Arguably periodontitis is the most prevalent bacteria-driven chronic inflammatory disease of the humankind. The incidence of periodontal disease has been reported to range from 30% of the population in developed countries (154) to over 70% of the population in developing countries (198) with severe disease inflicting 7% to 15% of human population worldwide (41, 67, 154). Periodontitis is a major cause of loss of teeth, and is also implicated in the onset, development and/or progression of systemic diseases such as cardiovascular diseases (68), rheumatoid arthritis (27, 207, 224) and Alzheimer disease (87-89).
The involvement of bacteria in the etiology of periodontitis is unquestionable but the composition of the causative species is still being debated. Presently, over 500 bacterial species have been detected in periodontal plaque (160) but only a handful, including Porphyromonas gingivalis has been implicated as major pathogens of chronic periodontitis (208), the most prevailing form of periodontal disease. This bacterium produces a variety of virulence factors including, bioactive metabolic products, fimbriae, and an array of proteolytic enzymes (107). Among the proteolytic enzymes, cysteine proteases referred to as gingipains are responsible for at least 85% of total proteolytic activity exerted by various strains of P. gingivalis (171) and are considered major contributors to the pathogenic potential of P. gingivalis. Indeed, three lines of evidence, including immunization, analysis of knockout strains, and testing of specific protease inhibitors using murine models of infection or bone loss clearly indicate that gingipains are absolutely essential for bacterial survival/proliferation in vivo and for the pathological outcome of the experimental infection (Table 1).
Table 1.
Gingipains are essential virulence factor of P. gingivalis indispensable for pathological outcome of experimental infection.
| Type of study | Experimental model | Outcome | Ref. |
|---|---|---|---|
| Immunization with a peptide domain of Rgp | Murine subcutaneous chamber model | Immunization confers immunity to P. gingivalis infection | (39) |
| Intradermal immunization with plasmid DNA carrying rgpA | Murine skin abscess model | Induction of protective immunity to an invasive P. gingivalis W50 challenge | (234) |
| Immunization with RgpA-Kgp complex and repeat motifs in the hemagglutinin-adhesin domain | Murine skin abscess model | Induction of protective immunity against challenge with invasive and non-invasive strains of P. gingivalis | (141) |
| Immunization with plasmids DNA carrying coding sequence of the Kgp and Rgp catalytic domains | Mice intraperitoneal inoculation model | Vaccination prevented inflammatory response and prolonged the survival rate of immunized animals | (104) |
| Immunization with RgpA | Murine periodontitis model | Prevention of P. gingivalis-induced oral bone loss | (44) |
| Immunization with the RgpA-Kgp proteinase-adhesin complex | Rat periodontitis model | Protection against P. gingivalis colonization and periodontal bone loss | (176) |
| Immunization with wild type P. gingivalis gingipain-deficient mutants and re-infection | Murine skin abscess model | Protection against re-infection elicited by the wild-type strains devoid of gingipains | (232) |
| Immunization with selected functional gingipain-derived domains including the adhesin binding motifs (ABMs) and the proteases' active sites | Murine periodontitis model | Protection against periodontal bone loss was associated with ABM2 sequence: EGLATATTFEEDGVA: ABM3: GTPNPNPNPNPNP NPGT; and the active sites sequence | (142) |
| Immunization with plasmid carrying coding sequence of rgpA | Murine skin abscess model | Reduced lethality in the rgpA DNA vaccine-immunized mice | (235) |
| Immunization with gingipain complexes | Murine skin abscess model | Immunization with detergent extracted gingipain complex elicited a better protective response than other methods of gingipain extraction | (163) |
| Inoculation with individual gingipain-deficient isogenic mutants in W50 background | Murine skin abscess model | Gingipain-deficient strains have severely attenuated virulence in the following order: Kgp≫RgpB≥RgpA | (140) |
| Mixed infection of Tannerella forsythia and P. gingivalis strains, including gingipain deficient mutants | Murine skin abscess model | Gingipains are important in a synergistic effect of mixed infection. | (231) |
| Inoculation with individual gingipain-deficient isogenic P. gingivalis W50 mutants | Murine periodontitis model | Kgp and RgpB but not RgpA are important for P. gingivalis induced bone loss. The gingipains contribute to the virulence of P. gingivalis in following order: Kgp≥RgpB≫RgpA | (161) |
| Pretreatment of P. gingivalis W50 with Rgp-specific inhibitor | Murine subcutaneous chamber model | Inactivation of Rgp attenuates virulence | (40) |
| The abdominal and the nasal cavity immunization with rgpA DNA vaccine | Murine periodontitis model | Immunization via the nasal cavity effectively prevented alveolar bone loos | (125) |
| Inoculation with individual gingipain-deficient isogenic P. gingivalis W50 mutants and pretreatment with Kgp-specific inhibitor | Murine abscess model | Decreased virulence of Kgp-deficient strain in comparison to the parental strain and a significant reduction in virulence induced by inhibitor pretreatment. | (25) |
| Inoculation of (i) bacteria alone, (ii) simultaneously with inhibitor (leupeptin) and (iii) administration of leupeptin 6 weeks after bacteria inoculation | Ligature-induced gingivitis in rats | Leupeptin inhibited P. gingivalis-induced gingivitis when leupeptin was administrated 6 weeks after bacteria inoculation | (98) |
| Passive immunization of human subjects | Gel containing anti-gingipains egg yolk Abs (IgY-GP) was applied into periodontal pockets | IgY-GP treated group had reduced level of P. gingivalis, showed significant reduction in probing depth and decreased bleeding on probing in comparison to the control group | (230) |
Gingipains, including arginine-specific gingipains (Arg-gingipain-A, RgpA and Arg-gingipain-B, RgpB) and lysine-specific gingipain (Lys-gingipain, Kgp), are encoded by three different genes referred to as rgpA, rgpB and kgp, which are conserved among laboratory and clinical strains of P. gingivalis (26). The rgpA and rgpB genes translation products, RgpA and RgpB, share a basically identical caspase-like protease domain with specificity restricted to Arg-Xaa peptide bonds and an Ig-like domain. In RgpA, the protease and the Ig-like domains are followed by a large C-terminal extension known as the hemagglutinin-adhesin domains. Similarly, the kgp gene translation product, Kgp, consists of a catalytic domain with selectivity for Lys-Xaa peptide bonds followed by a C-terminal extension similar to that in RgpA. Except for a short C-terminal domain shared with RgpA and Kgp and several other secretory proteins (136, 189), the hemagglutinin-adhesin domains are absent in RgpB. During the secretion process, progingipains are subjected to extensive posttranslational proteolytic processing and glycosylation. Finally, they are secreted either as a monomeric form (RgpB) or as a non-covalent but stable complexes of protease and hemagglutinin-adhesin domains (RgpA and Kgp), which are either predominantly attached to the bacterial surface or released into the medium in a soluble form depending on P. gingivalis strain (Fig. 1). The varied external localization of these multi-functional gingipain complexes makes them the perfect tool for manipulation of the host immune system by P. gingivalis.
Fig. 1.
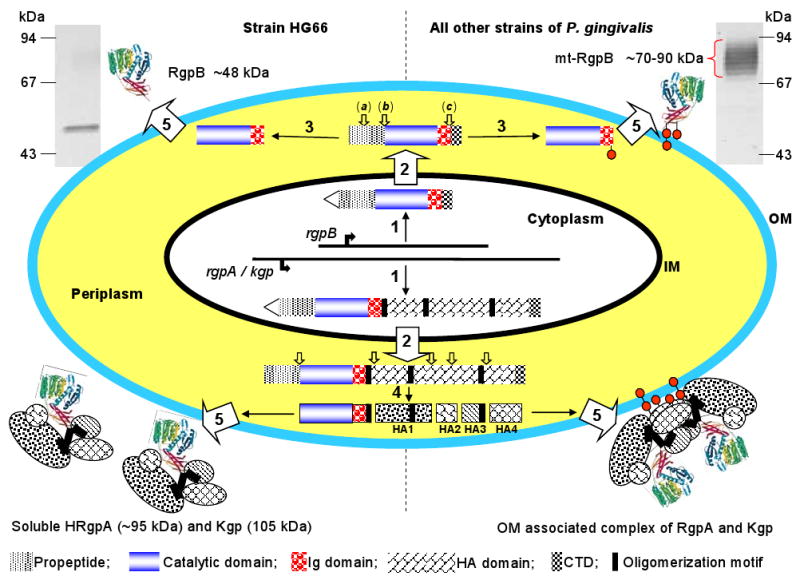
Schematic diagram of discrete steps in gingipain secretion, processing, posttranslational maturation, and assembly of extracellular gingipain complexes. Western blot analysis (upper corners) of the whole culture of P. gingivalis strain HG66 and W83 using anti-RgpB mAb illustrates the difference in the molecular mass between soluble and membrane-attached RgpB. [1] Gingipain genes are translated into a polypeptide chain composed of the classical signal peptide (triangle), a propeptide, a catalytic domain, an Ig-like domain, a hemagglutinin-adhesin domain (missing in RgpB) and a C-terminal domain. Black bars in Kgp/RgpA denote oligomerization motifs involved in non-covalent association of individual domains of the HRgpA/Kgp complex (200). [2] Nascent translation products are translocated across the inner membrane (IM) by the Sec system with simultaneous cleavage of the signal peptide. [3] In the periplasm, pro-RgpB undergoes three steps of sequential processing (from a to c, open arrows). First N-terminal propeptide is cleaved off in two steps (a and b) then the C-terminal domain is removed (c). Each processing step requires the previous step leading to incremental enhancement of the activity in a stepwise manner (123). The intact C-terminal domain, prior to its processing, is essential for RgpB maturation and secretion (136, 189). In strain HG66, non-glycosylated RgpB is released into extracellular milieu in the soluble form; in all other strains, RgpB is glycosylated and remain bound to the cell-surface. Small circles indicate lipopolysaccharide-like glycan moieties for membrane anchorage. [4] RgpA and Kgp processing is more complicated than that of RgpB. In addition to the propeptide removal, several cleavages within hemagglutinin-adhesin (HA) (open arrows) liberate individual functional domains which remain non-covalently associated. Concurrent with the proteolytic processing or the outer membrane (OM) translocation, a glycan moiety is attached to the HA4 domain for membrane anchorage. [5] Gingipains traverse the outer membrane using a unique secretion pathway apparently via a novel outer membrane translocase apparatus (185).
In this review, we will discuss how gingipains participate in sequential phases of the disease process starting from (i) adherence and colonization, through (ii) nutrient acquisition, (iii) neutralization of host defenses and manipulation of inflammatory response, to (iv) tissue destruction, invasion and dissemination to systemic sites. Over the past 15 years, gingipains have been the subject of more than 20 reviews, including a few truly outstanding comprehensive compendiums on the role of these enzymes as virulence factors of P. gingivalis (35, 84, 193). Consequently, in this paper, we will focus on the most recent investigations and cite the previous reviews where appropriate. For references on the early work on the gingipains, we invite the reader to peruse reviews by Imamura (69, 73), Kadowaki & Yamamoto (84), Potempa and colleagues (169, 172), Nakayama (131), and O'Brien-Simpson and colleagues (144).
Gingipains in adherence and colonization
P. gingivalis can be found in low numbers in healthy mouth at various locations but it is associated with disease only if it accumulates in the subgingival dental plaque and starts to proliferate in high numbers (53, 156). As in the case of other mucosal pathogens, the initial colonization is facilitated by P. gingivalis' ability to adhere to a wide variety of biotic host surfaces including epithelial cells, extracellular matrix and other bacteria. This adherence is mediated by fimbrial and non-fimbrial adhesions, and the gingipains, either directly or indirectly, are involved in this process and therefore they are indispensable for the initial stages of periodontal infection with P. gingivalis.
Specific co-aggregation among different species of oral bacteria dictates an orderly build-up of the dental plaque (100, 181) and it is believed to play an important role in the formation and maturation of a biofilm that facilitates the adherence and colonization by periodontal pathogens. P. gingivalis is a late colonizer of the bacterial biofilm on the tooth surface of subgingival sites. A variety of surface components, including lipopolysaccharides, carbohydrates, gingipain complexes and fimbriae have been implicated in co-aggregation with the latter two being the best characterized.
Several lines of evidence indicate that discrete hemagglutinin-adhesin domains of RgpA and Kgp play the direct role in coaggregation (1, 86, 227). These domains can also be derived from HagA. However, for P. gingivalis to acquire aggregative phenotype, polypeptide chains of pro-RgpA, pro-Kgp and pro-HagA need to be proteolytically processed to liberate individual hemagglutinin-adhesin subdomains which assembled into multifunctional, multi-domain complexes on the bacterial surface (Fig. 1). Therefore, P. gingivalis triple-knockout mutants devoid of gingipains (rgpA rgpB kgp-) or devoid of proteins bearing hemagglutinin-adhesin domains (hagA rgpA kgp-) exhibit no coaggregative activity (1). Furthermore, the lack or significantly decreased coaggregative phenotype in rgpA rgpB double-knockout corroborates the important role of the Rgp activity in processing other P. gingivalis proteins, including pro-fimbrilin. Although for coaggregation with Prevotella intermedia and Actinomyces viscosus fimbriae are dispensable, they have a role in coaggregation with other oral bacteria including Treponema denticola, Streptococcus oralis and Streptococcus gordonii (56).
