Abstract
Protein fluorescence and small-angle X-ray scattering (SAXS) have been used to monitor effector affinity and conformational changes previously associated with allosteric regulation in rabbit muscle pyruvate kinase (M1-PYK). In the absence of substrate (phosphoenolpyruvate; PEP), SAXS-monitored conformational changes in M1-PYK elicited by the binding of phenylalanine (an allosteric inhibitor that reduces the affinity of M1-PYK for PEP) are similar to those observed upon binding of alanine or 2-aminobutyric acid. Under the current assay conditions, these small amino acids bind to the protein, but elicit a minimal change in the affinity of the protein for PEP. Therefore, if changes in scattering signatures represent cleft closure via domain rotation as previously interpreted, it can be concluded that these motions are not sufficient to elicit allosteric inhibition. Additionally, although PEP has similar affinities for the free enzyme and the M1-PYK/small-amino-acid complexes (i.e. the small amino acids have minimal allosteric effects), PEP binding elicits different changes in the SAXS signature of the free enzyme vs. the M1-PYK/small-amino-acid complexes.
The pyruvate kinase isozyme found in brain and muscle (M1-PYK) is inhibited by phenylalanine. This inhibition results in a decreased affinity of the enzyme for its substrate, phosphoenolpyruvate (PEP) (1, 2). Although the physiological relevance of this regulation is not fully understood, this system has become a model for the study of heterotropic allostery (i.e. the impact that binding of one ligand to a protein has on the affinity of the protein for a second, chemically non-identical ligand (3)). High resolution structures determined by X-ray crystallography demonstrate that each subunit of the homotetramer contains a single active site and a single amino acid binding site (4). Each subunit contains three domains; the active site lays between the two most N-terminal domains (i.e. A and B domains). In an attempt to gain insight into how the allosteric and active sites communicate, a previous study using neutron scattering identified a contraction/expansion of M1-PYK upon binding of substrate or effector (5). These results were interpreted as indicating a cleft-closure resulting from the rotation of the B-domain with respect to the rest of the monomer (i.e. cleft-closure of the active site). Similarly, comparisons of protein structures of multiple PYK isozymes with and without various ligands bound have been used to support a mechanism by which rigid domains rotate with respect to each other in the allosteric mechanism of PYK isozymes (6–8).
In a previous study, we demonstrated that for the interaction between M1-PYK and amino acid effectors, the L-2-aminopropanaldehyde moiety of the amino acid ligand is most important for binding to the enzyme (4). The length of the hydrophobic side chain is responsible for determining the magnitude of the amino acid-dependent shift in the affinity of the protein for PEP. Small amino acids like alanine and serine can bind to the allosteric site on the protein, but are unable to modify the affinity of the protein for PEP. In the current work, the phenylalanine versus small amino acid comparison has been used in combination with fluorescence intensity and small-angle X-ray scattering (SAXS) measurements to further investigate what role the amino acid-elicited conformational changes might play in the allosteric mechanism.
Materials and Methods
Materials
M1-PYK purified from rabbit muscle was purchased from Roche Applied Science. Phenylalanine and the potassium salt of PEP were purchased from Fluka. Alanine and serine were purchased from Fisher Scientific. 2-Aminobutyric acid was produced by Aldrich. Other buffer components were from Fisher Scientific or Sigma.
Ligand Induced Changes in Protein Fluorescence
The apparent affinity of M1-PYK for PEP was determined by monitoring intrinsic protein fluorescence while varying PEP concentration. Intensity of M1-PYK fluorescence, at 2mg/ml, was measured in a 0.4 cm2 cuvette using a PTI steady-state fluorometer equilibrated at 11°C. Excitation was at 300 nm and emission was collected as area-under-the-peak from 310–460 nm. Titration experiments were performed in 50 mM Tris-HCl (pH 9.0), 10 mM MgCl2, 0.1 mM EDTA and with a range of concentrations of the respective amino acid as designated. Stock solutions of amino acids and PEP were adjusted to the pH 9.0 before addition. KCl was added to reach 500 mM K+ in the cuvette before the addition of titrant (Originally 500 mM K+ was used both to maintain some level of consistency with previous experiments (4) and to minimize the percent change in K+ due to additions with ligands; however, attempts to reduce the K+/ionic strength caused the protein to show signs of aggregation in SAXS studies.). By adding the respective amino acid concentration to all PEP additions, the amino acid concentration was maintained constant over the PEP titration. Total PEP was corrected for bound PEP to derive free PEP. Blank readings of buffer alone were subtracted from measured values.
Data were fit to appropriate equations using the nonlinear least-squares fitting analysis of Kaleidagraph (Synergy) software. The apparent affinity of M1-PYK for PEP (Kapp-PEP) was obtained by fitting fluorescent intensity (F) as a function of free PEP to:
| Equation 1 |
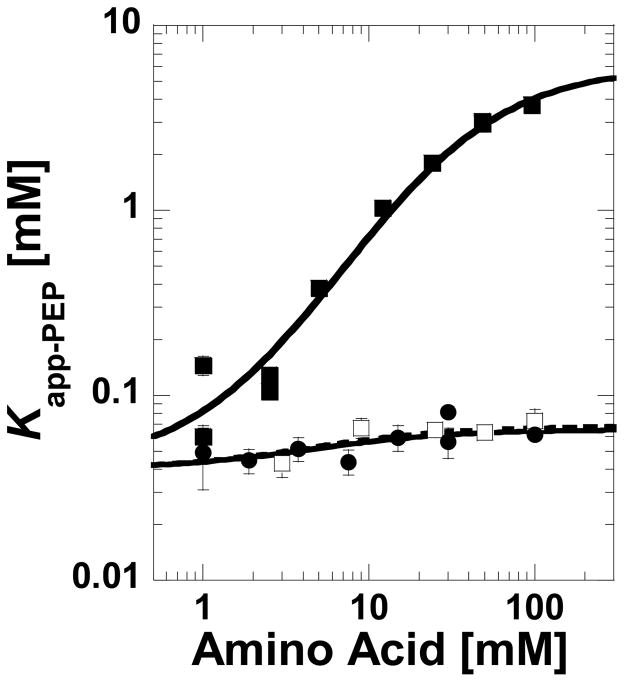
where F is intensity, F0 is intensity in the absence of effector, ΔF=maximum change in intensity, Kapp-PEP is the concentration of PEP that yields a change in intensity equal to one half ΔF, and n is the Hill coefficient. Midpoint values are referred to as Kapp-PEP, rather than Kd-PEP, since changes in fluorescence intensity are not always directly proportional to ligand-protein complex formation with this enzyme (9). For the same reason, only qualitative conclusions are drawn from Kapp-PEP data. Kapp-PEP values were plotted as a function of amino acid concentration to display general data trends. When presenting this data on a log-log graph (e.g. Figure 2), the allosteric coupling between PEP and the effector is the difference between the two data plateaus at low and high concentrations of amino acids (10–13).
