of clones (D1). Mistargeting to ventromedial (VM) quadrant occurs in 44% of clones (D2). A total of 9% showed dorsolateral (DL) targeting (data not shown). n = 32. White arrowhead denotes cell body, yellow dashed circle indicates location of the DL1 glomerulus, white arrow denotes mistargeted dendrites. E, Representative images of Rpd3303 mutant single-cell clones: 82% mistargeting to DM (E1), 14% mistargeting to VM (E2). A total of 4% showed DL targeting (data not shown). n = 22. F, Representative images of Rpd3def24 mutant single-cell clones: 61% mistargeting to DM (F1), 25% mistargeting to VM (F2). A total of 14% showed DL targeting (data not shown). n = 28. G, Postmitotic expression of UAS-Rpd3 in a single DL1 PN rescues the Rpd312–37 dendrite phenotype. H, Postmitotic expression of UAS-Rpd3 in a single DL1 PN rescues the Rpd3303 dendrite phenotype. I, Quantification of DL targeting for genotypes in C, D, and G. Number of analyzed clones left to right is 35, 32, 22, 19, 24. J, At 24hAPF, Rpd3 is expressed ubiquitously in all nuclei, including PNs. K, Rpd312–37 single-cell MARCM clone (green) lacks Rpd3 immunoreactivity. L, MARCM expression of UAS-Rpd3 restores Rpd3 immunoreactivity in an Rpd312–37 single-cell clone. Green marks mCD8-GFP-labeled PN dendrites (C–H) and cell bodies (J–L) generated by MARCM and labeled using GH146-GAL4. C–H show confocal stacks, magenta is the presynaptic marker nc82. J–L show single confocal sections, magenta is anti-Rpd3. Scale bars: C (for C–H), L (for J–L), 20 μm.
Materials and Methods
Identification of the Rpd3 mutation.
MARCM-based mosaic screen and SNP mapping leading to the identification of Rpd312–37 were analogous to procedures previously described (Chihara et al., 2007). After narrowing the region to ∼67 kb, all open reading frames and splice acceptor and donor sites were sequenced.
Identification of the prospero mutation.
pros38 was isolated from a MARCM-based P-element forward genetic screen. pros38 possesses a p[mCD8-GFP, y+] transposon inserted early in the open reading frame of prospero, at amino acid 188. The insertion site was confirmed by inverse PCR.
Immunostaining.
MARCM was performed as described previously (Wu and Luo, 2006b). Fly brains of both genders were dissected, fixed, and stained as described previously (Wu and Luo, 2006a). Antibody conditions: rabbit anti-Rpd3 1:1000 (gift from J. T. Kadonaga, University of California San Diego, San Diego, CA), rabbit anti-acetylated lysine 1:1000 (Cell Signaling Technology #9441), mouse anti-Prospero 1:4 (Developmental Studies Hybridoma Bank #MR1A; C. Q. Doe, University of Oregon, Eugene, OR), rat anti-mCD8α 1:100 (Invitrogen Caltag #RM2200), and mouse anti-nc82 1:40 (Developmental Studies Hybridoma Bank #nc82; E. Buchner, University of Würzburg, Würzburg, Germany).
Quantification of DL1 PN dendrite targeting.
DL targeting (see Figs. 1I, 5D, 6C) includes full innervation of the DL1 glomerulus, as well as minor dendrite spillover into nearby glomeruli in the dorsolateral quadrant of the antennal lobe.
Figure 5.
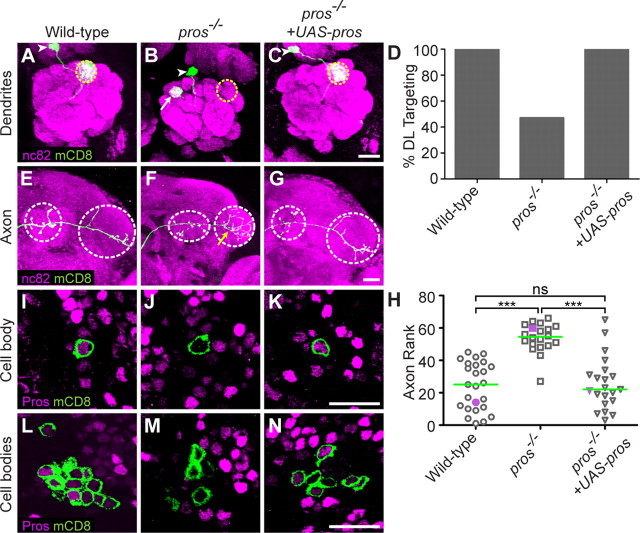
Prospero acts in postmitotic neurons to regulate PN dendrite targeting and axon terminal arborization. A, WT DL1 PN dendrites target specifically to the glomerulus DL1, same as Figure 1C. B, pros−/− DL1 PN dendrites mistarget to medial glomeruli (white arrow). C, Postmitotic expression of UAS-pros rescues the pros−/− DL1 PN dendrite phenotype. D, Quantification of DL targeting for the genotypes in A–C. Number of clones is 25, 45, 34. E, WT DL1 axon, same as Figure 2A. F, pros−/− DL1 PN axon exhibits an overbranching phenotype in the lateral horn (yellow arrow). G, Postmitotic expression of UAS-pros rescues the pros−/− overbranching phenotype. H, Rank order of axon overbranching phenotype of different genotypes as indicated. Pairwise Mann–Whitney U tests were performed to determine significance. Number of clones is 25, 20, 21. I, At 24hAPF, Pros is expressed at varying levels in different PN nuclei. The DL1 PN (green) expresses an intermediate level of Pros protein. J, pros−/− single-cell MARCM clone lacks Pros immunoreactivity. K, MARCM expression of UAS-pros restores Pros immunoreactivity in a single PN. L, At 24hAPF, Pros is expressed at varying levels in PN cell bodies. PNs can express high, medium, low, or undetectable levels of Pros. This variable expression persists throughout development and adulthood (data not shown). M, Cell bodies of pros−/− neuroblast MARCM clones (green) lack Pros immunoreactivity. N, MARCM expression of UAS-pros restores Pros immunoreactivity in neuroblast clone PNs at 24hAPF. The level of UAS-pros expression varies similarly to endogenous Pros expression, suggesting a posttranscriptional mechanism in regulating Pros protein level. Green marks mCD8-GFP-labeled PN dendrites (A–C), axons (E–G), and cell bodies (I–N) generated by MARCM and labeled using GH146-GAL4. A–C and E–G show confocal stacks; magenta is the presynaptic marker nc82. I–N show single confocal sections; magenta is anti-Prospero staining. Symbols are as in Figures 1 and 2. Scale bars: C (for A–C), G (for E–G), K (for I–K), N (for L–N), 20 μm.