The co-aggregation mediated by RgpA-, Kgp- and HagA-derived hemagglutinin-adhesin subdomain is inhibited by L-arginine, L-lysine and gingipain inhibitors (83). The effect of inhibitors seems to indicate direct involvement of gingipain activity in co-aggregation but it must be keep in mind that gingipain inhibitors invariably contains L-arginine or L-lysine. The most likely scenario is that the tested inhibitory compounds exert anti-coaggregative activity by binding to yet-to-be-defined site(s) in hemagglutinin-adhesin domains directly involved in recognition of ligands on an aggregative partner. This is in keeping with the fact that RgpA and Kgp, but not RgpB, avidly binds to arginine-Sepharose and are eluted with either free lysine (Kgp) or arginine (RgpA and Kgp) (167). Blocking the active site of gingipains with specific inhibitors does not affect enzymes binding to arginine-Sepharose (author's unpublished data).
Apart from the involvement of fimbrial adhesins in P. gingivalis co-aggregation with selected oral bacteria (159), the initial step of P. gingivalis attachment to the oral tissue is fimbriae-mediated. Long (FimA) and short (Mfa) fimbriae recognize large numbers of host proteins, including salivary components, extracellular matrix proteins, complement receptor 3, α5β1 integrin, Toll-like receptor 2, and Cluster of differentiation 14 (also known as CD14) and are responsible for cell invasion, induction of bone-eroding inflammatory responses and attenuation of innate immune clearance of bacteria (5). Thus, fimbriae can be considered a major virulence factor of P. gingivalis but it must be bear in mind that the maturation of the long fimbriae requires the Rgp activity (133). FimA is secreted onto the P. gingivalis surface in a precursor form which requires precise proteolytic processing by Rgps which removes a diacylglyceride-modified N-terminus apparently anchoring the FimA precursor in the outer membrane thus allowing the processed FimA to assemble into a filamentous structure of the mature fimbriae (197). In the absence of Rgps in the gingipain-null mutant, the precursor form of FimA accumulates on the cell surface but, interestingly, it can be converted into biologically active polymeric fimbriae by exogenous gingipains (94). The same pathway of assembly of short filaments on the P. gingivalis surface was also described for 75 kDa protein which in its polymerized form may also contribute to adherence and cytokine induction (197).
The gingipains are themselves, potent non-fimbrial adhesins avidly binding several extracellular matrix proteins such as fibrinogen, fibronectin, laminin and collagen type V (163). They also apparently mediate a tight adherence to epithelial cells and gingival fibroblasts (20, 21) with Kgp being implicated as providing most of the binding (164, 165). The adhesion activity is exerted by hemagglutinin-adhesin domains, apparently the same as those involved in erythrocytes agglutination (see below). Significantly, RgpA/Kgp-derived adhesins preferentially bind immobilized forms of matrix proteins. In experimental conditions, they mediated tight binding of live P. gingivalis cells even in the presence of soluble forms of the same protein (120). This can be essential for adhesion of P. gingivalis in the fluid environment of the oral cavity, where preferential binding of matrix-located proteins over soluble forms facilitates colonization of the host.
The third way gingipains participate in P. gingivalis adherence is through generation of cryptic ligands. Apparently, degradation of extracellular matrix proteins by Rgps exposes peptides with C-terminal arginine for which long fimbriae exhibits specific affinity (92, 101). This process enhances P. gingivalis binding to cells and extracellular matrix.
To sum up, it is beyond any doubt that gingipains are essential virulence factors, either directly and indirectly, in the initial phase of infection. Working as non-fimbrial adhesins or enabling the assembly of fimbriae through several mechanisms, gingipains mediate adherence of P. gingivalis to different sites within the oral cavity and facilitate colonization of the bacterial biofilm in the gingival crevice. Once a “beach-head” has been established in the oral cavity, P. gingivalis, like any other pathogen must acquire nutrients for growth and neutralize host defenses to start an infection process.
Gingipains in nutrient acquisition
P. gingivalis is a fastidious asaccharolytic microorganism acquiring both energy and carbon through fermentation of amino acid residues. In addition, it is auxotrophic with respect to porphyrins and therefore heme is an essential growth factor which simultaneously provides essential iron and porphyrin. To satisfy its nutritious needs, P. gingivalis evolved an entire system to efficiently exploit different sources of heme and iron in a human host. This system is based, in a large part, on the gingipains (151).
Hemagglutination and hemolysis
The largest reserve of heme and iron in the host are the red blood cells and to tap this source, P. gingivalis first efficiently agglutinates erythrocytes and then slowly lyses them to release hemoglobin. Although several different genes annotated in P. gingivalis genome are referred to as hemagglutinins, from analysis of isogenic knock-out strains, it is clear that basically all hemagglutinin activity is related to hemagglutinin-adhesin domains of RgpA, Kgp, and HagA (196). This activity is mapped to discrete sequential motifs, one of which is found on the hemagglutinin-adhesin-1 subdomain targeting glycophorin A: a major erythrocyte membrane sialoglycoprotein. The binding of recombinant hemagglutinin-adhesin-1 domains to asialoglycophorin is specific and avid since it occurs with the dissociation constant of 3 × 10-7M (184).
The mechanism of slow hemolysis by P. gingivalis is still poorly understood. Although proteolytic activity has been implicated in this process (23), it is clear that purified gingipains are not directly involved. Conversely, the finding that Kgp-null mutants show about half of the hemolytic activity of the wild-type strains or complemented mutants argues for the role of at least Kgp in hemolysis (109). Recently, based on bioinformatic analysis, the topology of hemagglutinin-adhesin subdomains of RgpA, Kgp and HagA was redefined and verified by crystallization and structure determination of a domain referred to as K2 (110). The K2 domain spans the conserved hemagglutinin-adhesin-2 domain (also known as HGP15 or hemoglobin receptor, HbR) and continues for an additional 44 residues downstream into a neighboring domain previously assigned as hemagglutinin-adhesin-3. Interestingly, the recombinant K2, as defined, exerts hemolysis of erythrocytes in a dose-dependent manner and degradation of glycophorin A by RgpB sensitizes erythrocytes to the hemolytic activity of K2 (110). The activity is lost by proteolysis at Lys1291-Pro1292 and/or Lys 1276-Gly1277 peptide bonds at the region boundary despite the fact that both fragments remain tightly associated by non-covalent means. Of note, this tight association may represent a mode of hemagglutinin-adhesin-2 association with the rest of the mature gingipain complex. Putting aside an argument if the hemolytic activity of the unprocessed recombinant K2 domain is physiologically relevant, it is obvious that P. gingivalis is able to hold erythrocytes by agglutination and lyse them to release hemoglobin.
Hemoglobin binding and heme acquisition
The role of Kgp and RgpA in hemoglobin binding and heme utilization is well investigated. First, both gingipains bind hemoglobin with high affinity (Kd = 3.72 nM and 3.92 nM for RgpA and Kgp, respectively), which is even higher for the RgpA-Kgp complex (Kd = 1.81 nM) (163). The binding is mediated by the 15 kDa domain referred to as the hemagglutinin-adhesin-2 or hemoglobin receptor which is absolutely conserved in RgpA, Kgp and HagA (132). To exert binding activity, the domain must be processed from the single polypeptide chain of the initial translation products, which require a concerted action of both Kgp and Rgp (144, 169, 172). For gingipains on the bacterial surface, binding of hemoglobin by hemagglutinin-adhesin-2 immobilizes the substrate next to the protease domain. For gingipains in solution, hemagglutinin-adhesin-2 can work as a “homing device” directing enzymes to hemoglobin. This facilitates hemoglobin degradation carried out especially efficiently by Kgp (109, 210). However, it needs to be kept in mind that in vivo, a concerted action of both gingipains is required since oxyhemoglobin, a form of hemoglobin released from erythrocytes, is resistant to degradation by Kgp (205). Apparently, gingipains exerts a sequential action on which Rgps converts oxyhemoglobin to methemoglobin, which renders the hemoglobin more susceptible to degradation by Kgp (204). The occurrence of gingipains in large complexes is a very cleaver design to facilitate hemoglobin degradation and the capture of the released heme is accomplished with high affinity by hemagglutinin-adhesin-2 (157). Subsequently, hemagglutinin-adhesin-2 mediates micro-oxo bisheme formation by converting FeIIIPPIX.OH monomers into [Fe(III)PPIX]2O and promoting their aggregation, thus, leading to the black pigmentation of P. gingivalis colonies grown on blood-containing media (203, 205, 206). This apparently is a very efficient system allowing for storage of excess growth factors (heme and iron) on the cell surface in a form that is non-toxic for the cell membrane (137). Apart from heme/iron storage, the micro-oxo bisheme layer on the bacterium surface may provide protection against oxidative bactericidal activity of neutrophils (202).
Degrading hemoglobin and releasing heme, gingipains are a part of the Hmu-mediated heme/iron uptake system by P. gingivalis (151, 152), composed of proteins encoded by the recently identified heme uptake (hmu) locus. In this system, gingipains may function as hemophore-like proteins shuttling captured heme to a hemoglobin receptor (HmuR) in the outer membrane. The reduced ability of a double hmuR kgp mutant to grow with either hemin or hemoglobin as an iron source (199) and gingipains' physical interaction with HmuR supports (150) this suggestion. Alternatively, heme is first transferred onto HmuY and then deposited on HmuR. The exceptional resistance of this protein to proteolysis (225) makes HmuY a good partner for the gingipains in the pathway of heme/iron sequestration by P. gingivalis. Finally, coordinately regulated transcription of kgp, rgpA and hmuR genes may further facilitate high efficiency of heme utilization (111).
Degradation of heme/iron-containing proteins
The majority of extracellular iron is bound by transferrin in serum and lactoferrin present within mucosal surfaces. In addition, any hemoglobin which leaks out of erythrocytes is immediately scavenged with high affinity by haptoglobin, while released heme is extremely efficiently captured by hemopexin and albumin (Kd ∼ 10-12M and ∼ 10-8M, respectively). As a result of the action of these scavenging proteins, the concentration of free iron and heme in physiological fluids in human host is at very low levels, down to ∼10-24 M in the case of Fe3+(180). To no surprise, pathogenic bacteria often use scavenger molecules known as siderophores to wrestle out the essential iron from host iron carriers. On the other hand, P. gingivalis use gingipains to degrade haptoglobin, transferrin, and hemopexin to enables this siderophore-lacking organism to grow on a minimal media with normal human serum as a source of heme/iron (210). Iron released from proteolytically degraded transferrin is assimilated by P. gingivalis grown in a chemically defined medium containing iron-saturated transferrin as the only source of iron (46). The role of gingipains in this process in highlighted by the fact that Kgp-deficient and gingipain null-mutants were unable to grow in such medium while the growth of Rgp-deficient strain was severely hindered. Furthermore, gingipain inhibitors were found to totally inhibit P. gingivalis growth in a media supplemented in transferrin as the source of iron but not in a medium containing hemin (16). Significantly, the use of transferrin as an iron source may occur in vivo since gingival crevicular fluid samples from periodontitis sites contained fragments of transferrin but not samples from healthy controls (46).
Production of nutritious peptides
The growth of asaccharolytic P. gingivalis depends entirely on the availability of peptides as a source of carbon and nitrogen since the bacterium is unable to utilize free amino acids (124). To fulfill this nutritional need, P. gingivalis apparently use its elaborate proteolytic system composed of endopeptidases (gingipains, periodontain, PrtT protease and Tpr protease), oligopeptidase, and di- and tripeptidyl peptidases in a cascade-like manner (169). Gingipains as the most “aggressive” endopeptidases initiate this cascade by degrading serum and tissue-derived proteins. This is supported by the inability of gingipain knock-out mutants to grow on chemically defined media with the sole carbon and nitrogen source provided by serum proteins (49), or specifically albumin, immunoglobulin and transferrin (145). Of note, an alternative source of nutrients could be proteins derived from fellow biofilm inhabitants such as Tannerella forsythia, since the lysate from this bacterium can stimulate P. gingivalis growth in a gingipain-dependent manner (233). The gingipain-generated protein fragments are finally subjected to the action of di- and tripeptidyl peptidases to release di- and tripeptides to be transported into the cell and used in P. gingivalis carbon and energy metabolism. The functioning of this cascade is strongly supported by findings that the triple-knockout mutant for dipeptidylpeptidase-4, dipeptidylpeptidase-7, and prolyltripeptidylpeptidase showed dramatically reduced growth on media supplemented with albumin and IgG as the only carbon source and the growth block was reverted by addition of purified exopeptidases (146). In vivo, the foraging proteolytic system of P. gingivalis is certainly reinforced by endogenous proteolytic activity enhanced in the inflammatory focus by activation of host matrix metalloproteases and simultaneous inactivation of protease inhibitors by gingipains (see tissue destruction section).