Figure 2.
Kapp-PEP determined by ligand-induced changes in protein fluorescence and plotted as a function of the concentration of phenylalanine (■), alanine (●), or 2-aminobutyric acid (□). In this presentation, the magnitude of the allosteric coupling is the distance between the upper and lower plateaus. The minimal fluorescence change caused by PEP binding in the absence of amino acids limits the ability to determine Kapp-PEP at amino acid concentrations below 1 mM. Lines represent data trends.
SAXS Data Acquisition and Analysis
Samples for SAXS measurement were prepared in 50 mM Tris-HCl (pH 9.0; 11°C), 10 mM MgCl2, and 0.1 mM EDTA. When present, 100 mM of the desired amino acid or 2.0 mM of K-PEP was included; indications of saturating conditions for each ligand are from Figure 2 and our previous study (4). As in fluorescence studies, KCl was added such that the total K+ in the solution was 500 mM. After the M1-PYK was dialyzed into the desired condition, protein concentration values as determined by absorbance (Absorbance at 280 of 1mg/ml at 1cm = 0.54 (14)) were 12–20 mg/ml. The dialysates were used for measurement of solvent blanks to be subtracted from the scattering from each protein sample in order to obtain the SAXS signal due to the proteins. Data acquisition times for all samples and solvent blanks were 1 h with the temperature maintained at 11°C
SAXS data were collected using the small-angle instrument at the University of Utah (described in (15)) and were reduced to I(q) versus q and analyzed as previously described (15). I(q) is the scattered X-ray intensity per unit solid angle and q is the amplitude of the scattering vector, given by 4π(sinθ)/λ, where 2θ is the scattering angle and λ is the wavelength of the scattered X-rays (1.54 Å).
Scattering data were initially subject to Guinier analysis to check for the expected linearity of the Guinier plots for monodisperse samples, then P(r) analyses were performed using the program GNOM (16) with corrections for the slit geometry of the scattering instrument. P(r) is the frequency of vector lengths connecting small-volume elements within the entire volume of the scattering particle, weighted by their scattering contrast. For a uniform scattering density object, P(r) goes to zero at the maximum dimension, dmax, of the object. Radius of gyration values (Rg) were calculated as the second moment of P(r). Analysis of the forward scattering, I(0), values normalized by protein concentration, in mg/ml, volume and mean contrast was used to check that the scattering particles were monodisperse. As an additional check on the monodispersity of the samples, estimates for the volume of the scattering particles were obtained using the Porod invariant (17) and compared to expected particle volumes. All errors quoted are based on propagated counting statistics.
Results
Allostery as Monitored by Ligand Induced Changes in Protein Fluorescence
Comparative studies reported herein were motivated by our previous findings that monitored the (initial velocity derived) apparent affinity of M1-PYK for PEP over a concentration range of various amino acids (4). The most relevant results from that study can be summarized as follows: 1) Phenylalanine allosterically decreases the affinity of M1-PYK for its substrate, PEP. 2) Small amino acids like alanine and serine bind competitively with phenylalanine, but elicit negligible change in the affinity of the protein for PEP. 3) 2-Aminobutyric acid binds competitively with phenylalanine and elicits a small allosteric response.
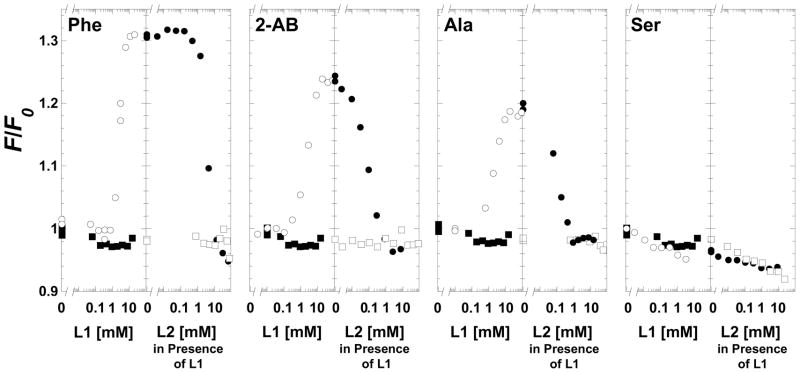
The previous study was carried out at 30°C and, due to the use of initial velocity measurements, in the presence of the second substrate, MgADP. To stabilize the protein during data collection, SAXS measurements reported herein were collected at 11°C and in the absence of ADP. As a means of verifying the previous conclusions under the same conditions used to collect SAXS data, ligand-induced changes in protein fluorescence intensity were used to monitor ligand binding. When binding to free M1-PYK, amino acids caused an increase in fluorescence intensity that was dependent on the type of amino acids (Figure 1). This increase followed the previous data trend of the ability of the amino acids to influence PEP affinity (4), with phenylalanine eliciting the largest increase in fluorescence intensity (32%), followed by 2-aminobutyric acid (24%), and then alanine (19%). Serine did not appreciably alter fluorescence intensity. Binding of PEP to free M1-PYK caused little, if any, change in fluorescence intensity. PEP titrated into M1-PYK with amino acid previously bound caused a decrease in intensity proportional to the increase in intensity that resulted from the addition of the amino acid. Regardless of the order of addition, the ternary complex is generated. The fluorescence intensity of this ternary complex with both amino acid and PEP bound to M1-PYK is very similar to that of the free M1-PYK protein alone.