Figure 6.
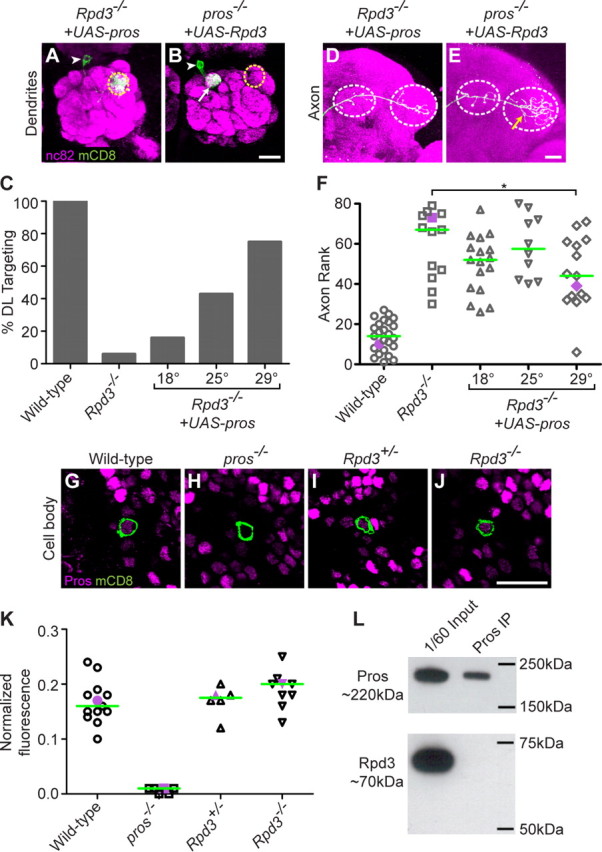
Postmitotic expression of Prospero suppresses Rpd3−/− PN phenotypes. A, Postmitotic expression of UAS-pros suppresses the Rpd3−/− PN phenotype, restoring DL1 PN dendrite targeting to the DL1 glomerulus. B, Postmitotic expression of UAS-Rpd3 does not suppress the pros−/−-mistargeting phenotype. C, Quantification of UAS-pros suppression of the Rpd3−/− phenotype at different temperatures. Number of clones is 25, 13, 20, 23, 16. D, Postmitotic expression of UAS-pros partially suppresses the Rpd3−/− axon overbranching phenotype. E, Postmitotic expression of UAS-Rpd3 does not suppress the pros−/− axon overbranching phenotype. F, Rank order of axon overbranching for different genotypes as indicated. Pairwise Mann–Whitney U tests were performed to determine significance. *p < 0.05 at 29°C; no significant difference at 18°C or 25°C. Number of clones is 25, 13, 17, 10, 15. G, At 24hAPF, Pros is expressed at an intermediate level in the DL1 PN (green). H, pros−/− DL1 MARCM clone lacks Pros immunoreactivity. I, Heterozygous Rpd3+/− MARCM clone shows a normal level of Pros immunoreactivity. J, Homozygous Rpd3−/− MARCM clone shows a normal level of Pros immunoreactivity. K, Normalized fluorescence of DL1 PNs for different genotypes as indicated. Purple symbols indicate the examples shown in G–J. Green line denotes median. Number of clones is 13, 6, 6, 9. Graph shows the ratio of DL1 to highest intensity. L, Immunoprecipitation of Prospero does not result in coimmunoprecipitation of Rpd3. Top shows 1/60 input and final immunoprecipitation of Pros blotted with anti-Pros. Bottom shows 1/60 input and denatured immunoprecipitate blotted with anti-Rpd3. Green marks mCD8-GFP-labeled PN dendrites (A, B), axons (D, E), and cell bodies (G–J) generated by MARCM and labeled using GH146-GAL4. (A, B, D, E) show confocal stacks; magenta is the presynaptic marker nc82. G–J show single confocal sections; magenta is anti-Prospero staining. Symbols are as in Figures 1 and 2. Scale bars: B (for A, B), E (for D, E), J (for G–J), 20 μm.
Axon rank order.
Confocal stacks of axons were blindly rank ordered by two independent observers with consistent results. Axons were ranked by degree of axon overbranching, with the lowest score denoting the least branching (closest to wild type) (see Figs. 2D, 5H, 6F).
Figure 2.