Gingipains in neutralization of host defenses
Successful colonization and local proliferation of a mucosal pathogen can be achieved in two ways: either by avoiding detection by host surveillance systems or by resisting attack by formidable forces of the innate immunity. Although oral epithelial cells invasion and intracellular growth can be considered a stealth approach to survive in the host, the overall strategy of P. gingivalis is to stand up to, or even to enhance, a full confrontation with host defense system. This initial response is composed of the innate system consisting of antibacterial peptides and proteins, complement system, neutrophils, and tissue-resident macrophages. Complement activation and neutrophil influx initiates an acute inflammatory reaction but P. gingivalis imbedded in biofilm is mostly impenetrable to this attack, so the second line of defense, the induced immunity, is mobilized to combat the pathogen using antibodies, activated macrophages and cytotoxic T cells. Both defense systems interact and are synchronized by the cytokine network. Unfortunately, this synergistic attempt to eliminate P. gingivalis is futile and the frustrated immune system, especially the innate branch, causes extensive collateral damage to the surrounding gingival and periodontal tissues which manifests as pathological changes associated with the development of periodontitis. There is a large amount of experimental data available to indicate that gingipains are key factors providing resistance to attack by host defense systems, and more importantly, frustrating and even manipulating the system so its activity is beneficial for P. gingivalis growth at the expense of the host. This aspect of the gingipain pathogenic potential will be discussed in the following sections of this review.
Degradation of antibacterial peptides
Cationic antimicrobial peptides are the very first line of mucosal innate defense against bacterial invaders. In the oral cavity, including gingival tissues, this defense is predominantly exerted by human β-defensins, the cathelicidin-type peptide LL-37 and neutrophil-derived α-defensins which together form a formidable barrier maintaining healthy homeostasis despite constant exposure to a variety of bacteria (29). In gingival crevicular fluid from chronic periodontitis patients, levels of α-defensins and LL-37 were found significantly elevated in comparison to healthy controls (175, 218). The fact that without therapeutic intervention, chronic periodontitis does not resolve spontaneously suggests that bacteria in subgingival microbial biofilm bathing in gingival crevicular fluid must be resistant to killing by cationic antimicrobial peptides. In the case of P. gingivalis, the level of sensitivity to killing by LL-37 and human β-defensinss depends on the strain: with W50, W83 and ATCC 49417 strains being less susceptible than ATCC 33277 (80, 81, 155). Interestingly, in the latter strain, as well as in other P. gingivalis strains, the high resistance to bactericidal activity of human β-defensins can be induced by pretreatment with sublethal levels of defensins and heat or peroxide stress (195). Taking into account that the latter treatments enhance expression of the rgpA and rgpB genes (194) and gingipains efficiently degrade defensins (18, 51, 106, 147) it is likely that these proteases, at least partially, provide P. gingivalis with protection against cationic antimicrobial peptides. Furthermore, it is tempting to speculate that in densely populated biofilm, gingipains as well as proteases released by other periodontopathogens (91, 173) can proteolytically inactivate cationic antimicrobial peptides to enable the survival of other bacterial species which are highly sensitive to them. Finally, one must keep in mind that degradation of cationic antimicrobial peptides also inactivates cationic antimicrobial peptides' ability to neutralize lipopolysaccharides (102) which may lead to exacerbated, sustain production of proinflammatory cytokines.
Exploiting complement
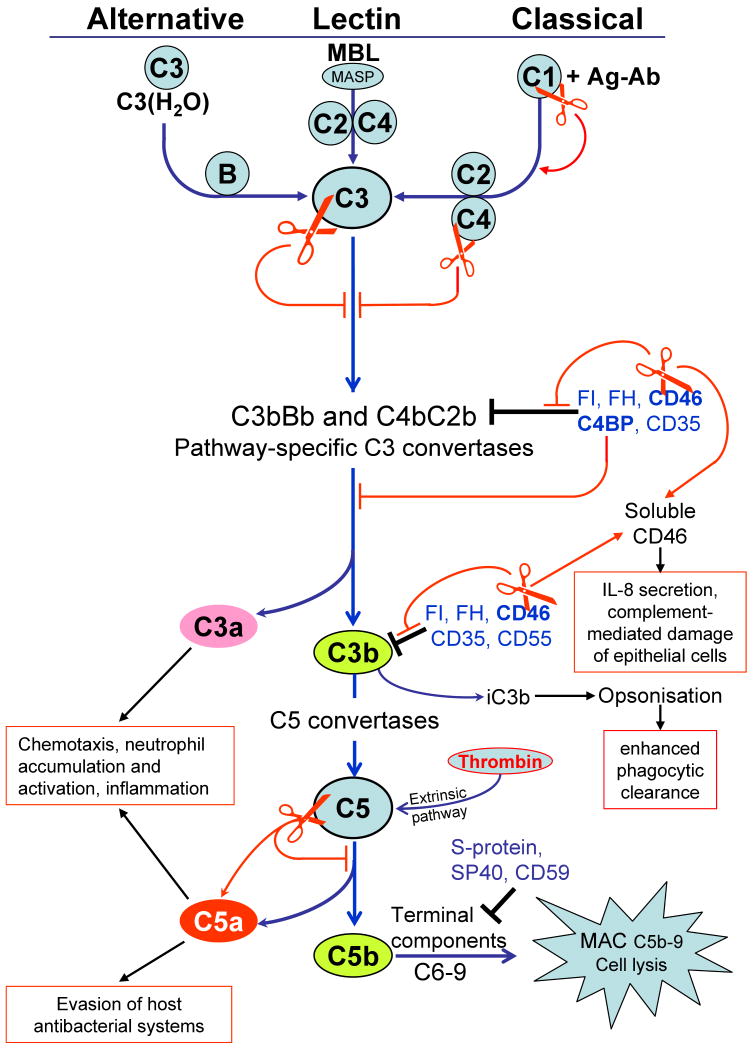
Complement is a major arm of the innate immunity defense system with a pivotal role to recognize and destroy microorganisms. Three pathways of human complement activation guarantee that literally any non-host biotic surface is recognized as hostile (Fig. 2). The classical pathway is usually mediated by binding of the C1 complex to immunoglobulin complexes (immunoglobulin-antigen) opsonizing microorganism. The lectin pathway recognizes polysaccharides normally present only on microbial surfaces by circulating mannose-binding lectins. Lastly, complement can be activated via the alternative pathway due to a failure to appropriately control the constant low-level spontaneous deposition of hydrolyzed C3 [C3(H2O)] on foreign surfaces. All three pathways converge at a central step involving C3 activation by pathway specific C3 convertases. The subsequent downstream proteolytic cascade releases the anaphylatoxins C3a and C3b which direct influx of professional phagocytes and their activation as well as the opsonin iC3b promoting phagocytosis of coated bacteria. Finally, C5b initiates the assembly of the C5b-9 membrane attack complex causing lysis of the foreign cells and directly accounting for the bactericidal activity of human serum. Therefore all successful human pathogens developed measures to circumvent complement, with the most common strategies being: (i) hijacking endogenous regulators of complement which protects host cells and tissue surfaces from initiation of complement activation; (ii) inhibition of complement proteins by direct interaction with specific pathogen-derived proteins; or (iii) proteolytic inactivation of complement components.
Fig. 2.
Activation pathways of the complement system and points of gingipain intervention. Complement is initiated by three major pathways. The alternative pathway is triggered due to a failure to appropriately regulate constant low-level spontaneous deposition of activated C3 on foreign surfaces. Spontaneously hydrolyzed C3 forms a complex with factor B (fB) leading to formation of the initial alternative pathway C3 convertase (C3bBb). The lectin pathway is initiated by the binding of mannan binding lectin (MBL) to mannose residues on the surface of microorganisms which activates mannan-binding lectin serine proteases (MASP), which then cleave C4 and C2. The classical pathway is activated when antibodies bind to their corresponding antigen and serine proteases (C1s and C1r) in C1 complex are activated which then cleave C4 and C2. As a result, in both pathways the same C3 convertase (C4bC2b) is formed. All three pathways converge at a central step, involving activation of the third component of complement (C3) leading to the generation of the anaphylatoxin C3a and opsonins C3b and iC3b. In the terminal pathway, C5b initiates the assembly of the C5b–9 membrane attack complex (MAC), which in turn induces microbial cell lysis. Host surfaces are protected from spontaneous complement activation by complement regulators acting at several stages of complement activation pathway (black end lines). The complement regulators are either cell-surface associated proteins, such as CD35 (Complement Receptor 1, CR1), CD46 (membrane cofactor protein, MCP), CD55 (decay accelerating factor, DAF) and CD59 or soluble regulators circulating in blood, including factor H (FH), factor I (FI), C4 binding protein (C4BP), S-protein (vitronectin), and SP-40 (clusterin) (245). Targets for gingipain attack are depicted by red scissors resulting either in the inhibition of the pathway (red end lines) or stimulation of the pathway (thin red arrows).
The human complement system is present at up to 70% of serum concentration in gingival crevicular fluid therefore it is not that surprising that P. gingivalis is resistant to killing by the human complement system. In a large part, this resistance is dependent on proteolytic activity of gingipains (49) degrading different components of complement (reviewed in refs (169, 201)). In addition, it is now clear that gingipains also contribute to proteolysis-independent protection of P. gingivalis against complement-mediated lysis (174). This is achieved through the capture of the human complement inhibitor C4b-binding protein, thus, hindering deposition of the membrane attack complex on the P. gingivalis surface (Fig.2). The C4b-binding protein binding was mediated specifically by RgpA and accordingly, the P. gingivalis rgpA- mutant showed significantly lower binding capacity than other gingipain mutants. Furthermore, since RgpB exerted negligible interaction with the C4b-binding protein despite its identity to the RgpA catalytic domain, it is apparent that the C4b-binding protein binding site is located within the hemagglutinin-adhesin domain unique for RgpA (174). In the context of the C4b-binding protein capture for bacterial cells protection, it is important to note that at the same time gingipains are proteolytically shedding the complement regulatory protein CD46 expressed by oral epithelial cells which can make these host cells vulnerable to complement-mediated lysis (116).
Although all three gingipains can destroy the bactericidal and hemolytic activity of normal human serum, Rgps are far more effective than Kgp. This is accomplished by very efficient inactivation of the classical pathway via degradation of multiple complement components, including C3, C4, and C5 (Fig. 2), at the enzyme concentration in the range found in the gingival crevicular fluid (168). Complement is also inactivated by interpain A, a cysteine protease produce by P. intermedia which significantly contributes to the resistance of this organism to the bactericidal activity of human serum (173). By efficient degradation of the α-chain of C3, the factor at which all three complement activation pathways converge, interpain A inhibits all three pathways. From a pathological point of view, it is important to note that interpain A synergizes with gingipains in complement inactivation (173) and since the two organisms have been known to co-aggregate in the dental plaque (85), together both bacteria may forge a very efficient defense strategy against complement.
It is interesting to note that at low concentrations of both gingipains and interpain A, they are able to activate the C1 complex in serum, resulting in the deposition of C1 not only on an inert surface of a microtitration plate but also on the surfaces on bacteria themselves. It is very likely that the action requires a precise proteolytic cleavage at specific peptide bonds, not only in C1q, but also in the protease zymogens C1r and C1s of the C1 complex. Furthermore, gingipains and interpain A at low concentrations appear to generate C5a and C3a which are chemoattractants for neutrophils and monocytes. Taken together, the following scenario outlined in details by Krauss and colleagues (103) can be envisioned: P. gingivalis and P. intermedia activate complement when present in low numbers, resulting in a local inflammatory reaction. Since P. gingivalis and P. intermedia are resistant to complement-mediated lysis, they survive the attack which may eliminate other bacteria which could otherwise compete for space and nutrients. At the later stages of infection, the concentration of proteases proximal to the enzyme-secreting bacteria is high enough to destroy C3, C4 and C5 thus inhibiting complement activation and promoting the survival of the entire biofilm community by helping bystander bacteria evade complement killing. However, it can be assumed that at some distance from the biofilm where enzymes are more diluted, gingipains and interpain A can still activate complement. This will propagate the inflammatory state fueling bacteria with nutrients from inflammatory plasma exudate and causing damage of the periodontal tissue. The presence of gingipain gradient across periodontal tissue (216) lends credit to such scenario.
Targeting innate immunity effector cells
At the periodontitis sites, P. gingivalis is besieged by phagocytic cells, neutrophils and macrophages. Therefore, the ability to survive a relentless attack by these immune cells is pivotal for survival in a hostile environment of the inflamed tissue. The bacterium is remarkably resistant to oxidative killing by phagocytes (128) and can survive phagocytosis by macrophages (55). It is clear that P. gingivalis is able to deter, sabotage and/or subvert the phagocyte-dependent regulation of the innate immunity (54). The gingipains play an important role in this evasion by exerting their protective effect either directly by degrading/shedding receptors and cytokines essential for phagocyte functions, or indirectly, by C5a which is used by P. gingivalis to undermine Toll-like receptor 2-dependent immunity (223).
As summarized in Table 2, gingipains proteolytically attack and destroy a number of host cell surface receptors on variety of cell types including immune cells, fibroblasts, epithelial and endothelial cells. The pathophysiological outcome of these gingipain activities is to cripple the communication network of the immune system in order to evade it. The net effect is also to sustain the local chronic inflammatory reaction to ensure a continual delivery of nutrients to the plaque community.
Table 2.