Figure 1.
Changes in the steady-state intrinsic fluorescence intensity of M1-PYK induced by amino acids and PEP. Panels are labeled by the amino acid type (2-AB is 2-aminobutyric acid). On the left of each data panel free M1-PYK was titrated with ligand 1 (L1), where L1 was either amino acid (○) or PEP (■). On the right of each panel M1-PYK previously saturated with L1 was titrated with ligand 2 (L2). When L2 was amino acid and M1-PYK was pre-saturated with 10mM PEP, data is represented by (□). When L2 was PEP and M1-PYK was pre-saturated with amino acid, data is represented by (●); concentrations of amino acids used in these titrations were: 96 mM phenylalanine (solubility limited, is not completely saturating in the presence of PEP, see Figure 2), 25 mM 2-aminobutyric acid, 30 mM alanine, and 40 mM serine.
Changes in protein fluorescence intensity caused by PEP titrations were used to evaluate PEP affinity over a range of amino acid concentrations (Figure 2). As previously observed (4), phenylalanine reduces the affinity of M1-PYK for PEP, but alanine causes a minimal change in the Kapp-PEP. Under the current conditions, the response of Kapp-PEP to 2-aminobutyric acid is not different from the response to alanine, a result that differs from the previous observations (4). The Hill numbers associated with PEP binding increased from 1 to 2.3 as phenylalanine concentration increased, but remained constant (= 1) over the concentration range of either alanine or 2-aminobutyric acid. As serine (and therefore PEP titrated in the presence of serine) does not elicit a change in protein fluorescence intensity, the response of Kapp-PEP to this amino acid was not determined. Note that due to the solubility limits of phenylalanine, even the highest obtainable concentrations of phenylalanine were not completely saturating in the presence of PEP (i.e. at the highest phenylalanine concentration used in Figure 2, the upper plateau is not fully formed).
SAXS Profiles of Enzyme Complexes
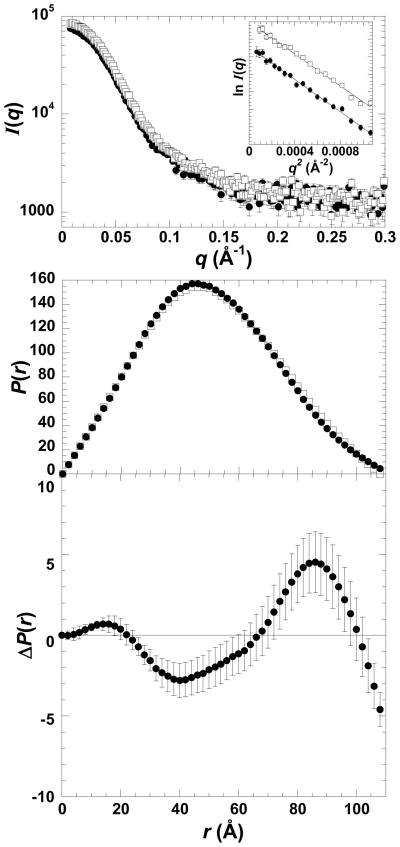
The measured I(q) profile and corresponding P(r) profile (calculated as the inverse Fourier transform of I(q)) for M1-PYK with and without phenylalanine are shown in Figure 3 and the basic structural parameters derived for the protein in each ligand-bound state (as well as other enzyme complexes discussed below) are given in Table 1. P(r) is simply the probable distribution of distances between scattering centers within the scattering particle and goes to zero at the maximum linear dimension, dmax. Of note, the variations in the determined dmax values are insignificant (all in the range of 108–112 Å with estimated uncertainties of 2–3 Å, based on the fact that dmax values in this range showed no statistically significant difference in quality of fit to the scattering data as estimated by χ2 values). The Rg values show only small variations and there are subtle yet statistically significant variations in the distribution of the frequency of vectors lengths in the P(r) functions. These latter variations are most easily seen in the difference P(r) functions (ΔP(r)) that reveal shifts in the distribution of the frequency of vector lengths that are characteristic of a small redistribution of mass within the tetramer as would result from domain or subunit rotations. The small scale of these ligand-induced changes is similar to those observed with neutron scattering, i.e. those previously interpreted as “cleft-closure” of the active site (5). As the P(r) differences are small, it is important to note that when the SAXS experiments were repeated using independent sample preparations, the same trends were observed.
Figure 3.
SAXS data for M1-PYK. The top panel is the I(q) vs. q function of free M1-PYK (●) and the enzyme-Phe complex (□). The Guinier plot for the q range satisfying qRg<1.3 is included in the inset (arbitrary y-axis units are not included). Middle panel: P(r) function derived from the SAXS profile of free M1-PYK (●) and the enzyme-Phe complex (□). The bottom panel shows the ΔP(r) between the free enzyme and the enzyme–Phe complex. Errors are based on propagated counting statistics; error bars are displayed for all data and when not apparent are smaller than symbols.
Table 1.