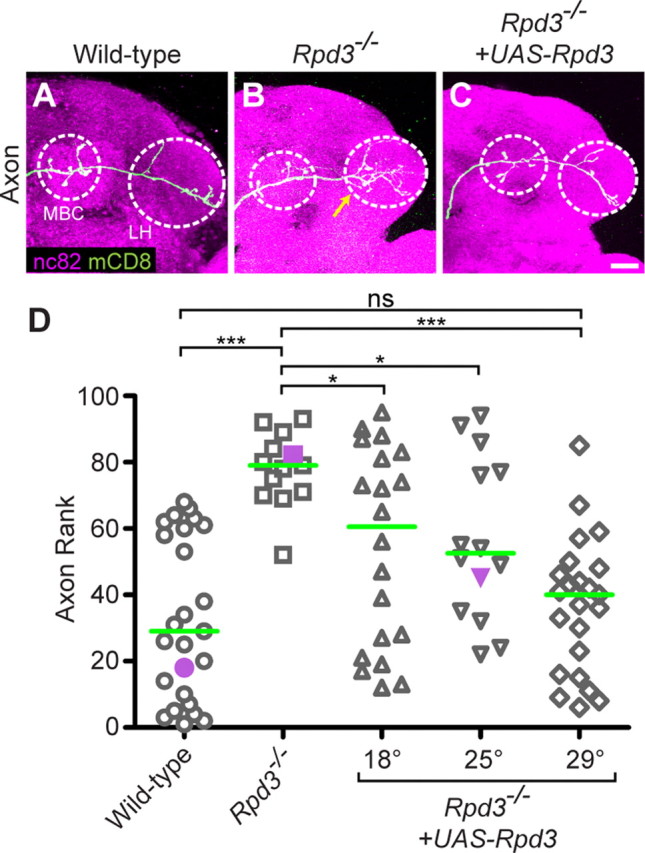
Rpd3 limits PN axon terminal arborization. A, WT DL1 axons show an L-shaped pattern in the lateral horn (LH), with additional branches in the mushroom body calyx (MBC), both outlined with white dashed circle. B, Rpd3−/− DL1 PN axons exhibit an overbranching phenotype in the lateral horn (yellow arrow). C, Postmitotic expression of UAS-Rpd3 in a single DL1 PN rescues the Rpd3−/− overbranching phenotype. D, Rank order of axon overbranching for different genotypes as indicated. Pairwise Mann–Whitney U tests were performed to determine significance. ns, not significant. *p < 0.05. ***p < 0.0001. Purple symbols indicate the examples shown in A–C. Green line denotes median. Number of clones is 25, 13, 20, 14, 23. Green marks mCD8-GFP-labeled PN axons (A–C) generated by MARCM and labeled using GH146-GAL4. A–C show confocal stacks; magenta is the presynaptic marker nc82. Scale bar, 20 μm.
Calculation of normalized fluorescence for Pros protein level.
Confocal images were taken with all pixel values in the linear range. Background fluorescence was subtracted from the average pixel intensities in the DL1 PN nucleus and the highest Pros expressing nucleus in the section. Ratio of DL1 to highest intensity was calculated as (DL1-background)/(highest Pros-background) (see Fig. 6K).
Construction of UAS-Rpd3-V5.
To generate UAS-Rpd3-V5, we obtained Rpd3-V5 cDNA from K. T. Min (National Institutes of Health, Bethesda, MD) (Cho et al., 2005). The Rpd3-V5 fragment was amplified using the following primers (5′-3′): GGGGTACCCCAAAATGCAGTCTCACAGCAAAAAGCGCG and GACTAGTCTACGTAGAATCGAGACCGAGGAGAGGGTTAGG. The first primer adds a KpnI site and a Kozak sequence, and the second primer adds a stop codon and an SpeI site to the amplified Rpd3-V5 fragment. The PCR product was subcloned into the pUAST vector (KpnI and XbaI). Germline transformation was performed using standard P-element transformation; an insertion on the second chromosome was used in all experiments.
Construction of UAS-HDAC3-3xFLAG.
To generate UAS-HDAC3-3xFLAG, a full-length cDNA (LD23745, Berkeley Drosophila Genome Project Gold cDNA, Drosophila Genomics Resource Center, Bloomington, IN) was amplified using the following primers (5′-3′): CACCCAAAATGACGGACCGTAGGGTGTC and CTACTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCACTTTCTGCCGAATCGGGCTTGTCTTG. The first primer amplifies from the 5′ end and adds a CACC overhang for the TOPO reaction and a Kozak sequence. The second primer adds a C-terminal 3xFLAG tag. The PCR product was subcloned into pENTR-D/TOPO (Invitrogen) and recombined into the destination vector pUAST-Gateway-attB (described below) using LR Clonase II (Invitrogen).
Another UAS-HDAC3 was constructed similarly with a C-terminal V5 tag an alternative second primer (5′-3′): CTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCACTTTCTGCCGAATCGGGCTTGTCTTG. This construct yielded similar results to the 3xFLAG tagged construct (data not shown).
Both UAS-HDAC3 constructs were integrated into the P24 landing site (Markstein et al., 2008) on the second chromosome.
Construction of pUAST-Gateway-attB.
The pUAST-Gateway-attB destination vector was created by PCR amplifying the Gateway cassette from pBPGUw (Pfeiffer et al., 2008) using the following primers (5′-3′): CACCTCGAGGTATCACGAGGCCCTTTC and CACTCTAGACTCGGCCGGCCGTTTATCAC. The first primer amplifies from the 5′ end and adds an XhoI site. The second primer adds an XbaI site to the 3′ end. The PCR product was cut and ligated into pUAST-attB (XhoI and XbaI) (Bischof et al., 2007).
Construction of UAS-3xFLAG-pros-L.