Cell surface receptors and proteins targeted by gingipains.
| Molecule | Targeted cells | Outcome | Putative pathophysiological outcome | Ref. |
|---|---|---|---|---|
| C5a receptor (C5aR, CD88) | Neutrophils | C5aR cleavage at the Lys18-Asp19 and Lys28-Thr29 peptide bonds by Kgp | Suppressed neutrophil migration and activation | (79) |
| CD14 | Human monocytes | Reduced tumor necrosis factor-α production in response to lipopolysaccharide | Attenuation of the cellular recognition of bacteria may help P. gingivalis to evade the host immune system and contribute to sustained chronic inflammation | (212) |
| CD14 | Human gingival fibroblasts | Reduced lipopolysaccharide-induced interleukin-8 production | (214) | |
| CD14 | U937 macrophage-like cells | Suppressed tumor necrosis factor-α production in response to lipopolysaccharide | (32) | |
| CD4 and CD8 | Human T cells | Decreased mitogenic response to phytohemagglutinin or concanavalin A | Impediment of T cell functions leading to impair responses at periodontitis sites | (97) |
| CD2 and CD4 | Human CD4+ T cells | Decreased binding of anti-CD2 and anti-CD4 mAbs | (238) | |
| Intracellular adhesion molecula-1 (ICAM-1) | Epithelial cell lines (KB and USC) | Hindered ICAM-1 dependent adhesion of neutrophils | Disruption of neutrophil-oral epithelium interaction | (213) |
| Platelet endothelial cell adhesion molecule-1 (PECAM-1, CD31) | Endothelial cell monolayers | Reduced intercellular junctional CD31 expression and gap formation | Increased vascular permeability and neutrophil influx, enhanced local inflammation | (239) |
| CD31 | Human neutrophils | Removal of antiphagocytic signal (CD31) together with the generation of a novel “eat-me” signal leads to recognition of healthy neutrophils as apoptotic and their phagocytosis by macrophages | Evasion of neutrophils antibacterial potential and effect on local inflammatory process | (52) |
| Tumor necrosis factor-α | Fibroblasts stably trensfected with cDNA of human protumor necrosis factor-α | Suppressed activation of NFκB and lost of ability to induce apoptosis in HL-60 cells | Dysregulation of the cytokine network | (121) |
| CD99 | Human endothelial cell monolayers | Inhibition of CD99 ligation-dependent expression of leukocyte adhesion | Hindered leukocyte recruitment to infected site | (243) |
| CD27 and CD70 | Resting human T cells and activated B cells, respectively | A reduction in CD27-ligation dependent co-stimulatory CD40L expression | Downregulation of CD27/CD70-mediated lymphocytes activation | (237) |
| Thrombomodulin (TM) | Human endothelial cell | Reduced thrombin-dependent activation of protein C | Promotion of vascular coagulation and inflammation | (77) |
Gingipains in manipulation of the inflammatory response
Deregulation of cytokine signaling network
Inflammation as a response to an incoming foreign agent involves a complex interplay of signaling molecules known as cytokines and their receptors expressed by the host tissues and immune cells. Consequently, these cytokine mediators are often targets for the invading microorganisms to manipulate and to disrupt the immune response to their advantage. In the case of P. gingivalis, the gingipains play a key role in the dysregulation of this system. They have been known to be able to disrupt various components of the cytokine and their receptor networks for many years with targets including interleukin-1β (190), interleukin-6 and its receptor (11, 153), interleukin-8 (122), interleukin-12 (240, 241), interleukin-4 (236), interferon-γ (242), tumor necrosis factor-α (17), CD4/CD8 (97), CD14 (212) and Intercelluar adhesion molecular-1 (CD54) (213). More detailed review of these work have been reported previously (73, 162, 169).
More recent investigations in this field have revealed additional targets for the gingipains including membrane-bound tumor necrosis factor-α (121). As a proinflammatory cytokine, both soluble and membrane-bound forms of tumor necrosis factor-α induce apoptosis of various cell types (33, 127) and both stimulate T-cell activation and proliferation with the membrane-bound tumor necrosis factor-α being more potent (48). Since soluble forms of tumor necrosis factor-α have previously been reported to be susceptible to degradation by the gingipains (17), questions were raised whether the membrane-embedded forms of membrane-bound tumor necrosis factor-α were protected from proteolytic cleavage. This was found not to be the case as both forms were readily inactivated by the gingipains, resulting in the loss of ability to induce apoptosis (121). In contrast, further dissection of interleukin-8 hydrolysis by the gingipains has revealed a more complex picture. Interleukin-8 is known to have varying functions from promoting neutrophil chemotaxis (66) to respiratory burst priming of neutrophils (50). Although the major form of interleukin-8 with 72 amino acids (interleukin-872aa) as produced by resident inflammatory cells is readily degraded by the gingipains (122), a less common and less active form with 77 amino acids (interleukin-877aa) as produced by epithelial cells, fibroblasts and endothelial cells (45) was found to be converted into a more active form of interleukin-8 upon exposure to the gingipains (30). Prolonged exposure to the gingipains resulted in inactivation of both forms of interleukin-8 suggesting that a gradient of gingipain exposure within the periodontal tissues may elicit differing immune effects: nearest to the periodontal pocket where gingipains are present at high concentrations, the net effect is stunted neutrophilic response due to interleukin-8 degradation along with other cytokines but further away from the pocket wall, lower concentrations of gingipains may serve to activate tissue-derived interleukin-877aa to induce an inflammatory response to promote vascular leakage and delivery of nutrients to the subgingival plaque population (143).
Other recent reports have also detailed gingipain-mediated degradation of various cytokines and receptors such as CD27/CD70 system (237) which plays a key role in regulating B-cell activation and immunoglobulin synthesis, CD46 (116) which is a complement regulatory protein and has a role in protecting the host cells from damage by complement, and interleukin-5 (216) which is a Th2 response cytokine being responsible for B-cell proliferation and immunoglobulin synthesis. Finally, RgpA and Kgp, but not RgpB mediate in a proteolytic activity-independent manner strong enhancement of production of pro-inflammatory cytokines in macrophages via a yet unknown mechanism (34). Together, it is clear that through the gingipains, P. gingivalis is able to manipulate the cytokine network through many facets in order to evade the host defence and to change the local environment to suit their needs.
Manipulation of host proteolytic cascades
Blood coagulation, clot lysis, complement activation, kinin release and apoptotic cell death are the best examples of important physiological systems activated in the cascade-like manner. This allows for powerful amplification of the initial signal and provides means for very precise regulation at the multiple levels. Unfortunately, several pathogens developed strategies to take advantage of these cascade pathways. In many respect, P. gingivalis is a champion in this group, being highly efficient at taking advantage of the coagulation cascade and the kallikrein/kinin pathway.
Participation of the coagulation cascade in the host response to bacterial infection is a recognized pathophysiological reaction that plays a pivotal role in both confinement of infection and enhancement of phagocytosis. The cascade is extremely tightly regulated and any inherited “glitches” in the system results in coagullopathies. Interestingly, Rgps can activate the cascade at several different levels with precision and efficiency resembling endogenous activators (reviewed in refs. (73, 169)). Actually, in the case of factor X activation, the RgpA catalyzed reaction mimics the endogenous one with respect to stimulation by calcium and phospholipids (71). Apart from factor X, gingipains can also activate factor IX and prothrombin (factor II) (70, 72). Significantly, these reactions occur efficiently in human plasma leading to clot formation. However, the formed clot is loose due to simultaneous destruction of fibrinogen clotability by Kgp. Thrombin, the final protease of the coagulation cascade exerts a multitude of diverse biological activities that include the stimulation of prostaglandin and platelet-activating factor synthesis and production of interleukin-1 by endothelial cells and macrophages (19, 108). These compounds are regarded as important in the tissue destruction process of periodontal disease. Furthermore, both thrombin and other activated clotting factors are endogenous agonists for protease activated receptors. Together with gingipains, these activated clotting factors can drive havoc in the periodontal tissue by excessive stimulation of the protease activated receptors. This includes protease activated receptor-2, which was found to play a pivotal role in cellular mechanisms involved in experimental periodontitis (see below). Finally, a recent finding showed a direct shunt between clotting and complement, executed by thrombin acting as a C5 convertase (65) to cause C5a generation, resulting in the paralysis of the host defense systems (223).
Contact activation system, also known as the kallikrein-kinin pathway (36), is the proteolytic cascade activated by gingipains with an efficiency exceeding endogenously occurring reactions. In human plasma, Rgps convert prekallikrain to kallikrain, the reaction normally catalyzed by Hageman factor (activated factor XII, XIIa) which in turn produce a nonapeptide, bradykinin, from high molecular weight kininogen. Conversely, Rgps and Kgp working in concert can directly release the decapeptide, Met-bradykinin, referred to as kallidin. Kinins exert powerful biological activity. At the site of infection/inflammation, they are responsible for pain and local extravasation leading to edema. At the systemic level, they enhance development of hypotension and shock. At periodontitis sites, kinins contribute to the generation of gingival crevicular fluid and, most importantly, they may participate in alveolar bone erosion through activation of prostaglandin synthesis in human osteoblasts, endothelium and periodontal ligament cells. In addition, kinins synergistically potentiate bone resorption induced by interleukin-1 and tumor necrosis factor-α (14) and this process is driven by enhanced kinin receptor expression as induced by these pro-inflammatory cytokines (15). Furthermore, Rgp-mediated kinins can forge a trans-cellular cross-talk between Toll-like receptor-2 and kinin receptor B2R driving modulation of T cell responses which may affect the severity of chronic periodontitis (126). Finally, activation of the kinin system was shown to be essential for breaching the vascular barrier and dissemination of P. gingivalis (64).
Taken together, it is apparent that P. gingivalis is taking considerable advantage of the host proteolytic cascade systems. Illegitimate activation of the coagulation and kallikrain/kinin pathways to drive the local inflammation provides the bacterium with nutrients and “leaky” blood vessels facilitating escape and dissemination. At the same time, excessive generation of C5a contributes to local paralysis of the bactericidal activity of phagocytic cells: the main adversaries of P. gingivalis in the infected periodontal tissue.
Signaling through protease activated receptor receptors
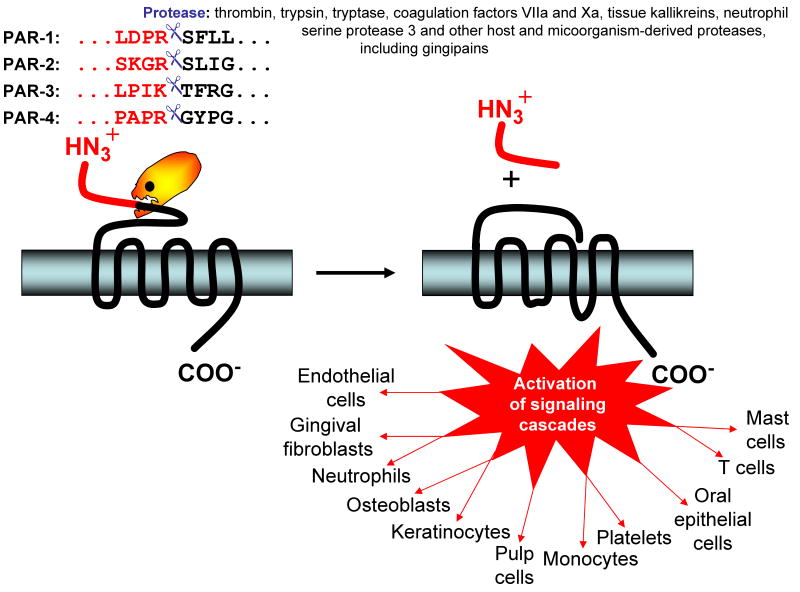
Human protease activated receptors belong to the subfamily of G-protein coupled receptors expressed on many cell types, including among the others, osteoblasts, immune cells and epithelial and endothelial cells. Protease activated receptors are activated by proteolytic cleavage at the N-terminus which uncovers a tethered ligand that interacts with an external loop of the receptor to induce a conformational change for signal transduction into the cell (57, 59, 177). There are four known protease activated receptors: protease activated receptor-1, -3, and -4 have thrombin as their usual physiological activator, whereas protease activated receptor-2 is activated by variety of endogenous proteases, including trypsin, tryptase, coagulation factors VIIa and Xa, tissue kallikreins, and neutrophil serine protease 3 (Fig. 3). Collectively, protease activated receptors participate in several pathophysiological processes, such as hemostasis, thrombosis, regulation of vascular permeability, mediation of vascular smooth muscle contraction, proliferation and hypertrophy, bone cell differentiation and proliferation (60).
Fig. 3.
Scheme of the signaling via protease activated receptors (PAR) and cell in the periodontium affected by the gingipain-exerted PAR activation. The inactive status of PARs is associated with a hidden N-terminal-tethered ligand sequence. The tethered ligand part cannot interact with the body of the receptor until it is cleaved off at peptide bond depicted by scissors. Gingipains via cleaving these bonds in different PAR receptors activate signaling pathways in various cell types leading to large variety of pathophysiological responses (for details see Table 3).