Structural Parameters Derived from the SAXS Data
| Rg (Å) | dmax (Å) | Porod volume (×10−3) (Å3) | |
|---|---|---|---|
| M1-PYK | 38.6 ± 0.1 | 108 | 189 ± 3 |
| M1-PYK-PEP | 37.6 ± 0.3 | 110 | 188 ± 4 |
| M1-PYK-Phe | 38.5 ± 0.2 | 112 | 192 ± 3 |
| M1-PYK-Ser | 38.0 ± 0.2 | 108 | 194 ± 4 |
| M1-PYK-Ser-PEP | 38.4 ± 0.2 | 108 | 191 ± 4 |
| M1-PYK-Ala | 38.3 ± 0.2 | 110 | 184 ± 4 |
| M1-PYK-Ala-PEP | 38.1 ± 0.2 | 110 | 179 ± 4 |
| M1-PYK-2AB | 38.0 ± 0.3 | 112 | 185 ± 4 |
| M1-PYK-2AB-PEP | 38.4 ± 0.27 | 110 | 180 ± 4 |
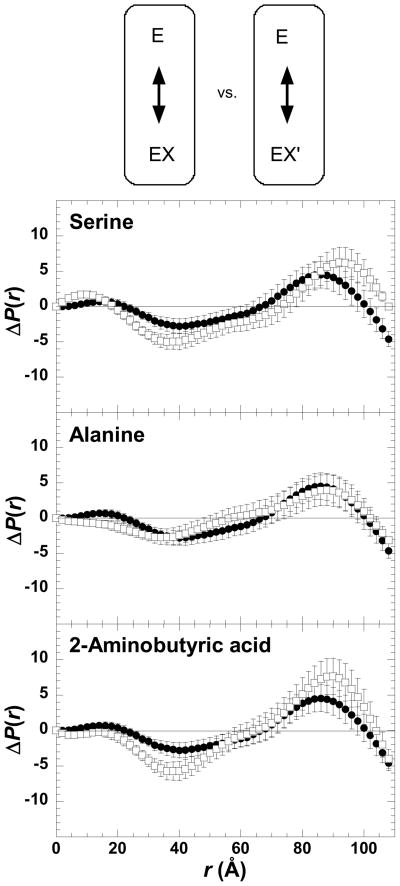
Binding of phenylalanine to the free enzyme causes a conformational change. This change has been observed using a variety of techniques, including differential absorbance spectroscopy, fluorescence, protease sensitivity, sensitivity to sulfhydryl modifying reagents, analytical gel chromatography, and hydrodynamic measurements (5, 18–23). To further analyze the role of these conformational changes in the allosteric mechanism, we compared the impact that alanine, 2-aminobutyric acid and serine binding had on the SAXS signature of M1-PYK (Figure 4). Binding of these three small amino acids elicit conformational changes very similar to those changes caused by phenylalanine binding.
Figure 4.
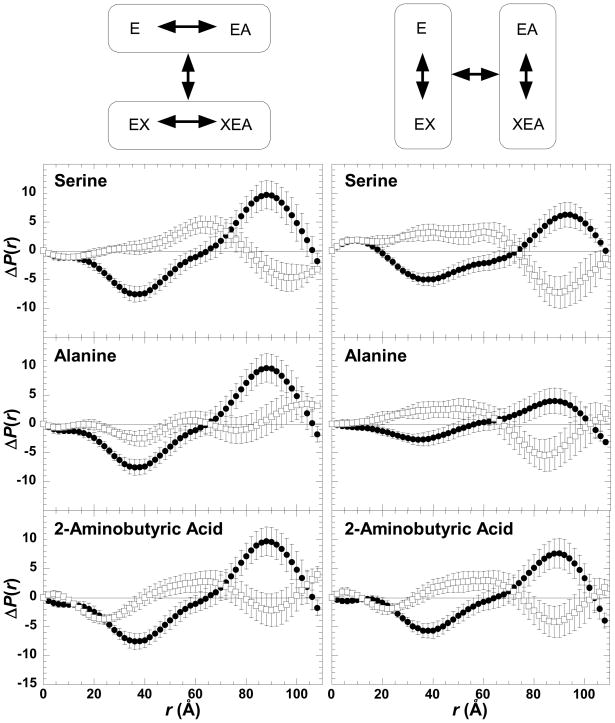
Comparisons of SAXS characterized changes in the average conformation of M1-PYK (E) elicited by binding of phenylalanine (X), versus alanine, 2-aminobutyric acid, or serine (X′). In each panel the ΔP(r) between the free enzyme and the enzyme–Phe complex is in filled circles (●) and ΔP(r) that results from the binding of a second amino acid is in open squares (□). Panels are labeled by the second amino acid included in the comparison. A schematic of the representative comparisons is shown above data panels; ΔP(r) data included in data panels are from comparisons within each circle in the schematic. In the schematic, E represents enzyme, X represent phenylalanine, and X′ represent one of the small amino acids. Arrows within the schematic represent either the comparisons used to generate ΔP(r) data (within circles) or the comparisons made in each data panel (between circles). Errors are based on propagated counting statistics; error bars are displayed for all data and when not apparent are smaller than symbols.
As previously reported (5), PEP binding causes conformational changes in M1-PYK (Figure 5). SAXS data were also measured for the ternary complexes containing one of the small amino acids and PEP. However, a study of the ternary complex including phenylalanine and PEP was not possible due to the inability to simultaneously saturate the enzyme with both ligands (see Figure 2) within the solubility limit of phenylalanine. For alanine, 2-aminobutyric acid, and serine, conformational comparisons can be represented as 1) how the substrate (A) binds to the protein (E) in the absence versus in the presence of the effector (X); i.e. compare the ΔP(r) for (E minus EA) and the ΔP(r) for (EX minus XEA) or 2) how the effector binds to the protein in the absence versus in the presence of substrate; i.e. compare the ΔP(r) for (E minus EX) and the ΔP(r) for (EA minus XEA) (3). Following either comparative approach (Figure 5), it is clear that the ternary complexes of M1-PYK with an amino acid and PEP are not equivalent to protein complexes with only PEP bound or with only an amino acid bound. This finding suggests that each of these ligands (PEP and a small amino acid) influence how the other ligand binds to M1-PYK.