UAS-1xFLAG-pros-L DNA was obtained from F. Matsuzaki (RIKEN Center for Developmental Biology, Japan, Kobe, Japan). We amplified ∼4.5kb of the 5′ side of pros-L using the following primers (5′-3′): GAAGATCTTCATGAGTAGCGATTACAAGGATGATG and CTCCCGCAGAGTCGATTCGACCACG. The first primer adds a BglII site to the 5′ end. The amplified fragment was digested with BglII and KpnI. The fragment was then subcloned into a pUAST vector (BglII and KpnI), which already contained the N-terminal 3xFLAG fragment between EcoRI and BglII sites. Then we amplified ∼2.5kb of the 3′ side of pros-L, digested the fragment with KpnI and XbaI, and ligated with the plasmid above to obtain UAS-3xFLAG-pros-L.
UAS-3xFLAG-pros-L was used for transfection of S2 cell culture (see below).
For all genetic experiments, flies containing an insertion of UAS-pros-L on the X chromosome were kindly provided by C. Q. Doe (University of Oregon, Eugene, OR).
Pros immunoprecipitation from wild-type embryos.
Fly embryos were collected on grape plates for 14 h, washed, and homogenized using a Dounce homogenizer in RIPA buffer (150 mm NaCl, 0.5% deoxycholic acid, 0.1% SDS, 50 mm Tris pH 8.0, protease inhibitor cocktail (1:100, Sigma-Aldrich #P8340), 150 nm trichostatin A (Sigma-Aldrich #T1952), 5 mm sodium butyrate (Sigma-Aldrich #B5887), 10 mm niacinamide (Sigma-Aldrich #N0636), and with or without 1% Triton-X). Immunoprecipitation was performed using anti-Pros antibody bound to Protein G beads.
The final precipitate was denatured in SDS sample buffer and protein was run on a standard SDS polyacrylamide gel, transferred, and blotted using anti-Rpd3 or anti-acetylated lysine antibody.
Pros immunoprecipitation from S2 cell culture.
S2 cells were raised in Shields and Sang M3 Insect Medium (Sigma-Aldrich #S8398) supplemented with potassium bicarbonate, penicillin, streptomycin, and 10% fetal bovine serum. Cells were split at 24 h prior and immediately before cotransfection of tubP-GAL4 and UAS-3xFLAG-pros-L using Effectene Transfection Reagent (Qiagen #301425). S2 cells endogenously express Rpd3.
Forty-eight hours after transfection, HDAC inhibitors were added for 4 h at the following final concentrations: trichostatin A 150 nm and sodium butyrate 5 mm. Cells were lysed in the following immunoprecipitation buffer: 150 mm NaCl, 1% Triton-X, 2 mm EDTA, 50 mm Tris pH 8.0, 10% glycerol, protease inhibitor cocktail 1:100, 150 nm trichostatin A, and 5 mm sodium butyrate. Immunoprecipitation was performed using anti-Pros antibody bound to Protein G beads.
The final precipitate was denatured in SDS sample buffer, run on a standard SDS polyacrylamide gel, transferred, and blotted using anti-Pros, anti-Rpd3, or anti-acetylated lysine antibody. Alternatively, the final precipitate was denatured in SDS sample buffer, run on a standard SDS polyacrylamide gel, stained using GelCode Blue Stain Reagent (Thermo Scientific #24590), and the Pros band cut from the gel for mass spectrometry. Mass spectrometry was performed by NextGen Sciences to determine the acetylation state of Pros.
Results
Rpd3 is required in postmitotic projection neurons for dendrite targeting
To identify genes that are essential for dendrite wiring specificity in Drosophila olfactory projection neurons (PNs), we performed a MARCM-based forward genetic screen using ethyl methanesulfonate as mutagen (Chihara et al., 2007). MARCM allows visualization and genetic manipulation of single-cell or neuroblast clones in an otherwise heterozygous animal, allowing the study of essential genes in mosaic animals (Fig. 1A) (Lee and Luo, 1999). We used GH146-GAL4 to label a single PN born at newly hatched larva (Jefferis et al., 2001), which in wild-type (WT) animals always projects its dendrites to the dorsolateral glomerulus DL1 in the antennal lobe (Fig. 1C). We identified a mutant, called 12–37, in which DL1 PNs mistargeted toward dorsomedial or ventromedial regions of the antennal lobe (Fig. 1D). SNP and deletion mapping identified the causal gene to be Rpd3, encoding a homolog of mammalian HDAC1 and HDAC2. The mutant 12–37 causes a G136R missense mutation in the catalytic domain; the glycine at this position is conserved from yeast to humans (Fig. 1B).
We confirmed that the mistargeting phenotype was caused by the mutation in Rpd3 using the following two criteria. First, MARCM single-cell clones of two previously existing Rpd3 mutants (Fig. 1B) (Mottus et al., 2000) gave similar mistargeting phenotypes (Fig. 1E,F). Second, MARCM expression of UAS-Rpd3 (see Materials and Methods) in DL1 single-cell clones using GH146-GAL4 significantly rescued the phenotype for all three Rpd3 alleles (Fig. 1G,H; data for Rpd3def24 not shown), especially at higher temperatures where GAL4 activity increases (Fig. 1I). Because GH146-GAL4 expresses UAS-Rpd3 only in postmitotic PNs (Spletter et al., 2007), we conclude that Rpd3 plays an essential role in postmitotic PNs to regulate dendrite targeting.