Within the periodontal and gingival tissues, protease activated receptors are expressed on a variety of cells including oral epithelial cells, gingival fibroblasts, mast cells, osteoblasts, vascular endothelial cells, macrophages and infiltrating immune system cells, such as neutrophils, monocytes and T cells. With respect to periodontal disease, protease activated receptor involvement in innate immunity and bone cells metabolism seems to be of paramount importance. For maintaining homeostasis of the periodontium, signaling through protease activated receptor needs to be precisely regulated and this is achieved by the tight control of host proteases within cascade systems, by compartmentalization and through endogenous protease inhibitors. Once the periodontal plaque has been colonized by periodontal pathogens, the fragile balance of maintaining proteolytic activity in check is disturbed by bacteria-derived proteases which activate proteolytic cascades while simultaneously inactivating endogenous inhibitors of proteases (see below). The ensuing inflammation further contributes to the imbalance due to the local infiltration of neutrophils loaded with proteolytic enzymes. It is easy to imagine that in such an environment, thrombin, factor Xa and protease 3 will become available to activate protease activated receptors. Protease activated receptors are also likely subjected to activation by bacteria-derived proteases, and indeed, several in vitro investigation have shown that Rgps can activate and signal through all four protease activated receptors. The effect of the signaling depends on the type of protease activated receptor cleaved in the given cellular context (Table 2). Analyzing the table content, one must be aware that the described response does not cover all effects exerted by activation of specific protease activated receptor signaling on a specific cell.
Setting aside discussion of individual responses, the analysis of the available data clearly indicates that the excessive protease activated receptor activation by either endogenous host proteases or gingipains must have significant impact on disease development. Signaling via protease activated receptor-1 and -2 stimulates oral epithelial cells to produce an array of proinflammatory cytokines, including interleukin-1α, interleukin-1β, interleukin-6, tumor necrosis factor-α, and interleukin-8. The enhanced secretion of a similar set of cytokines, supplemented with monocyte chemoattractant protein-1, is also exerted by treatment of monocytes with Rgps. Although not reported for Rgps-induced signaling via protease activated receptor-1 on monocytes (221), it is known that in these cells, activation of the protease activated receptor pathway leads to secretion of regulated upon activation, normal T-cell expressed, and secreted cytokine (RANTES) - a pivotal cytokine in bone remodeling (12, 187). Similarly, Rgp-dependent neutrophil activation via protease activated receptor-2 cleavage (112) will be accompanied by enhanced released of interleukin-1β, interleukin-6 and interleukin-8. Together with hepatocyte growth factor (a factor known to induce osteoclast formation) produced by oral fibroblasts stimulated via protease activated receptor-1 and -2, locally secreted cytokines not only fuel inflammation but also contribute to bone resorption. In parallel, the bone resorptive process driven by cytokines may synergize with likely protease activated receptor-dependent gingipain effect on receptor activator for nuclear factor κ B ligand (RANKL) / osteoprotegrin expression ratio in osteoblasts (149). In healthy bone tissue, osteoblasts control osteoclastogenesis by expressing RANKL and osteoprotegrin, with the latter (a decoy receptor of RANKL) being a strong inhibitor of osteoclast formation by osteotropic factors, including RANKL. P. gingivalis disturbs this ratio in favor of RANKL by Kgp proteolytic degradation of osteoprotegrin (229).
Taking into account that in normal circumstances the inflammatory reaction is indispensable to eliminate invading pathogens, a picture emerges that in the periodontal tissue, protease activated receptors plays an important role in the surveillance system. Proteolytic activation of protease activated receptors on monocytes, neutrophils, epithelial and endothelial cells by gingipains and, as a matter of fact, by any other bacteria-derived proteases, activates an alarm. Activated cells start to secrete chemokines to bring in phagocytes and bactericidal peptides to enhance mucosal antibacterial defense and cytokines to orchestrate an assault on the invaders. Unfortunately, periodontal pathogens are resistant to protease activated receptor-mobilized innate immunity defense forces. As indicated by several in vivo studies, protease activated receptor-2 plays a pivotal role in this process. First, rats challenged orally with a protease activated receptor-2 agonist not only exacerbated existing periodontitis but caused the development of the disease by itself as manifested by alveolar bone loss and gingival granulocyte infiltration accompanied by overexpression of matrix metalloprotease-2, matrix metalloprotease-9, cyclooxygenase 1 (COX-1) and COX-2 in gingival tissues (62). Second, protease activated receptor-2 knockout mice orally infected with P. gingivalis showed significantly reduced (61) or displayed no periodontal bone loss as compared to wild-type infected mice and protease activated receptor-1-null infected mice (226). In stark contrast to control animals, lack of development of periodontitis in protease activated receptor-2-null mice was correlated with a large decrease in mast cells infiltration of the periodontal tissues and significantly lower expression of Type 1 helper T cells (Th1)-dependent inflammatory cytokines. Taken together, it was concluded that the absence of protease activated receptor-2 decreases proinflammatory mechanisms in T cells and mast cells which help to protect protease activated receptor-2-null animals from bone lost induced by the P. gingivalis oral infection. In this respect, gingipain stimulation of protease activated receptor-2 expression in human gingival fibroblasts and T cells and their subsequent activation may play a central role in the development of periodontitis (13).
In comparison to other cellular activation via protease activated receptor signaling, platelet activation and aggregation elicited by proteolysis of protease activated receptor-1 and -4 by RgpB and RgpA has no obvious effect on the pathogenicity of periodontal disease. Nevertheless, the phenomenon is interesting because, firstly, RgpA is an extremely efficient protease activated receptor-1 agonist, in some respect, exceeding the efficiency of thrombin, secondly, platelet aggregation may have a direct bearing on a link between periodontitis and atherosclerosis (114). The superior potency of RgpA in comparison to RgpB on platelet activation/aggregation despite the near identity of their respective catalytic domain suggests that the hemagglutinin-adhesin domains also play a role in platelet activation. This suggestion finds support in a paper describing the hemagglutinin-adhesin-1 domain present in RgpA, Kgp and HagA was able to exert proteolysis-independent platelet activation/aggregation via interaction with FcγRII receptors and to lesser extent GPIbα receptors on platelets (129). Apparently, hemagglutinin-adhesin binding to these receptors guides the protease domain of the gingipains to close proximity to the cleavage sites in protease activated receptor-1 and/or -4 for protease activated receptor proteolysis.
Gingipains in tissue destruction
Morphological changes associated with advanced periodontal disease include alveolar bone resorption and periodontal ligament destruction leading to attachment loss and periodontal pocket formation. Pathological periodontal pockets are lined with altered epithelial cells which are different from those which form the junctional epithelium in the healthy periodontium. All these tissue remodelling require proteolytic degradation of several structural elements like collagen fibers forming the periodontal ligament, proteins involved in cellular junctions and extracellular matrix proteins. As described below, gingipains are important, directly and indirectly, in the pathological tissue remodeling associated with development and progression of periodontitis.
Direct degradation of extracellular matrix proteins
Gingipains efficiently degrade several extracellular matrix proteins in vitro (for review see ref. (169)). However, it is still controversial if gingipains can cleave native type I collagen: the triple helical protease-resistant protein abundant in periodontal ligaments and dentin. It has been shown that purified soluble RgpA, RgpB and Kgp are unable to cleave native type I collagen and special care must be taken to eliminate artificial degradation which occurs during preparing samples for sodium dodecyl sulfate polyacrylamide gel electrophoresis (170). This is in conflict with an observation that collagenolytic activity of whole P. gingivalis cells is dependent on the Rgp activity (63). There are two possibilities to explain this discrepancy. First, Rgps are indispensable to activate a “true collagenase” and in the Rgp-null mutant, collagenolytic activity is not expressed. The presence of several protease genes encoding proproteases in P. gingivalis which may be activated by profragment cleavage at Arg-Xaa peptide bonds lends credit to this scenario. The second possibility is that only membrane-attached gingipain possesses collagenolytic activity. This option is supported by the finding that multidomain complexes of RgpA and Kgp containing lipopolysaccharide as purified from the P. gingivalis surface was able to degrade human collagen type I (215). For this activity, however, an enzyme co-purifying or associated with the gingipain complex may be responsible. Putting aside argument about Rgp's collagenolytic activity, it is clear that gingipains can potentially do a lot of direct damage to the connective tissue at the infection site. The only question is whether they need to do that themselves? In our opinion, gingipains can accomplish a lot more harm indirectly by disturbing the protease-protease inhibitor balance and allowing the host's “professional” endogenous matrix degrading enzymes free reign do the dirty job.
The imbalance, or excessive, broad-spectrum proteolytic activity at the local inflammatory sites can arise due to several mechanisms including: (i) excessive expression of matrix metalloproteases over their inhibitors (TIMPs); (ii) release of proteases from necrotic cells and activated neutrophils; (iii) inactivation of endogenous protease inhibitors; and (iv) uncontrolled conversion of zymogenic matrix metalloproteases into their proteolytically active forms. Apparently, all these mechanisms occur in the infected periodontitis sites and as the presence of active matrix metalloproteases (matrix metalloprotease-8, matrix metalloprotease-9) in gingival cervicular fluid (209) or even in saliva (178) have been reported to be the most reliable predictor of active periodontitis.
Discussing in detail the vast literature on the effect of P. gingivalis on the expression of matrix metalloproteases/TIMPs by different cell types is beyond the scope of this review. Briefly, the consensus picture is that P. gingivalis cell, cell extracts, spent media or lipopolysaccharide can stimulate the secretion of matrix metalloproteases at a higher level than TIMPs in dendritic cells (82), human periodontal ligament cells (186), human gingival fibroblasts (244) and an engineered human oral mucosa (8), just to give few examples. At least in the case of human gingival fibroblasts, it was shown that matrix metalloprotease-1 expression was stimulated by Rgp activity (119). On top of that, differentially regulated expression of matrix metalloproteases and TIMPs by a variety of growth factors, cytokines, chemokines (24) and neutrophil degranulation will further drive imbalance between matrix metalloproteases and TIMPs. Matrix metalloproteases are secreted as zymogens which can be activated by limited proteolysis of the profragment. In this context, it needs to be stressed that latent matrix metalloproteases can be directly activated by gingipains (28, 47). This finding is corroborated by data showing that Rgps stimulate the degradation of type I collagen by human gingival fibroblasts cultured on collagen-coated plates (4). The excessive release and uncontrolled activation of matrix metalloproteases unopposed by TIMPs is the most likely explanation for destruction of collagen fibers in periodontal ligament and pathological remodeling of the periodontal tissues.
Neutrophils infiltrating en masse into inflamed periodontal tissues are the main source of serine proteases (elastase, cathepsin G, and protease 3) and metalloproteases (matrix metalloprotease-8, and -9) packed into intracellular granules. The enzymes are released into the environment by frustrated, activated neutrophils or due to necrotic death of these short lived cells where they are usually neutralised by endogenous serine protease inhibitors, derived from blood plasma or produced locally. Several studies have shown that serine proteinase inhibitors, including α1-proteinase inhibitor (135), α1-antichymotrypsin (170), SLPI (78) and elafin (90) are proteolytically inactivated by P. gingivalis-derived proteases. If this happens in vivo, the powerful activated host proteases are unopposed and are free to attack a myriad of targets in the periodontium. Such scenario is corroborated by the presence of the neutrophil elastase activity in the gingival crevicular fluid (117). So far, however, no correlation between P. gingivalis presence at discrete periodontitis sites and neutrophil elastase in gingival crevicular fluid has been described.
To sum up, it is clear that gingipains despite their general proteolytic activity are probably not involved directly in causing damage to periodontal tissues. It is far more likely that these enzymes in synergy with other mechanisms used by P. gingivalis are able disturb proteolytic balance between host proteases, especially those derived from neutrophils, and endogenous protease inhibitors. Being released from tight control, host proteases, especially matrix metalloproteases and neutrophil elastase, are apparently the main force driving pathological remodeling of the periodontium.
Cell death, invasion and dissemination into the periodontium
The ability to invade host cells is an important strategy used by many pathogens to evade the host immune response, escape from initial colonization site, cross tissue barriers and disseminate to other sites in the infected host. The first line of host defense against this stealth tactic is the evolution of a cellular suicide mechanism known as apoptosis, where the infected host cell undergoes a highly regulated silent cell death. Perversely, pathogenic intracellular bacteria counteract the apoptotic machinery to keep invaded cells alive long enough to allow for their intracellular proliferation. Alternatively, pathogens can induce host cell death to destroy the physical barrier limiting their expansion. In the case of P. gingivalis, all these strategies are used and the gingipains are the deadly weapons in this war between host cells and the bacterium.
Gingipains can readily attack cell adhesion molecules which host cells employ to attach themselves to the underlying extracellular matrix proteins and cause the cells to undergo apoptosis. In the case of human gingival fibroblasts, Rgps were able to disrupt fibronectin-integrin interactions resulting in the detachment of the cells with subsequent induction of apoptosis (9, 49). Similarly, Rgp and Kgp were shown to cooperatively disrupt the adhesion activity of human umbilical vein endothelial cells and decreased their viability (10) most likely via activation of the apoptotic program (99). The lost of attachment was apparently due to cleavage of β1 integrin and N- and VE-cadherins and was followed by apoptosis (191, 192). Furthermore, gingipains were also able to induce invasion-independent epithelial cell rounding and detachment (138). In this phenomenon, gingipains caused the degradation of syndecan-1 (CD138) (7), as well as the cell-cell adhesion proteins in the epithelial adherens junction, including N-cadherin (21) or E-cadherin and catenins (58, 95).