Figure 5.
Comparisons of SAXS characterized changes amongst the protein complexes in an allosteric energy cycle. As represented by the schematic above data panels, this energy cycle includes four enzyme complexes. Therefore, comparisons can be made in two ways. In panels/schematic on the left, the ΔP(r) function associated with the binding of PEP are compared when amino acid is absent (closed circles; ●) or is present (open squares; □). In the panels/schematic on the right; the ΔP(r) function associated with the binding of amino acid are compared when PEP is absent (closed circles; ●) or is present (open squares; □). Data panels are labeled by the respective amino acid. Schematics of representative comparisons are shown above data panels; ΔP(r) data included in data panels are from comparisons within each circle within the schematic. In the schematic as in the data panels, E represents enzyme and A represents PEP. In contrast to Figure 4, X represents any amino acid ligand. Arrows within the schematic represent either the comparisons used to generate ΔP(r) data (within circles) or the comparisons made in each data panel (between circles). Errors are based on propagated counting statistics; error bars are displayed for all data and when not apparent are smaller than symbols.
The SAXS signatures in Figures 4 and 5 are similar, but not identical when the various amino acids are used. Therefore, consistent with the differences in fluorescence changes caused by the amino acids, small variations in conformation are likely. In Figure 5, it is also noteworthy that the changes caused by amino acid binding appear similar to the changes caused by PEP binding. Interestingly, this may suggest that the EA and EX complexes are more similar to each other than either is to the free enzyme, a conclusion that is in contrast to that made in the previous study (5). We note that the previous neutron scattering study did not include the high salt concentrations that were required in our preparations to prevent protein aggregation, and that the neutron scattering experiments were performed in D2O, which is known to alter the solubility properties of proteins.
Discussion
Ligand binding often modifies protein conformation independent of whether the protein demonstrates an allosteric regulation. In spite of this observation, when a protein is known to be allosterically regulated, there is a general trend to consider any and all conformational changes observed upon effector binding as having a role in the allosteric mechanism (3). Following this trend, the previous scattering studies of M1-PYK were interpreted as if an all-or-none conformational change elicited by phenylalanine binding was the basis of the allosteric inhibition of PEP affinity (5, 24). To further test this interpretation, we have monitored conformational changes with SAXS, both when phenylalanine binds and when the small amino acids bind. The data presented in Figure 4 demonstrate that the small amino acids are able to cause global conformational changes similar to those elicited by the allosteric effector, phenylalanine. Therefore, the global conformational changes of M1-PYK observed upon amino acid ligand binding do not correlate with the ability of the amino acid to modify the affinity of the protein for PEP. Following the previous interpretation that scattering data monitors rotations of domains in M1-PYK (5), domain rotations must be important for amino acid ligand binding. However, as the small amino acids that minimally modify PEP affinity elicit a similar SAXS-monitored conformational change as that caused by phenylalanine, clearly these rotations are not sufficient to describe the allosteric mechanism.
Rather than interpreting data in the context of an all-or-none change in global conformations, data interpretations can focus on identifying response signatures (no matter how subtle) that are uniquely associated with individual amino acids. As one example, amino acid binding to free enzyme elicits a range of fluorescence responses (Figure 1). In the structure of M1-PYK the nearest tryptophan residue is more than 20 Å away from the amino acid binding site (4). Therefore, the variation in fluorescence intensity responses to the amino acids must reflect subtle conformational differences in the enzyme complexes when the various amino acids are bound. These subtle amino acid-dependent conformational differences may also be reflected in the observation that ΔP(r) functions, although very similar, are not completely identical (Figures 4 and 5). It then appears that each enzyme-ligand(s) complex may have distinct conformational features.
Distinct conformational features for each enzyme-ligand(s) complex are clearly inconsistent with the previously proposed two-state model (24). Unlike two-state mechanisms, a linked-equilibrium analysis (3, 10–13) defines allostery as how the enzyme (E) binds its substrate (A) differently when the allosteric effector (X) is present versus is not present. In the absence of the effector, substrate binding involves two enzyme complexes (E and EA). Likewise, when the effector is present, two protein complexes are involved in substrate binding (EX, and XEA). Each of the four enzyme complexes (E, EA, EX, and XEA) can have unique conformational and/or dynamic properties. Because linked-equilibrium considers enzyme complexes rather than designated conformations, no restrictions/assumptions are made about conformational differences/similarities between, for example, M1-PYK-phenylalanine and M1-PYK-alanine. Therefore, the data presented herein are consistent with a linked-equilibrium view of allostery.
Based on linked-equilibrium, allostery does not derive from a conformational transition resulting from the binding of a single ligand. Instead, how the binding of A (E compared to EA) changes when X is present (EX versus XEA), requires a comparison of four enzyme complexes, in order to identify allosterically relevant changes (3, 10–13). Unfortunately, the inability to completely saturate the effector binding site with phenylalanine when PEP is present (Figure 2), prevents a full evaluation of the ternary complex of M1-PYK with PEP and phenylalanine bound.
We were able to characterize the four relevant enzyme complexes when the effector ligand was one of the small amino acids (Figure 5). The impact of PEP binding on the SAXS signature is different when one of the small amino acids is present versus is not present. These structural observations are consistent with each ligand (PEP and amino acid) mutually influencing how the other ligand binds to M1-PYK. We found this result surprising given that the affinity of M1-PYK for PEP is minimally altered by binding of the small amino acids. Taken together and using Ala as an example, it appears that PEP elicits different structural changes when binding to M1-PYK vs. M1-PYK/Ala, although both enzyme species bind this substrate with similar affinity. This result might indicate some form of compensation within the allosteric mechanism. Both enthalpy/entropy compensation and compensation between multiple allosteric pathways have been identified in other allosteric proteins (27–31). However, confirming a compensatory mechanism was beyond the scope of the current study.