Rpd3 is ubiquitously expressed and located in the nucleus of all cells surrounding the developing antennal lobe at 24 h after puparium formation (24hAPF) (Fig. 1J), when wiring specificity in the antennal lobe is being established. Rpd3 immunostaining is absent in single-cell Rpd312–37 MARCM clones (Fig. 1K), suggesting that the G136R mutation destabilizes the Rpd3 protein. Rpd3 protein expression is restored by MARCM-mediated activation of UAS-Rpd3 (Fig. 1L). Neuroblast clones homozygous mutant for Rpd3 resulted in a 50% reduction in the number of PNs (WT cell bodies 33.8 ± 2.0; Rpd312–37 14.9 ± 1.2, mean ± SD, n = 8 each), indicating that Rpd3 is essential for the proliferation or survival of neuroblasts. Postmitotic expression of Rpd3 does not rescue the reduced cell number phenotype. This is consistent with a previously described role for Rpd3 in cell proliferation (Zhu et al., 2008).
Rpd3 limits PN axon terminal arborization
To further study the role of Rpd3 in postmitotic neuronal development, we examined the terminal arborization patterns of DL1 PN axons in the lateral horn. Wild-type DL1 PNs have a stereotypical L-shaped axon projection in the lateral horn, with one dorsal collateral and a major branch extending laterally (Fig. 2A) (Marin et al., 2002). Rpd312–37 (hereafter referred to as Rpd3−/−) DL1 PNs exhibit an axon overbranching phenotype in the lateral horn (Fig. 2B), regardless of where dendrites mistarget. This is evidence against the possibility that loss of Rpd3 simply causes a PN fate switch. This axon overbranching phenotype can be rescued by postmitotic expression of UAS-Rpd3 (Fig. 2C,D). Thus, Rpd3 functions cell autonomously in PNs to limit the arborization of axon terminal branches.
HDAC3 does not regulate PN dendrite targeting or axon arborization
To test whether the only other class I HDAC in Drosophila is also involved in PN development, we generated MARCM clones mutant for HDAC3N, an early stop and presumably null mutation (Zhu et al., 2008). HDAC3−/− neuroblast clones exhibited a 20% reduction in cell number (26.4 ± 1.8, n = 8), suggesting that HDAC3 is also required for PN neuroblast proliferation or survival. This is consistent with previous findings that HDAC3 is involved in suppression of apoptosis (Zhu et al., 2008). However, HDAC3−/− DL1 PNs exhibited normal dendrite targeting (Fig. 3A) and axon arborization in the lateral horn (Fig. 3B), indicating that HDAC3 is not required in postmitotic PNs for dendrite targeting or axon arborization.
Figure 3.
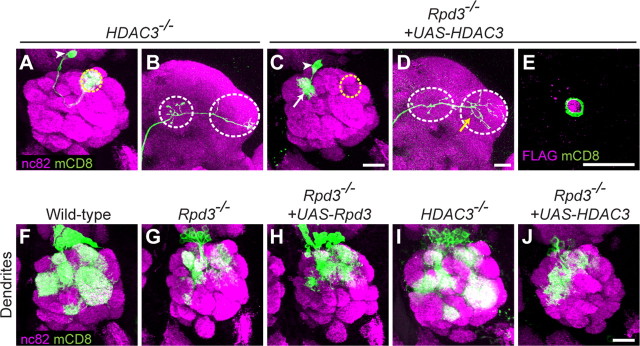
HDAC3 does not regulate PN targeting or arborization. A, Dendrites of HDAC3−/− DL1 PN target normally to the DL1 glomerulus. B, HDAC3−/− PN axon exhibits a normal branching pattern in the lateral horn. C, Postmitotic expression of UAS-HDAC3 does not suppress the Rpd3−/−-mistargeting phenotype (compare with Fig. 1D,G). D, Postmitotic expression of UAS-HDAC3 does not suppress the Rpd3−/− axon overbranching phenotype (compare with Fig. 2B,C). E, UAS-HDAC3 is expressed in a MARCM-labeled Rpd3−/− cell body at 24hAPF, as visualized by anti-FLAG staining of HDAC3–3xFLAG. F, Wild-type PN neuroblast clones target their dendrites to stereotyped glomeruli in the antennal lobe. G, Rpd3−/− neuroblast clones show a reduced dendritic mass due at least partially to a 50% decrease in cell number, and those dendrites do not target their correct glomeruli. H, The reduced cell number phenotype of Rpd3−/− neuroblast clones is not rescued by postmitotic expression of UAS-Rpd3. Because of the reduction of cell number and labeling of multiple neurons, it cannot be determined whether the dendrite-mistargeting phenotype is rescued by UAS-Rpd3. I, HDAC3−/− neuroblast clones show a 20% reduction in cell number, and their dendrites appear to target normally. J, The reduced cell number phenotype of Rpd3−/− neuroblast clones cannot be rescued by postmitotic expression of UAS-HDAC3. Because there are multiple neurons labeled, it cannot be determined whether the dendrite-mistargeting phenotype is rescued by UAS-HDAC3. Green marks mCD8-GFP-labeled PN dendrites (A, C, F–J) axons (B, D), and cell body (E) generated by MARCM and labeled using GH146-GAL4. A–D and F–J show confocal stacks; magenta is the presynaptic marker nc82. E, shows a single confocal section, magenta is anti-FLAG staining. Symbols are as in Figures 1 and 2. Scale bars: C (for A, C), D (for B, D), E, J (for F–J), 20 μm.
To determine whether Rpd3 and HDAC3 have distinct roles, we tested the rescue of Rpd3−/− PN phenotypes by increasing HDAC3 expression. MARCM-mediated expression of UAS-HDAC3 was confirmed by nuclear staining of HDAC3-3xFLAG as revealed by anti-FLAG staining (Fig. 3E, see Materials and Methods), but HDAC3 expression did not rescue the dendrite-targeting phenotype (Fig. 3C, compared with Fig. 1D1) or the axon arborization phenotype (Fig. 3D, compared with Fig. 2B) of Rpd3−/− DL1 PNs. Similar results were obtained for PN neuroblast clones (Fig. 3F–J). Thus, HDAC3 cannot replace Rpd3 function in PN dendrite targeting and axon arborization.