Although the majority of P. gingivalis are present in the subgingival plaque, live bacteria have been identified within host cells, including epithelial (115, 179, 182, 183) and endothelial cells (228) and deep inside connective tissue distant from the biofilm. The finding that P. gingivalis strains isolated from periodontitis sites had stronger invasion and dissemination ability than those isolated from healthy oral sites is a feature which may help P. gingivalis to cross the physical barrier of the mucosa membrane and spread within the periodontal tissues.
As with many other pathogens, P. gingivalis exploit host cell signaling pathways to actively invade the cell. The majority of available data indicates that fimbriae play the major role in stimulating the non-phagocytic cells to engulf the bacterium. In the case of epithelial cells, integrin receptor activation by fimbriae seems to initiate at least one of the pathway leading to bacterial internalization. Since all cellular events and mechanism involved in P. gingivalis uptake by epithelial cells are described in details in a comprehensive review by Andrian and colleagues (6), here, we will only discuss what is little is know about intracellular activity of gingipains.
During the internalization process, components of cellular focal adhesion - a multiprotein complex integrating cellular adhesion molecules with cytoskeleton and signaling pathways including paxillin and focal adhesion kinase - are apparently degraded by the gingipains (130). Degradation of these two signaling molecules involved in cellular anchorage and directional migration together with transferring receptor were observed in epithelial cells engulfing P. gingivalis outer membrane vesicles via an endocytic pathway (37, 38). Transferring receptor and the signaling molecules were reported to be degraded by vesicular Rgp and Kgp, respectively. Proteolysis of the transferring receptor correlated with an impaired uptake of extracellular transferrin and affected cell migration and proliferation in stark contrast to degradation of paxillin and focal adhesion kinase which had no apparent effect on cell function (37). Similarly, cleavage of paxillin along with signal-regulated kinase 1 and 2 (ERK1/2) was also observed in P. gingivalis infected periodontal ligament cells without an apparent effect on the cell phenotype (74). Clearly, the biological effect of proteolysis of focal adhesion kinase, paxillin and ERK1/2 by gingipains on the life-death decision of the infected cells require more detailed studies.
Apart from inducing cell death by the disruption of cell-cell and cell-extracellular matrix communications, gingipain also can cause apoptosis of infected cells from within. Live P. gingivalis have been reported to invoke apoptosis in human gingival epithelial cells via a gingipain-dependent mechanism (211). This finding is in conflict with an observation that although gingival epithelial cells containing internalized P. gingivalis appear rounded and detached from the substratum, they do not undergo apoptosis and maintain their integrity for expended periods (134). This discrepancy is most likely a result of the experimental set up. From the available data, a following picture can be suggested. By proteolytic damage of cell to matrix adhesion, gingipain activates a specific type of apoptosis known as “anoikis” (22). However, P. gingivalis in direct contact with gingival epithelial cells have been reported to down regulate gingipain production thereby reducing the risk of killing the host cell (3, 158). Nevertheless, the cell will be dead if P. gingivalis is not promptly internalized. If engulfment occurred efficiently, live bacterium inside the cell induces expression of antiapoptotic genes to prolong the lifetime of the infected cells. This allows for intracellular proliferation of P. gingivalis that finally leads to activation of the proapoptotic genes and the ensuing cell death. Such hypothetical scenario is corroborated by the finding that P. gingivalis in infected gingival epithelial cells induced pro-survival phenotype (118). Similarly in gingival fibroblasts, intracellular P. gingivalis initially activated cellular survival response which later shifted to apoptosis. In this system, the involvement of gingipains in the manipulation of cell survival was inferred from the observation that gingipain-null P. gingivalis mutant induced cellular changes far less efficiently than the parental wild-type strain (222).
Apoptosis is interconnected with cell cycle progression on several levels and arrest in G1 often leads to apoptotic cell death. Invasion of human trophoblasts and osteoblastoma/stromal cells by P. gingivalis leads to inhibition of these cells to proliferation by inducing G1 arrest with ensuing apoptosis (75, 93). At least in the case of osteoblastoma/stromal cells, the arrest was exerted by gingipain-dependent reduction of cyclins - proteins which control the cell cycle progression (93). To date, this is the best documented example of the direct gingipain effect from within the host cell to induce its survival. It is important to note that the effect of P. gingivalis internalization is dependent on the cell type since in gingival epithelial cells, the invaders accelerate cell cycle progression (105).
Taken together, the role of gingipains in invasion and intracellular life of P. gingivalis is ambiguous. While some studies claim that gingipain of internalized bacteria contribute to host cell apoptosis, the others present results to the contrary (96). Nevertheless, there is consensus that gingipains aid in the intracellular survival of P. gingivalis but through what mechanism, we do not know (228). The finding that the hemagglutinin-adhesin domains are involved in nuclear targeting of soluble gingipains in epithelial cells in vitro (188) corroborates with the observation of gingipain localization near the perinuclear region of periodontal epithelial cells in clinical samples is as fascinating as it is puzzling (179). Even without knowing the precise role of gingipains, the ability of P. gingivalis to invade different cells and manipulate their life cycle has a direct bearing on the pathomechanism of periodontal disease development and progression.
Conclusions
It is unambiguous that gingipains are the major and primary virulence factors of P. gingivalis since gingipain activity is essential at every step of infection: from attachment and colonization, to nutrient acquisition, to evasion of host defenses and to tissue invasion. The spectrum of host proteins targeted by gingipains is very broad, with some being degraded while others being a subject of very selective, limited proteolysis. The specificity for the Arg-Xaa peptide bonds of Rgp enable these proteases to interfere with the proteolytic cascades of coagulation, complement, and kinin generation which are usually regulated by very specific host proteases via limited proteolytic steps. In many regards, these pathways have been hijacked by P. gingivalis far more efficiently than by any other pathogen. The proteolytic processing of clotting factors, prekallikrein and signaling via protease activated receptors illuminate the surgical precision of the gingipain action. At the same time, gingipains can degrade proteins indiscriminately with brutal efficiency. Fascinatingly, gingipains often show the Janus face: at low concentrations they enhance specific proinflammatory mechanisms of host immune system, while they are able to destroy these pathways at high concentrations. Hence, the finesse performance of the gingipains may have a paramount significance in the life or death not only for P. gingivalis, but also for the entire dental plaque biofilm community.
Table 3.
Protease activated receptors (PAR) targeted by gingipains and the consequence of their activation
| Targeted cells | Targeted PAR / gingipain | Response to PAR signaling | Pathophysiological consequence | Ref. |
|---|---|---|---|---|
| Neutrophils | PAR-2 Rgp | Increase in the intracellular Ca2+ concentration | Neutrophil activation | (112) |
| Osteoblasts | PAR-2 Rgp | Increase in the intracellular Ca2+ concentration | Activation of osteoblast-mediated osteoclastic bone resorption | (2) |
| Platelets | PAR-1, -4 RgpA, RgpB | Platelets aggregation | A putative link between periodontitis and cardiovascular disease | (114) |
| Oral epithelial cells | PAR-1, -2 | Stimulation of interleukin-6 expression | Inflammatory events associated with periodontal disease | (113) |
| Platelets | PARs Rgp enriched vesicles | Platelets aggregation | As above | (166) |
| Pulp cells | PAR-2 RgpB | Induced expression of neuropeptides: calcitonin gene-related peptide (CGRP) and substance P (SP) | A putative link between periodontitis and pulp inflammation | (217) |
| Gingival fibroblasts | PAR-1, -2 Rgp | Induced expression of hepatocyte growth factor | Effect on both inflammatory and tissue reparative processes | (220) |
| Rat kidney epithelial cells | PAR-2 P. gingivalis culture supernatants | Ca2+ mobilization | (61) | |
| Keratinocytes | PAR-1, -2 Rgp | Up-regulation of HIV-1 co receptor CCR5 | Promotion of HIV infection of oral keratinocytes | (43) |
| Oral epithelial cells | PAR-2 Rgp | Induced expression of human β-defensins-2 and CC chemokine ligand 20 (CCL20) | Enhancement of mucosal antibacterial defense | (31) |
| T cells | PAR-1, -2, -4 RgpA | Induced expression of CD25 and CD65 | T cell activation | (237) |
| Vascular endothelial cells | PAR RgpA | Weibel-Palade body exocytosis, enhanced production of interleukin-8 in response to lipopolysaccharide | Enhanced proinflammatory responses to P. gingivalis | (76) |
| Oral epithelial cells | PAR-1, -2 Rgp, Kgp | RgpB upregulate while Kgp and RgpA downregulate interleukin-8 expression | Attenuation of neutrophils migration and sustained chronic inflammation | (221) |
| Monocytic THP-1 cells | PARs Rgp, Kgp | Up-regulation of pro-inflammatory cytokines (interleukin-6, interleukin-8 and monocyte chemoattractant protein-1) | Synergistic effect of PAR-dependent and PAMPs receptor (Toll-like receptors or NOD1/2) signaling resulting in enhancement of mucosal antibacterial defense | (219) |
| ST2 mouse stromal cells and calvarial osteoblasts | PAR P. gingivalis cells | Proinflammatory responses independent of the Toll-like receptor/MyD88 pathway | Pathogen recognition: enhancement of mucosal antibacterial defense | (148) |
| Platelets | PARs Rgps | Platelet sensitization to aggregation by the stress hormone epinephrine | Mechanistic explanation of a direct connection between periodontitis and stress | (139) |
| Human oral keretinocytes | PAR-1, -2 P. gingivalis cells | PAR-1 cleavage by Rgps mediated up-regulation of pro-inflammatory cytokines (interleukin-1α, interleukin-1β, interleukin-6, tumor necrosis factor-α) | Osteoclasts differentiation and alveolar bone resorption | (42) |
Acknowledgments
This work was supported by grants from National Institutes of Health, USA (DE 09761), Polish Ministry of Science and Education (1642/B/P01/2008/35) and Foundation for Polish Science (Team/2009-4/8). The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University is a beneficiary of the structural funds from the European Union (grant No: POIG.02.01.00-12-064/08 – “Molecular biotechnology for health”).