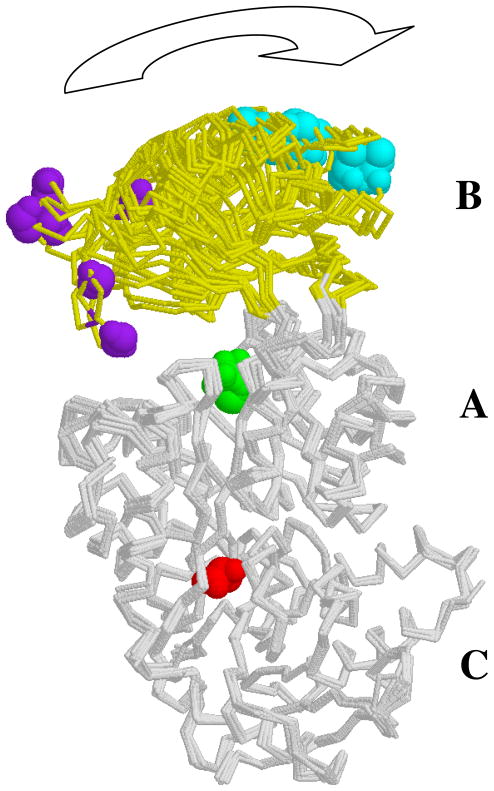
The goal of our study was to examine the relationship between allostery and the conformational changes that give rise to the signature changes in the P(r) functions that were previously interpreted as representing a cleft closure. The question arises as to whether it would be appropriate to use the now readily available three-dimensional modeling routines to attempt to generate a structural model of these changes and prove the earlier interpretation of domain rotation/cleft closure. We do not present such an analysis here as there are too many degrees of freedom in the tetrameric structure of M1-PYK to prove a single three-dimensional model of domain rotation correct using the inherently one-dimensional scattering data; there are multiple ways that the observed mass re-distribution could be accomplished. Importantly, we have used the simplest and most straightforward analysis that makes the fewest assumptions and does not suffer from the redundancy that is inherent in three-dimensional modeling of this multiple subunit protein. Our difference P(r) analysis definitively shows the lack of correlation between the SAXS signatures of conformational change and allostery. Nonetheless, we can consider the existing high resolution crystal structure data and how it informs our thinking and supports the domain rotation hypothesis. In the case of M1-PYK, only a limited number of changes are revealed by comparing X-ray crystallography determined structures of M1-PYK. The simplest summarization of these changes is that the B-domain rotates with respect to other domains (Figure 6). Due to the location of the active site between the A and B domains, it seems reasonable to speculate that PEP binding to the active site influences the B-domain rotation and is influenced by the B-domain rotation. However, the same structural comparisons do not give insight into a potential signaling mechanisms by which ligand binding to the allosteric amino acid binding site influences the active site (PEP binding or B-domain rotation) (Figure 6). Instead, previous speculations regarding signaling mechanisms are derived from comparisons of structures from many different isozymes (6–8); given that single mutations alter allosteric regulation (32), proposed allosteric mechanisms derived from structural comparisons among isozymes should be viewed with caution. Of particular interest, the structure with alanine bound (4) does not reveal changes in the vicinity of the effector binding site or global conformational changes that have not already been identified by comparisons among structures with no effector present (Figure 6). One potential reason might be constraints due to crystal lattice formation. However, it is also possible that the conditions used for crystal growth result in a structure that is not “allosterically competent,” used here in the same way crystallographic data for enzymes are often discussed as being in enzymatically competent forms. It is important to note that solution studies of M1-PYK most generally include Mg2+ to satisfy the divalent cation requirement for catalysis. However, with only a few exceptions, structures of M1-PYK have been determined with Mn2+ present in the active site (4, 33–37). The allosteric functions of both yeast and human liver pyruvate kinase isozymes are dependent on the type of divalent cation present in the active site (38–42). If the same trend is true in M1-PYK, then this effect would have to be taken into account in comparing high resolution structures and solution studies.
Figure 6.
Overlay of M1-PYK subunits as determined by X-ray crystallography. Of the 28 M1-PYK subunits represented in various pdb files, subunits shown represent the ten unique backbone traces (4, 33–37). Each subunit has three domains (labeled A, B, and C). The allosteric amino acid effector (red) binds between the A- and C-domain; this ligand is present in only one of the represented subunits. PEP (or a PEP analogue: pyruvate, oxalate, or phospholactate; in green) binds in the active site, between the A- and B-domain; most but not all subunits shown have a ligand present in the PEP binding site. Here, the B-domain is in yellow. To emphasize the motion of the B-domain, two residues on the B-domain are also colored (purple or cyan). However, this overlay does not give evidence for global structural changes at the effector binding site; no additional global changes are observed when comparing tetramers (data not shown).
In summary, amino acid binding elicits conformational changes in M1-PYK, independent of whether this binding event alters Kapp-PEP. All amino acids tested elicit similar, but not identical conformational changes. Both the similar global responses to the amino acids that do and do not modify Kapp-PEP, and the subtle variations among the complexes investigated are inconsistent with the previously proposed two-state mechanism (24). However, these results can be accounted for in a linked-equilibrium view of allostery that allows each enzyme complex to have unique conformational properties (3). This view of allostery also allows compensating mechanism to contribute to the observed free energy associated with allostery. The finding that the small amino acids influence the conformational changes that PEP causes when it binds to M1-PYK, allows us to speculate that the small amino acids do not modify Kapp-PEP due to a compensating mechanism.
Acknowledgments
We thank Drs. Antonio Artigues and Cy Jeffries for critically reading an early version of the manuscript and making very helpful suggestions. Initial subunit overlays were generated as a project directed by Randy Dix in the Biotechnology/Life Sciences Program at Olathe North High School, Olathe, Kansas; we appreciate contributions from Mr. Dix and his high school students.