Rpd3 is the major histone deacetylase in PNs
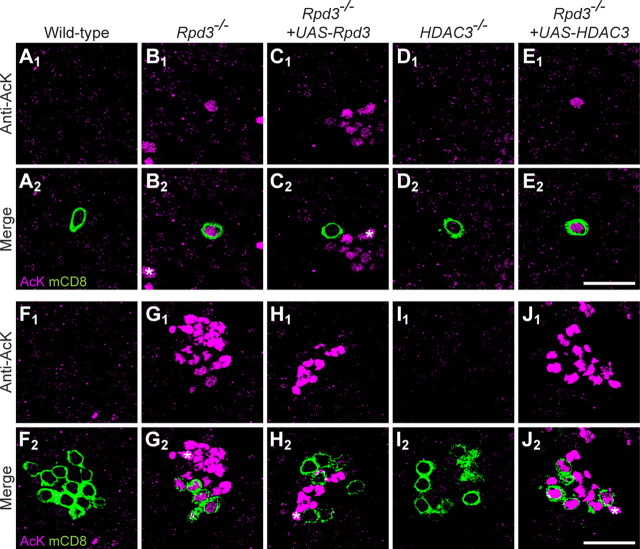
Histone deacetylases are best studied for their roles in deacetylating lysine residues on histones, which are the most abundant substrates for HDACs (Thiagalingam et al., 2003). To examine the effect of Rpd3 on lysine acetylation in PNs, we compared the levels of acetylated lysine immunoreactivity in nuclei of PNs, which are labeled by mCD8-GFP via MARCM, with nuclei of neighboring cells. In wild-type controls, we found anti-acetylated lysine immunoreactivity at a low level in all nuclei (Fig. 4A). In contrast, Rpd3−/− PN nuclei exhibit a much higher level of acetylated lysine compared with neighboring nuclei (Fig. 4B). This finding indicates that Rpd3 plays a predominant role in deacetylation of lysines in PNs. The elevated level of acetylated lysine in Rpd3−/− PNs can be reduced to wild-type low levels upon postmitotic expression of Rpd3 (Fig. 4C). Interestingly, HDAC3−/− PN cell bodies exhibited levels of acetylated lysine indistinguishable from control cells (Fig. 4D), suggesting a minor role of HDAC3 in PN histone deacetylation. Expression of HDAC3 cannot detectably compensate for loss of Rpd3, as exhibited by the remaining high level of acetylated lysine in the nucleus (Fig. 4E). Similar results were obtained for PN neuroblast clones (Fig. 4F–J). Together these data indicate that Rpd3 is the major histone deacetylase in PNs, and that its function cannot be compensated for by the only other class I HDAC in the fly.
Figure 4.
Rpd3 is the major histone deacetylase in PNs. A, A single confocal section in WT 24hAPF pupa showing the DL1 PN (green in A2) stained with an antibody against acetylated lysine (AcK; magenta). A low level of acetylated lysine is found throughout all cells. B, AcK is increased in an Rpd3−/− MARCM single-cell DL1 clone at 24hAPF. C, Postmitotic expression of UAS-Rpd3 restores wild-type acetylation levels in an Rpd3−/− MARCM single-cell DL1 clone. D, HDAC3−/− DL1 PN clone does not show an elevated level of acetylated lysine. E, Expression of HDAC3 cannot reduce the elevated acetylation in Rpd3−/− PNs. F, Single confocal section of WT 24hAPF pupa surrounding a PN neuroblast clone (green in F2) stained with an antibody against acetylated lysine (magenta). A low level of acetylated lysine is found throughout all cell bodies. G, A marked increase of acetylated lysine is evident in an Rpd3−/− MARCM neuroblast clone. H, Postmitotic expression of UAS-Rpd3 can restore wild-type acetylation levels in an Rpd3−/− neuroblast clone at 24hAPF. I, HDAC3−/− neuroblast clone does not show an elevated level of acetylated lysine. J, Expression of HDAC3 cannot reduce the elevated acetylation in Rpd3−/− PNs. Green marks mCD8-GFP-labeled PN cell body (A2–E2) or cell bodies (F2–J2) generated by MARCM and labeled using GH146-GAL4. A–J show single confocal sections; magenta is anti-acetylated lysine staining. *Increased acetylation in other cell bodies indicates additional unlabeled, GH146-negative, Rpd3−/− cells. Scale bars: E2 (for A1 to E2), J2 (for F1 to J2), 20 μm.
prospero mutant PNs exhibit phenotypes similar to Rpd3 mutant PNs
In a parallel screen for PN-mistargeting phenotypes, we found that a P-element insertion early in the open reading frame of prospero yielded a homozygous clonal phenotype similar to Rpd3 mutants. Comparing with wild-type DL1 PNs, pros−/− DL1 PNs also project to medial glomeruli (Fig. 5A,B,D). Interestingly, pros−/− DL1 PNs also exhibit an axon overbranching phenotype, similar to Rpd3−/− PNs (Fig. 5E,F,H). The similarity of the phenotypes of Rpd3−/− and pros−/− PNs suggests that the two proteins might function in the same pathway in PNs.