References
- 1.Abe N, Baba A, Takii R, Nakayama K, Kamaguchi A, Shibata Y, Abiko Y, Okamoto K, Kadowaki T, Yamamoto K. Roles of Arg- and Lys-gingipains in coaggregation of Porphyromonas gingivalis: identification of its responsible molecules in translation products of rgpA, kgp, and hagA genes. Biol Chem. 2004;385:1041–1047. doi: 10.1515/BC.2004.135. [DOI] [PubMed] [Google Scholar]
- 2.Abraham LA, Chinni C, Jenkins AL, Lourbakos A, Ally N, Pike RN, Mackie EJ. Expression of protease-activated receptor-2 by osteoblasts. Bone. 2000;26:7–14. doi: 10.1016/s8756-3282(99)00237-9. [DOI] [PubMed] [Google Scholar]
- 3.Agnani G, Tricot-Doleux S, Du L, Bonnaure-Mallet M. Adherence of Porphyromonas gingivalis to gingival epithelial cells: modulation of bacterial protein expression. Oral Microbiol Immunol. 2000;15:48–52. doi: 10.1034/j.1399-302x.2000.150108.x. [DOI] [PubMed] [Google Scholar]
- 4.Al-Shibani N, Windsor LJ. Effects of Porphyromonas gingivalis on human gingival fibroblasts from healthy and inflamed tissues. J Periodontal Res. 2008;43:465–470. doi: 10.1111/j.1600-0765.2008.01100.x. [DOI] [PubMed] [Google Scholar]
- 5.Amano A. Bacterial adhesins to host components in periodontitis. Periodontol 2000. 2010;52:12–37. doi: 10.1111/j.1600-0757.2009.00307.x. [DOI] [PubMed] [Google Scholar]
- 6.Andrian E, Grenier D, Rouabhia M. Porphyromonas gingivalis-epithelial cell interactions in periodontitis. J Dent Res. 2006;85:392–403. doi: 10.1177/154405910608500502. [DOI] [PubMed] [Google Scholar]
- 7.Andrian E, Grenier D, Rouabhia M. Porphyromonas gingivalis gingipains mediate the shedding of syndecan-1 from the surface of gingival epithelial cells. Oral Microbiol Immunol. 2006;21:123–128. doi: 10.1111/j.1399-302X.2006.00248.x. [DOI] [PubMed] [Google Scholar]
- 8.Andrian E, Mostefaoui Y, Rouabhia M, Grenier D. Regulation of matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases by Porphyromonas gingivalis in an engineered human oral mucosa model. J Cell Physiol. 2007;211:56–62. doi: 10.1002/jcp.20894. [DOI] [PubMed] [Google Scholar]
- 9.Baba A, Abe N, Kadowaki T, Nakanishi H, Ohishi M, Asao T, Yamamoto K. Arg-gingipain is responsible for the degradation of cell adhesion molecules of human gingival fibroblasts and their death induced by Porphyromonas gingivalis. Biol Chem. 2001;382:817–824. doi: 10.1515/BC.2001.099. [DOI] [PubMed] [Google Scholar]
- 10.Baba A, Kadowaki T, Asao T, Yamamoto K. Roles for Arg- and Lys-gingipains in the disruption of cytokine responses and loss of viability of human endothelial cells by Porphyromonas gingivalis infection. Biol Chem. 2002;383:1223–1230. doi: 10.1515/BC.2002.135. [DOI] [PubMed] [Google Scholar]
- 11.Banbula A, Bugno M, Kuster A, Heinrich PC, Travis J, Potempa J. Rapid and efficient inactivation of IL-6 gingipains, lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Biochem Biophys Res Commun. 1999;261:598–602. doi: 10.1006/bbrc.1999.1075. [DOI] [PubMed] [Google Scholar]
- 12.Barille-Nion S, Bataille R. New insights in myeloma-induced osteolysis. Leuk Lymphoma. 2003;44:1463–1467. doi: 10.3109/10428190309178765. [DOI] [PubMed] [Google Scholar]
- 13.Belibasakis GN, Bostanci N, Reddi D. Regulation of protease-activated receptor-2 expression in gingival fibroblasts and Jurkat T cells by Porphyromonas gingivalis. Cell Biol Int. 2010;34:287–292. doi: 10.1042/CBI20090290. [DOI] [PubMed] [Google Scholar]
- 14.Brechter AB, Lerner UH. Bradykinin potentiates cytokine-induced prostaglandin biosynthesis in osteoblasts by enhanced expression of cyclooxygenase 2, resulting in increased RANKL expression. Arthritis Rheum. 2007;56:910–923. doi: 10.1002/art.22445. [DOI] [PubMed] [Google Scholar]
- 15.Brechter AB, Persson E, Lundgren I, Lerner UH. Kinin B1 and B2 receptor expression in osteoblasts and fibroblasts is enhanced by interleukin-1 and tumour necrosis factor-alpha. Effects dependent on activation of NF-kappaB and MAP kinases. Bone. 2008;43:72–83. doi: 10.1016/j.bone.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 16.Brochu V, Grenier D, Nakayama K, Mayrand D. Acquisition of iron from human transferrin by Porphyromonas gingivalis: a role for Arg- and Lys-gingipain activities. Oral Microbiol Immunol. 2001;16:79–87. doi: 10.1034/j.1399-302x.2001.016002079.x. [DOI] [PubMed] [Google Scholar]
- 17.Calkins CC, Platt K, Potempa J, Travis J. Inactivation of tumor necrosis factor-alpha by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J Biol Chem. 1998;273:6611–6614. doi: 10.1074/jbc.273.12.6611. [DOI] [PubMed] [Google Scholar]
- 18.Carlisle MD, Srikantha RN, Brogden KA. Degradation of human alpha- and beta-defensins by culture supernatants of Porphyromonas gingivalis strain 381. J Innate Immun. 2009;1:118–122. doi: 10.1159/000181015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Dorling A. Critical roles for thrombin in acute and chronic inflammation. J Thromb Haemost. 2009;7 1:122–126. doi: 10.1111/j.1538-7836.2009.03413.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Duncan MJ. Gingipain adhesin domains mediate Porphyromonas gingivalis adherence to epithelial cells. Microb Pathog. 2004;36:205–209. doi: 10.1016/j.micpath.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Chen T, Nakayama K, Belliveau L, Duncan MJ. Porphyromonas gingivalis gingipains and adhesion to epithelial cells. Infect Immun. 2001;69:3048–3056. doi: 10.1128/IAI.69.5.3048-3056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiarugi P, Giannoni E. Anoikis: a necessary death program for anchorage-dependent cells. Biochem Pharmacol. 2008;76:1352–1364. doi: 10.1016/j.bcp.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 23.Chu L, Bramanti TE, Ebersole JL, Holt SC. Hemolytic activity in the periodontopathogen Porphyromonas gingivalis: kinetics of enzyme release and localization. Infect Immun. 1991;59:1932–1940. doi: 10.1128/iai.59.6.1932-1940.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark IM, Swingler TE, Sampieri CL, Edwards DR. The regulation of matrix metalloproteinases and their inhibitors. Int J Biochem Cell Biol. 2008;40:1362–1378. doi: 10.1016/j.biocel.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Curtis MA, Aduse Opoku J, Rangarajan M, Gallagher A, Sterne JA, Reid CR, Evans HE, Samuelsson B. Attenuation of the virulence of Porphyromonas gingivalis by using a specific synthetic Kgp protease inhibitor. Infect Immun. 2002;70:6968–6975. doi: 10.1128/IAI.70.12.6968-6975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis MA, Kuramitsu HK, Lantz M, Macrina FL, Nakayama K, Potempa J, Reynolds EC, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodontal Res. 1999;34:464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 27.de Pablo P, Chapple IL, Buckley CD, Dietrich T. Periodontitis in systemic rheumatic diseases. Nat Rev Rheumatol. 2009;5:218–224. doi: 10.1038/nrrheum.2009.28. [DOI] [PubMed] [Google Scholar]
- 28.DeCarlo AA, Jr, Windsor LJ, Bodden MK, Harber GJ, Birkedal-Hansen B, Birkedal-Hansen H. Activation and novel processing of matrix metalloproteinases by a thiol-proteinase from the oral anaerobe Porphyromonas gingivalis. J Dent Res. 1997;76:1260–1270. doi: 10.1177/00220345970760060501. [DOI] [PubMed] [Google Scholar]
- 29.Diamond G, Beckloff N, Ryan LK. Host defense peptides in the oral cavity and the lung: similarities and differences. J Dent Res. 2008;87:915–927. doi: 10.1177/154405910808701011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dias IH, Marshall L, Lambert PA, Chapple IL, Matthews JB, Griffiths HR. Gingipains from Porphyromonas gingivalis increase the chemotactic and respiratory burst-priming properties of the 77-amino-acid interleukin-8 variant. Infect Immun. 2008;76:317–323. doi: 10.1128/IAI.00618-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dommisch H, Chung WO, Rohani MG, Williams D, Rangarajan M, Curtis MA, Dale BA. Protease-activated receptor 2 mediates human beta-defensin 2 and CC chemokine ligand 20 mRNA expression in response to proteases secreted by Porphyromonas gingivalis. Infect Immun. 2007;75:4326–4333. doi: 10.1128/IAI.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duncan L, Yoshioka M, Chandad F, Grenier D. Loss of lipopolysaccharide receptor CD14 from the surface of human macrophage-like cells mediated by Porphyromonas gingivalis outer membrane vesicles. Microb Pathog. 2004;36:319–325. doi: 10.1016/j.micpath.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Eissner G, Kohlhuber F, Grell M, Ueffing M, Scheurich P, Hieke A, Multhoff G, Bornkamm GW, Holler E. Critical involvement of transmembrane tumor necrosis factor-alpha in endothelial programmed cell death mediated by ionizing radiation and bacterial endotoxin. Blood. 1995;86:4184–4193. [PubMed] [Google Scholar]
- 34.Fitzpatrick RE, Aprico A, Wijeyewickrema LC, Pagel CN, Wong DM, Potempa J, Mackie EJ, Pike RN. High molecular weight gingipains from Porphyromonas gingivalis induce cytokine responses from human macrophage-like cells via a nonproteolytic mechanism. J Innate Immun. 2009;1:109–117. doi: 10.1159/000181145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fitzpatrick RE, Wijeyewickrema LC, Pike RN. The gingipains: scissors and glue of the periodontal pathogen, Porphyromonas gingivalis. Future Microbiol. 2009;4:471–487. doi: 10.2217/fmb.09.18. [DOI] [PubMed] [Google Scholar]
- 36.Frick IM, Bjorck L, Herwald H. The dual role of the contact system in bacterial infectious disease. Thromb Haemost. 2007;98:497–502. [PubMed] [Google Scholar]
- 37.Furuta N, Takeuchi H, Amano A. Entry of Porphyromonas gingivalis outer membrane vesicles into epithelial cells causes cellular functional impairment. Infect Immun. 2009;77:4761–4770. doi: 10.1128/IAI.00841-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furuta N, Tsuda K, Omori H, Yoshimori T, Yoshimura F, Amano A. Porphyromonas gingivalis outer membrane vesicles enter human epithelial cells via an endocytic pathway and are sorted to lysosomal compartments. Infect Immun. 2009;77:4187–4196. doi: 10.1128/IAI.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Genco CA, Odusanya BM, Potempa J, Mikolajczyk-Pawlinska J, Travis J. A peptide domain on gingipain R which confers immunity against Porphyromonas gingivalis infection in mice. Infect Immun. 1998;66:4108–4114. doi: 10.1128/iai.66.9.4108-4114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genco CA, Potempa J, Mikolajczyk-Pawlinska J, Travis J. Role of gingipains R in the pathogenesis of Porphyromonas gingivalis-mediated periodontal disease. Clin Infect Dis. 1999;28:456–465. doi: 10.1086/515156. [DOI] [PubMed] [Google Scholar]
- 41.Gera I. Periodontal treatment needs in Central and Eastern Europe. J Int Acad Periodontol. 2000;2:120–128. [PubMed] [Google Scholar]
- 42.Giacaman RA, Asrani AC, Ross KF, Herzberg MC. Cleavage of protease-activated receptors on an immortalized oral epithelial cell line by Porphyromonas gingivalis gingipains. Microbiology. 2009 doi: 10.1099/mic.0.029132-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giacaman RA, Nobbs AH, Ross KF, Herzberg MC. Porphyromonas gingivalis selectively up-regulates the HIV-1 coreceptor CCR5 in oral keratinocytes. J Immunol. 2007;179:2542–2550. doi: 10.4049/jimmunol.179.4.2542. [DOI] [PubMed] [Google Scholar]
- 44.Gibson FC, 3rd, Genco CA. Prevention of Porphyromonas gingivalis-induced oral bone loss following immunization with gingipain R1. Infect Immun. 2001;69:7959–7963. doi: 10.1128/IAI.69.12.7959-7963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gimbrone MA, Jr, Obin MS, Brock AF, Luis EA, Hass PE, Hebert CA, Yip YK, Leung DW, Lowe DG, Kohr WJ, et al. Endothelial interleukin-8: a novel inhibitor of leukocyte-endothelial interactions. Science. 1989;246:1601–1603. doi: 10.1126/science.2688092. [DOI] [PubMed] [Google Scholar]
- 46.Goulet V, Britigan B, Nakayama K, Grenier D. Cleavage of human transferrin by Porphyromonas gingivalis gingipains promotes growth and formation of hydroxyl radicals. Infect Immun. 2004;72:4351–4356. doi: 10.1128/IAI.72.8.4351-4356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grayson R, Douglas C, Heath J, Rawlinson A, Evans G. Activation of human matrix metalloproteinase 2 by gingival crevicular fluid and Porphyromonas gingivalis. J Clin Periodontol. 2003;30:542–550. doi: 10.1034/j.1600-051x.2003.00301.x. [DOI] [PubMed] [Google Scholar]
- 48.Grell M, Douni E, Wajant H, Lohden M, Clauss M, Maxeiner B, Georgopoulos S, Lesslauer W, Kollias G, Pfizenmaier K, Scheurich P. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 49.Grenier D, Roy S, Chandad F, Plamondon P, Yoshioka M, Nakayama K, Mayrand D. Effect of inactivation of the Arg- and/or Lys-gingipain gene on selected virulence and physiological properties of Porphyromonas gingivalis. Infect Immun. 2003;71:4742–4748. doi: 10.1128/IAI.71.8.4742-4748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guichard C, Pedruzzi E, Dewas C, Fay M, Pouzet C, Bens M, Vandewalle A, Ogier-Denis E, Gougerot-Pocidalo MA, Elbim C. Interleukin-8-induced priming of neutrophil oxidative burst requires sequential recruitment of NADPH oxidase components into lipid rafts. J Biol Chem. 2005;280:37021–37032. doi: 10.1074/jbc.M506594200. [DOI] [PubMed] [Google Scholar]