Abbreviations
- PYK
pyruvate kinase
- M1-PYK
the pyruvate kinase isozyme found in mammal brain and muscle
- PEP
phosphoenolpyruvate
- E
free enzyme complex
- EA
substrate-enzyme complex
- EX
enzyme-effector
Footnotes
This work was supported by NIH grant DK78076 (to A.W.F.) and an Australian Research Council Federation Fellowship FF0457488 (to J.T.). The scattering experiments were performed at the University of Utah supported by U. S. Department of Energy, Grant No. DE-FG02-05ER64026 (to J.T.).
References
- 1.Carminatti H, Jimenez de Asua L, Leiderman B, Rozengurt E. Allosteric properties of skeletal muscle pyruvate kinase. J Biol Chem. 1971;246:7284–7288. [PubMed] [Google Scholar]
- 2.Kayne FJ, Price NC. Amino acid effector binding to rabbit muscle pyruvate kinase. Arch Biochem Biophys. 1973;159:292–296. doi: 10.1016/0003-9861(73)90455-4. [DOI] [PubMed] [Google Scholar]
- 3.Fenton AW. Allostery: an illustrated definition for the ‘second secret of life’. Trends Biochem Sci. 2008;33:420–425. doi: 10.1016/j.tibs.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams R, Holyoak T, McDonald G, Gui C, Fenton AW. Differentiating a Ligand’s Chemical Requirements for Allosteric Interactions from Those for Protein Binding. Phenylalanine Inhibition of Pyruvate Kinase(,) Biochemistry. 2006;45:5421–5429. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]
- 5.Consler TG, Uberbacher EC, Bunick GJ, Liebman MN, Lee JC. Domain interaction in rabbit muscle pyruvate kinase. II. Small angle neutron scattering and computer simulation. J Biol Chem. 1988;263:2794–2801. [PubMed] [Google Scholar]
- 6.Valentini G, Chiarelli LR, Fortin R, Dolzan M, Galizzi A, Abraham DJ, Wang C, Bianchi P, Zanella A, Mattevi A. Structure and function of human erythrocyte pyruvate kinase. Molecular basis of nonspherocytic hemolytic anemia. J Biol Chem. 2002;277:23807–23814. doi: 10.1074/jbc.M202107200. [DOI] [PubMed] [Google Scholar]
- 7.Jurica MS, Mesecar A, Heath PJ, Shi W, Nowak T, Stoddard BL. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 8.Rigden DJ, Phillips SE, Michels PA, Fothergill-Gilmore LA. The structure of pyruvate kinase from Leishmania mexicana reveals details of the allosteric transition and unusual effector specificity. J Mol Biol. 1999;291:615–635. doi: 10.1006/jmbi.1999.2918. [DOI] [PubMed] [Google Scholar]
- 9.Oberfelder RW, Lee JC. Measurement of ligand-protein interaction by electrophoretic and spectroscopic techniques. Methods Enzymol. 1985;117:381–399. doi: 10.1016/s0076-6879(85)17023-0. [DOI] [PubMed] [Google Scholar]
- 10.Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972;11:864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- 11.Reinhart GD. Quantitative analysis and interpretation of allosteric behavior. Methods Enzymol. 2004;380:187–203. doi: 10.1016/S0076-6879(04)80009-0. [DOI] [PubMed] [Google Scholar]
- 12.Reinhart GD. The determination of thermodynamic allosteric parameters of an enzyme undergoing steady-state turnover. Arch Biochem Biophys. 1983;224:389–401. doi: 10.1016/0003-9861(83)90225-4. [DOI] [PubMed] [Google Scholar]
- 13.Reinhart GD. Linked-function origins of cooperativity in a symmetrical dimer. Biophys Chem. 1988;30:159–172. doi: 10.1016/0301-4622(88)85013-0. [DOI] [PubMed] [Google Scholar]
- 14.Boyer PD. Pyruvate Kinase. In: Boyer PD, Lardy HA, Myrback K, editors. The Enzymes. 2. Academic Press; New York: 1962. pp. 95–113. [Google Scholar]
- 15.Heidorn DB, Trewhella J. Comparison of the crystal and solution structures of calmodulin and troponin C. Biochemistry. 1988;27:909–915. doi: 10.1021/bi00403a011. [DOI] [PubMed] [Google Scholar]
- 16.Svergun DI. Mathematical methods in small-angle scattering data analysis. J Appl Cryst. 1991;24:485–492. [Google Scholar]
- 17.Porod G. Die Roentgenkleinwinkel-Steuung von Dichtgepackten Kolloiden Systemen, I Teil. Kolloid Z Biol. 1951;124:83–111. [Google Scholar]
- 18.Oberfelder RW, Barisas BG, Lee JC. Thermodynamic linkages in rabbit muscle pyruvate kinase: analysis of experimental data by a two-state model. Biochemistry. 1984;23:3822–3826. doi: 10.1021/bi00312a005. [DOI] [PubMed] [Google Scholar]
- 19.Heyduk E, Heyduk T, Lee JC. Global conformational changes in allosteric proteins. A study of Escherichia coli cAMP receptor protein and muscle pyruvate kinase. J Biol Chem. 1992;267:3200–3204. [PubMed] [Google Scholar]
- 20.Consler TG, Lee JC. Domain interaction in rabbit muscle pyruvate kinase. I. Effects of ligands on protein denaturation induced by guanidine hydrochloride. J Biol Chem. 1988;263:2787–2793. [PubMed] [Google Scholar]
- 21.Kayne FJ, Price NC. Conformational changes in the allosteric inhibition of muscle pyruvate kinase by phenylalanine. Biochemistry. 1972;11:4415–4420. doi: 10.1021/bi00773a031. [DOI] [PubMed] [Google Scholar]