Pros acts in postmitotic PNs to regulate dendrite targeting and axon branching
Three lines of evidence indicate that Pros expression and function in the PN lineage differs from its function in the embryonic lineage previously described (see Introduction). First, Pros is clearly present in postmitotic PNs. Immunostaining for Pros at 24hAPF shows that wild-type postmitotic PNs express varying amounts of Pros protein (Fig. 5L); this variable expression persists throughout development and adulthood (data not shown). Wild-type DL1 PNs express an intermediate level of Pros protein (Fig. 5I). pros−/− DL1 or neuroblast clone PNs have an undetectable level of Pros protein in the cell body (Fig. 5J,M), confirming both antibody specificity and the nature of the loss-of-function mutation. Pros immunoreactivity can be restored by MARCM expression of UAS-pros in postmitotic PNs (Fig. 5K,N).
Second, the pros−/− single-cell clone phenotype indicates that pros mRNA must be transcribed in postmitotic neurons and/or ganglion mother cells; it cannot be transcribed only in neuroblasts. There are two scenarios in which a single-cell MARCM clone can be produced. The mitotic recombination could occur right before the ganglion mother cell divides to give rise to two postmitotic cells, and therefore only the postmitotic DL1 PN is pros−/− (Fig. 1A, left). Alternatively, because the sibling of the DL1 PN dies during development (Lin et al., 2010; Potter et al., 2010), the mitotic recombination could also occur in the neuroblast; in this case, the ganglion mother cell giving rise to the DL1 PN is pros−/− (Fig. 1A, center). In either case, the neuroblast remains pros+/−. Therefore pros must be transcribed either in the postmitotic PN or the ganglion mother cell to account for a single-cell phenotype.
Third, postmitotic UAS-pros expression can rescue the mistargeting defect of pros−/− DL1 dendrites (Fig. 5C,D) as well as overbranching of DL1 axons (Fig. 5G,H), indicating that Pros functions in postmitotic neurons to regulate PN dendrite targeting and axon branching.
Prospero overexpression suppresses Rpd3−/− dendrite-mistargeting phenotypes
Given the similarity of their phenotypes, we sought to test the relationship between Rpd3 and Pros using genetic interactions. We expressed UAS-pros in Rpd3−/− PNs using MARCM, and found that expression of Pros suppresses the dendrite-mistargeting phenotype of Rpd3 mutants (Fig. 6A,C). In contrast, expressing UAS-Rpd3 did not suppress the pros−/− PN phenotype (Fig. 6B). UAS-pros also partially but significantly suppresses the Rpd3−/− axon overbranching phenotype at higher temperatures where GAL4 activity increases (Fig. 6D,F). Conversely, UAS-Rpd3 cannot suppress the pros−/− axon overbranching phenotype (Fig. 6E). Overexpression of Pros in wild-type DL1 PNs does not cause any dendrite targeting defects, and Pros expression cannot suppress several other medially mistargeting mutants (data not shown). These data suggest that Pros acts downstream of Rpd3 to regulate PN dendrite targeting and to limit PN axon terminal arborization.
Discussion
It has long been thought that histone deacetylases play a general role in chromatin remodeling and transcriptional control, and many studies have examined genome-wide patterns of histone modifications (Millar and Grunstein, 2006). Yet recent studies have also suggested that “general” chromatin remodeling factors can have very specific roles (Ho and Crabtree, 2010). In this study, we show a new function for Rpd3, a ubiquitously expressed protein that is the major histone deacetylase in Drosophila olfactory projection neurons. Rpd3 plays a specific role in controlling dendrite targeting and axon terminal branching, and this function cannot be replaced by the only other class I HDAC. Furthermore, we show that the majority of its function in regulating dendrite targeting and a portion of its function in regulating axon branching are likely performed through the downstream transcription factor Prospero.
Although we cannot rule out the possibility that Rpd3 and Pros act in parallel pathways to regulate dendrite targeting and axon branching, several lines of evidence support the notion that Rpd3 acts via Pros to regulate these events. First, loss-of-function mutations in single-cell clones of Rpd3 and pros give similar dendrite targeting and axon branching phenotypes. Second, overexpression of Pros can largely suppress the dendrite-mistargeting phenotype of Rpd3−/− and can partially suppress the axon overbranching phenotype of Rpd3−/−. This suppression is specific, as overexpressing Pros does not cause defects in wild-type cells nor does it suppress mistargeting phenotypes due to a few other mutations. Conversely, overexpression of Rpd3 does not suppress pros mutant phenotypes. Because the suppression is more robust for dendrite mistargeting defects compared with axon overbranching defects, it is possible that Pros function accounts for more of Rpd3's activity in regulating dendrite targeting than axon branching.
To address the mechanism by which Rpd3 and Pros function together in regulating PN development, we tested several models of their interactions. One model is that Rpd3 functions to upregulate the expression of Pros, which predicts that Rpd3−/− would lead to a decrease in Pros protein. However, anti-Pros immunoreactivity in Rpd3−/− DL1 PN clones was not decreased compared with wild-type DL1 PN clones (Fig. 6G–K). A second model is that Rpd3 and Pros directly bind and work together to regulate the transcription of Pros target genes. Yet in wild-type embryos or in S2 cell culture, we could not detect a complex between Rpd3 and Pros via immunoprecipitation (Fig. 6L). A third model is that Rpd3 directly deacetylates the Pros protein to affect its function, but we could not detect Pros acetylation in the presence of HDAC inhibitors by immunostaining or mass spectrometry (data not shown). Together, these data suggest that Rpd3 indirectly affects the function of Pros. This effect may be through posttranslational modification of Pros to modify its activity. For example, Pros has previously been shown to be phosphorylated (Srinivasan et al., 1998). If posttranslational modification increases Pros activity, then Rpd3−/− would result in reduced Pros activity. Overexpression of Pros in Rpd3−/− may compensate for the reduced activity of unmodified Pros, and therefore suppress the Rpd3−/− phenotype.
Future studies will determine how Rpd3 regulates Prospero, how these factors act together with other transcription factors (Komiyama and Luo, 2007), and what transcriptional target genes they regulate to orchestrate the developmental program for precise wiring of the olfactory circuit.
Footnotes
This work was supported by a National Institutes of Health grant (R01-DC005982), a fellowship from the National Science Foundation (J.S.T.), the Grant-in-Aid for Scientific Research on Priority Areas “Molecular Brain Science” and “Dynamics of Extracellular Environments,” a grant from the Japan Society for the Promotion of Science, and the Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency program (T.C.). L.L. is a Howard Hughes Medical Institute Investigator. We thank J. T. Kadonaga and the Developmental Studies Hybridoma Bank for antibodies; C. Q. Doe, J. A. Simon, the Bloomington Drosophila Stock Center, and the Kyoto Drosophila Genetic Resource Center for fly stocks; K. T. Min and the Drosophila Genomics Resource Center for DNA; R. J. Watts, E. D. Hoopfer, and O. Schuldiner for collaboration on the EMS screen; D. Luginbuhl and E. J. Rao for technical assistance; S. Sekine and M. Miura for supporting T.C. to complete this work; and D. Berdnik, Y. H. Chou, V. S. Dani, W. Hong, L. Liang, M. L. Spletter, L. B. Sweeney, B. Tasic, and X. Yu for comments on the manuscript.
References
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus J, Fuerstenberg S, Doe CQ. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature. 1998;391:792–795. doi: 10.1038/35861. [DOI] [PubMed] [Google Scholar]
- Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nat Neurosci. 2007;10:828–837. doi: 10.1038/nn1910. [DOI] [PubMed] [Google Scholar]
- Cho Y, Griswold A, Campbell C, Min KT. Individual histone deacetylases in Drosophila modulate transcription of distinct genes. Genomics. 2005;86:606–617. doi: 10.1016/j.ygeno.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Wang X, Dobbin M, Tsai LH. Recovery of learning and memory is associated with chromatin remodelling. Nature. 2007;447:178–182. doi: 10.1038/nature05772. [DOI] [PubMed] [Google Scholar]
- Foglietti C, Filocamo G, Cundari E, De Rinaldis E, Lahm A, Cortese R, Steinkühler C. Dissecting the biological functions of Drosophila histone deacetylases by RNA interference and transcriptional profiling. J Biol Chem. 2006;281:17968–17976. doi: 10.1074/jbc.M511945200. [DOI] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenin L, Grosjean Y, Fraichard S, Acebes A, Baba-Aissa F, Ferveur JF. Spatio-temporal expression of Prospero is finely tuned to allow the correct development and function of the nervous system in Drosophila melanogaster. Dev Biol. 2007;304:62–74. doi: 10.1016/j.ydbio.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Guenin L, Raharijaona M, Houlgatte R, Baba-Aissa F. Expression profiling of prospero in the Drosophila larval chemosensory organ: between growth and outgrowth. BMC Genomics. 2010;11:47. doi: 10.1186/1471-2164-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- Johnson CA, Barlow AL, Turner BM. Molecular cloning of Drosophila melanogaster cDNAs that encode a novel histone deacetylase dHDAC3. Gene. 1998;221:127–134. doi: 10.1016/s0378-1119(98)00435-1. [DOI] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Luo L. Intrinsic control of precise dendritic targeting by an ensemble of transcription factors. Curr Biol. 2007;17:278–285. doi: 10.1016/j.cub.2006.11.067. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Johnson WA, Luo L, Jefferis GS. From lineage to wiring specificity: POU domain transcription factors control precise connections of Drosophila olfactory projection neurons. Cell. 2003;112:157–167. doi: 10.1016/s0092-8674(03)00030-8. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lin S, Lai SL, Yu HH, Chihara T, Luo L, Lee T. Lineage-specific effects of Notch/Numb signaling in post-embryonic development of the Drosophila brain. Development. 2010;137:43–51. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin EC, Jefferis GS, Komiyama T, Zhu H, Luo L. Representation of the glomerular olfactory map in the Drosophila brain. Cell. 2002;109:243–255. doi: 10.1016/s0092-8674(02)00700-6. [DOI] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- Mottus R, Sobel RE, Grigliatti TA. Mutational analysis of a histone deacetylase in Drosophila melanogaster: missense mutations suppress gene silencing associated with position effect variegation. Genetics. 2000;154:657–668. doi: 10.1093/genetics/154.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Spletter ML, Liu J, Liu J, Su H, Giniger E, Komiyama T, Quake S, Luo L. Lola regulates Drosophila olfactory projection neuron identity and targeting specificity. Neural Dev. 2007;2:14. doi: 10.1186/1749-8104-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Peng CY, Nair S, Skeath JB, Spana EP, Doe CQ. Biochemical analysis of Prospero protein during asymmetric cell division: cortical Prospero is highly phosphorylated relative to nuclear Prospero. Dev Biol. 1998;204:478–487. doi: 10.1006/dbio.1998.9079. [DOI] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, Kurokawa R, Housman DE, Jackson GR, Marsh JL, Thompson LM. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006a;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc. 2006b;1:2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- Zhu CC, Bornemann DJ, Zhitomirsky D, Miller EL, O'Connor MB, Simon JA. Drosophila histone deacetylase-3 controls imaginal disc size through suppression of apoptosis. PLoS Genet. 2008;4:e1000009. doi: 10.1371/journal.pgen.1000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–422. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]