- 22.Kwan CY, Davis RC. L-phenylalanine induced changes of sulfhydryl reactivity in rabbit muscle pyruvate kinase. Can J Biochem. 1981;59:92–99. doi: 10.1139/o81-014. [DOI] [PubMed] [Google Scholar]
- 23.Kwan CY, Davis RC. pH-dependent amino acid induced conformational changes of rabbit muscle pyruvate kinase. Can J Biochem. 1980;58:188–193. doi: 10.1139/o80-025. [DOI] [PubMed] [Google Scholar]
- 24.Lee JC. Modulation of allostery of pyruvate kinase by shifting of an ensemble of microstates. Acta biochimica et biophysica Sinica. 2008;40:663–669. doi: 10.1111/j.1745-7270.2008.00445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fenton AW, Hutchinson M. The pH dependence of the allosteric response of human liver pyruvate kinase to fructose-1,6-bisphosphate, ATP, and alanine. Arch Biochem Biophys. 2009;484:16–23. doi: 10.1016/j.abb.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fenton AW, Tang Q. An activating interaction between the unphosphorylated n-terminus of human liver pyruvate kinase and the main body of the protein is interrupted by phosphorylation. Biochemistry. 2009;48:3816–3818. doi: 10.1021/bi900421f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher HF, Tally J. Isoergonic cooperativity: a novel form of allostery. Methods Enzymol. 1998;295:331–349. doi: 10.1016/s0076-6879(98)95047-9. [DOI] [PubMed] [Google Scholar]
- 28.Fisher HF, Tally J. Isoergonic cooperativity in glutamate dehydrogenase complexes: a new form of allostery. Biochemistry. 1997;36:10807–10810. doi: 10.1021/bi9708388. [DOI] [PubMed] [Google Scholar]
- 29.Tlapak-Simmons VL, Reinhart GD. Obfuscation of allosteric structure-function relationships by enthalpy-entropy compensation. Biophys J. 1998;75:1010–1015. doi: 10.1016/S0006-3495(98)77589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braxton BL, Tlapak-Simmons VL, Reinhart GD. Temperature-induced inversion of allosteric phenomena. J Biol Chem. 1994;269:47–50. [PubMed] [Google Scholar]
- 31.Fenton AW, Reinhart GD. Disentangling the Web of Allosteric Communication in a Homotetramer: Heterotropic Inhibition in Phosphofructokinase from Escherichia coli. Biochemistry. 2009 doi: 10.1021/bi901456p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda Y, Tanaka T, Noguchi T. Conversion of non-allosteric pyruvate kinase isozyme into an allosteric enzyme by a single amino acid substitution. J Biol Chem. 1997;272:20495–20501. doi: 10.1074/jbc.272.33.20495. [DOI] [PubMed] [Google Scholar]
- 33.Larsen TM, Benning MM, Rayment I, Reed GH. Structure of the bis(Mg2+)-ATP-oxalate complex of the rabbit muscle pyruvate kinase at 2.1 A resolution: ATP binding over a barrel. Biochemistry. 1998;37:6247–6255. doi: 10.1021/bi980243s. [DOI] [PubMed] [Google Scholar]
- 34.Larsen TM, Benning MM, Wesenberg GE, Rayment I, Reed GH. Ligand-induced domain movement in pyruvate kinase: structure of the enzyme from rabbit muscle with Mg2+, K+, and L-phospholactate at 2.7 A resolution. Arch Biochem Biophys. 1997;345:199–206. doi: 10.1006/abbi.1997.0257. [DOI] [PubMed] [Google Scholar]
- 35.Larsen TM, Laughlin LT, Holden HM, Rayment I, Reed GH. Structure of rabbit muscle pyruvate kinase complexed with Mn2+, K+, and pyruvate. Biochemistry. 1994;33:6301–6309. doi: 10.1021/bi00186a033. [DOI] [PubMed] [Google Scholar]
- 36.Wooll JO, Friesen RH, White MA, Watowich SJ, Fox RO, Lee JC, Czerwinski EW. Structural and functional linkages between subunit interfaces in mammalian pyruvate kinase. J Mol Biol. 2001;312:525–540. doi: 10.1006/jmbi.2001.4978. [DOI] [PubMed] [Google Scholar]
- 37.Stuart DI, Levine M, Muirhead H, Stammers DK. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 A. J Mol Biol. 1979;134:109–142. doi: 10.1016/0022-2836(79)90416-9. [DOI] [PubMed] [Google Scholar]
- 38.Fenton AW. Chapter 5: The Impact of Ions on Allosteric Functions in Human Liver Pyruvate Kinase. Methods Enzymol. 2009;466:83–107. doi: 10.1016/S0076-6879(09)66005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bollenbach TJ, Nowak T. Kinetic linked-function analysis of the multiligand interactions on Mg(2+)-activated yeast pyruvate kinase. Biochemistry. 2001;40:13097–13106. doi: 10.1021/bi010126o. [DOI] [PubMed] [Google Scholar]
- 40.Bollenbach TJ, Nowak T. Thermodynamic linked-function analysis of Mg(2+)-activated yeast pyruvate kinase. Biochemistry. 2001;40:13088–13096. doi: 10.1021/bi010125w. [DOI] [PubMed] [Google Scholar]
- 41.Mesecar AD, Nowak T. Metal-ion-mediated allosteric triggering of yeast pyruvate kinase. 2. A multidimensional thermodynamic linked-function analysis. Biochemistry. 1997;36:6803–6813. doi: 10.1021/bi962870s. [DOI] [PubMed] [Google Scholar]
- 42.Mesecar AD, Nowak T. Metal-ion-mediated allosteric triggering of yeast pyruvate kinase. 1. A multidimensional kinetic linked-function analysis. Biochemistry. 1997;36:6792–6802. doi: 10.1021/bi962869t. [DOI] [PubMed] [Google Scholar]