- 51.Gutner M, Chaushu S, Balter D, Bachrach G. Saliva enables the antimicrobial activity of LL-37 in the presence of proteases of Porphyromonas gingivalis. Infect Immun. 2009;77:5558–5563. doi: 10.1128/IAI.00648-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guzik K, Bzowska M, Smagur J, Krupa O, Sieprawska M, Travis J, Potempa J. A new insight into phagocytosis of apoptotic cells: proteolytic enzymes divert the recognition and clearance of polymorphonuclear leukocytes by macrophages. Cell Death Differ. 2007;14:171–182. doi: 10.1038/sj.cdd.4401927. [DOI] [PubMed] [Google Scholar]
- 53.Haffajee AD, Cugini MA, Tanner A, Pollack RP, Smith C, Kent RL, Jr, Socransky SS. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 54.Hajishengallis G. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 2009;11:637–645. doi: 10.1016/j.micinf.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc Natl Acad Sci U S A. 2008;105:13532–13537. doi: 10.1073/pnas.0803852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamada S, Amano A, Kimura S, Nakagawa I, Kawabata S, Morisaki I. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol Immunol. 1998;13:129–138. doi: 10.1111/j.1399-302x.1998.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 57.Hansen KK, Oikonomopoulou K, Baruch A, Ramachandran R, Beck P, Diamandis EP, Hollenberg MD. Proteinases as hormones: targets and mechanisms for proteolytic signaling. Biol Chem. 2008 doi: 10.1515/BC.2008.120. [DOI] [PubMed] [Google Scholar]
- 58.Hintermann E, Haake SK, Christen U, Sharabi A, Quaranta V. Discrete proteolysis of focal contact and adherens junction components in Porphyromonas gingivalis-infected oral keratinocytes: a strategy for cell adhesion and migration disabling. Infect Immun. 2002;70:5846–5856. doi: 10.1128/IAI.70.10.5846-5856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hollenberg MD. Physiology and pathophysiology of proteinase-activated receptors (PARs): proteinases as hormone-like signal messengers: PARs and more. J Pharmacol Sci. 2005;97:8–13. doi: 10.1254/jphs.fmj04005x2. [DOI] [PubMed] [Google Scholar]
- 60.Hollenberg MD. Getting the message across: pathophysiology and signaling via receptors for polypeptide hormones and proteinases. Clin Invest Med. 2010;33:E133. doi: 10.25011/cim.v33i2.12352. [DOI] [PubMed] [Google Scholar]
- 61.Holzhausen M, Spolidorio LC, Ellen RP, Jobin MC, Steinhoff M, Andrade-Gordon P, Vergnolle N. Protease-activated receptor-2 activation: a major role in the pathogenesis of Porphyromonas gingivalis infection. Am J Pathol. 2006;168:1189–1199. doi: 10.2353/ajpath.2006.050658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holzhausen M, Spolidorio LC, Vergnolle N. Role of protease-activated receptor-2 in inflammation, and its possible implications as a putative mediator of periodontitis. Mem Inst Oswaldo Cruz. 2005;100 1:177–180. doi: 10.1590/s0074-02762005000900030. [DOI] [PubMed] [Google Scholar]
- 63.Houle MA, Grenier D, Plamondon P, Nakayama K. The collagenase activity of Porphyromonas gingivalis is due to Arg-gingipain. FEMS Microbiol Lett. 2003;221:181–185. doi: 10.1016/S0378-1097(03)00178-2. [DOI] [PubMed] [Google Scholar]
- 64.Hu SW, Huang CH, Huang HC, Lai YY, Lin YY. Transvascular dissemination of Porphyromonas gingivalis from a sequestered site is dependent upon activation of the kallikrein/kinin pathway. J Periodontal Res. 2006;41:200–207. doi: 10.1111/j.1600-0765.2005.00858.x. [DOI] [PubMed] [Google Scholar]
- 65.Huber-Lang M, Sarma JV, Zetoune FS, Rittirsch D, Neff TA, McGuire SR, Lambris JD, Warner RL, Flierl MA, Hoesel LM, Gebhard F, Younger JG, Drouin SM, Wetsel RA, Ward PA. Generation of C5a in the absence of C3: a new complement activation pathway. Nat Med. 2006;12:682–687. doi: 10.1038/nm1419. [DOI] [PubMed] [Google Scholar]
- 66.Huber AR, Kunkel SL, Todd RF, 3rd, Weiss SJ. Regulation of transendothelial neutrophil migration by endogenous interleukin-8. Science. 1991;254:99–102. doi: 10.1126/science.1718038. [DOI] [PubMed] [Google Scholar]
- 67.Hugoson A, Sjodin B, Norderyd O. Trends over 30 years, 1973-2003, in the prevalence and severity of periodontal disease. J Clin Periodontol. 2008;35:405–414. doi: 10.1111/j.1600-051X.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- 68.Humphrey LL, Fu R, Buckley DI, Freeman M, Helfand M. Periodontal disease and coronary heart disease incidence: a systematic review and meta-analysis. J Gen Intern Med. 2008;23:2079–2086. doi: 10.1007/s11606-008-0787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Imamura T. The role of gingipains in the pathogenesis of periodontal disease. J Periodontol. 2003;74:111–118. doi: 10.1902/jop.2003.74.1.111. [DOI] [PubMed] [Google Scholar]
- 70.Imamura T, Banbula A, Pereira PJ, Travis J, Potempa J. Activation of human prothrombin by arginine-specific cysteine proteinases (gingipains R) from Porphyromonas gingivalis. J Biol Chem. 2001;276:18984–18991. doi: 10.1074/jbc.M006760200. [DOI] [PubMed] [Google Scholar]
- 71.Imamura T, Potempa J, Tanase S, Travis J. Activation of blood coagulation factor X by arginine-specific cysteine proteinases (gingipain-Rs) from Porphyromonas gingivalis. J Biol Chem. 1997;272:16062–16067. doi: 10.1074/jbc.272.25.16062. [DOI] [PubMed] [Google Scholar]
- 72.Imamura T, Tanase S, Hamamoto T, Potempa J, Travis J. Activation of blood coagulation factor IX by gingipains R, arginine-specific cysteine proteinases from Porphyromonas gingivalis. Biochem J. 2001;353:325–331. doi: 10.1042/0264-6021:3530325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imamura T, Travis J, Potempa J. The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr Protein Pept Sci. 2003;4:443–450. doi: 10.2174/1389203033487027. [DOI] [PubMed] [Google Scholar]
- 74.Inaba H, Kawai S, Nakayama K, Okahashi N, Amano A. Effect of enamel matrix derivative on periodontal ligament cells in vitro is diminished by Porphyromonas gingivalis. J Periodontol. 2004;75:858–865. doi: 10.1902/jop.2004.75.6.858. [DOI] [PubMed] [Google Scholar]
- 75.Inaba H, Kuboniwa M, Bainbridge B, Yilmaz O, Katz J, Shiverick KT, Amano A, Lamont RJ. Porphyromonas gingivalis invades human trophoblasts and inhibits proliferation by inducing G1 arrest and apoptosis. Cell Microbiol. 2009;11:1517–1532. doi: 10.1111/j.1462-5822.2009.01344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inomata M, Into T, Ishihara Y, Nakashima M, Noguchi T, Matsushita K. Arginine-specific gingipain A from Porphyromonas gingivalis induces Weibel-Palade body exocytosis and enhanced activation of vascular endothelial cells through protease-activated receptors. Microbes Infect. 2007;9:1500–1506. doi: 10.1016/j.micinf.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 77.Inomata M, Ishihara Y, Matsuyama T, Imamura T, Maruyama I, Noguchi T, Matsushita K. Degradation of vascular endothelial thrombomodulin by arginine- and lysine-specific cysteine proteases from Porphyromonas gingivalis. J Periodontol. 2009;80:1511–1517. doi: 10.1902/jop.2009.090114. [DOI] [PubMed] [Google Scholar]
- 78.Into T, Inomata M, Kanno Y, Matsuyama T, Machigashira M, Izumi Y, Imamura T, Nakashima M, Noguchi T, Matsushita K. Arginine-specific gingipains from Porphyromonas gingivalis deprive protective functions of secretory leucocyte protease inhibitor in periodontal tissue. Clin Exp Immunol. 2006;145:545–554. doi: 10.1111/j.1365-2249.2006.03156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jagels MA, Travis J, Potempa J, Pike R, Hugli TE. Proteolytic inactivation of the leukocyte C5a receptor by proteinases derived from Porphyromonas gingivalis. Infection & Immunity. 1996;64:1984–1991. doi: 10.1128/iai.64.6.1984-1991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ji S, Hyun J, Park E, Lee BL, Kim KK, Choi Y. Susceptibility of various oral bacteria to antimicrobial peptides and to phagocytosis by neutrophils. J Periodontal Res. 2007;42:410–419. doi: 10.1111/j.1600-0765.2006.00962.x. [DOI] [PubMed] [Google Scholar]
- 81.Joly S, Maze C, McCray PB, Jr, Guthmiller JM. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J Clin Microbiol. 2004;42:1024–1029. doi: 10.1128/JCM.42.3.1024-1029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jotwani R, Eswaran SV, Moonga S, Cutler CW. MMP-9/TIMP-1 imbalance induced in human dendritic cells by Porphyromonas gingivalis. FEMS Immunol Med Microbiol. 2009;58:314–321. doi: 10.1111/j.1574-695X.2009.00637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kadowaki T, Baba A, Abe N, Takii R, Hashimoto M, Tsukuba T, Okazaki S, Suda Y, Asao T, Yamamoto K. Suppression of pathogenicity of Porphyromonas gingivalis by newly developed gingipain inhibitors. Mol Pharmacol. 2004;66:1599–1606. doi: 10.1124/mol.104.004366. [DOI] [PubMed] [Google Scholar]
- 84.Kadowaki T, Takii R, Yamatake K, Kawakubo T, Tsukuba T, Yamamoto K. A role for gingipains in cellular responses and bacterial survival in Porphyromonas gingivalis-infected cells. Front Biosci. 2007;12:4800–4809. doi: 10.2741/2428. [DOI] [PubMed] [Google Scholar]
- 85.Kamaguchi A, Nakayama K, Ohyama T, Watanabe T, Okamoto M, Baba H. Coaggregation of Porphyromonas gingivalis and Prevotella intermedia. Microbiol Immunol. 2001;45:649–656. doi: 10.1111/j.1348-0421.2001.tb01298.x. [DOI] [PubMed] [Google Scholar]
- 86.Kamaguchi A, Ohyama T, Sakai E, Nakamura R, Watanabe T, Baba H, Nakayama K. Adhesins encoded by the gingipain genes of Porphyromonas gingivalis are responsible for co-aggregation with Prevotella intermedia. Microbiology. 2003;149:1257–1264. doi: 10.1099/mic.0.25997-0. [DOI] [PubMed] [Google Scholar]
- 87.Kamer AR, Craig RG, Dasanayake AP, Brys M, Glodzik-Sobanska L, de Leon MJ. Inflammation and Alzheimer's disease: possible role of periodontal diseases. Alzheimers Dement. 2008;4:242–250. doi: 10.1016/j.jalz.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 88.Kamer AR, Craig RG, Pirraglia E, Dasanayake AP, Norman RG, Boylan RJ, Nehorayoff A, Glodzik L, Brys M, de Leon MJ. TNF-alpha and antibodies to periodontal bacteria discriminate between Alzheimer's disease patients and normal subjects. J Neuroimmunol. 2009;216:92–97. doi: 10.1016/j.jneuroim.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamer AR, Dasanayake AP, Craig RG, Glodzik-Sobanska L, Bry M, de Leon MJ. Alzheimer's disease and peripheral infections: the possible contribution from periodontal infections, model and hypothesis. J Alzheimers Dis. 2008;13:437–449. doi: 10.3233/jad-2008-13408. [DOI] [PubMed] [Google Scholar]
- 90.Kantyka T, Latendorf T, Wiedow O, Bartels J, Glaser R, Dubin G, Schroder JM, Potempa J, Meyer-Hoffert U. Elafin is specifically inactivated by RgpB from Porphyromonas gingivalis by distinct proteolytic cleavage. Biol Chem. 2009;390:1313–1320. doi: 10.1515/BC.2009.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karim AY, Kulczycka M, Kantyka T, Dubin G, Jabaiah A, Daugherty PS, Thogersen IB, Enghild JJ, Nguyen KA, Potempa J. A novel matrix metalloprotease-like enzyme (karilysin) of the periodontal pathogen Tannerella forsythia ATCC 43037. Biol Chem. 2010;391:105–117. doi: 10.1515/BC.2010.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kato T, Kawai S, Nakano K, Inaba H, Kuboniwa M, Nakagawa I, Tsuda K, Omori H, Ooshima T, Yoshimori T, Amano A. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol. 2007;9:753–765. doi: 10.1111/j.1462-5822.2006.00825.x. [DOI] [PubMed] [Google Scholar]
- 93.Kato T, Tsuda T, Inaba H, Kawai S, Okahashi N, Shibata Y, Abiko Y, Amano A. Porphyromonas gingivalis gingipains cause G(1) arrest in osteoblastic/stromal cells. Oral Microbiol Immunol. 2008;23:158–164. doi: 10.1111/j.1399-302X.2007.00405.x. [DOI] [PubMed] [Google Scholar]
- 94.Kato T, Tsuda T, Omori H, Yoshimori T, Amano A. Maturation of fimbria precursor protein by exogenous gingipains in Porphyromonas gingivalis gingipain-null mutant. FEMS Microbiol Lett. 2007;273:96–102. doi: 10.1111/j.1574-6968.2007.00779.x. [DOI] [PubMed] [Google Scholar]
- 95.Katz J, Yang QB, Zhang P, Potempa J, Travis J, Michalek SM, Balkovetz DF. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect Immun. 2002;70:2512–2518. doi: 10.1128/IAI.70.5.2512-2518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kinane DF, Galicia JC, Gorr SU, Stathopoulou PG, Benakanakere M. P. gingivalis interactions with epithelial cells. Front Biosci. 2008;13:966–984. doi: 10.2741/2736. [DOI] [PubMed] [Google Scholar]
- 97.Kitamura Y, Matono S, Aida Y, Hirofuji T, Maeda K. Gingipains in the culture supernatant of Porphyromonas gingivalis cleave CD4 and CD8 on human T cells. J Periodontal Res. 2002;37:464–468. doi: 10.1034/j.1600-0765.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- 98.Kitano S, Irimura K, Sasaki T, Abe N, Baba A, Miyake Y, Katunuma N, Yamamoto K. Suppression of gingival inflammation induced by Porphyromonas gingivalis in rats by leupeptin. Jpn J Pharmacol. 2001;85:84–91. doi: 10.1254/jjp.85.84. [DOI] [PubMed] [Google Scholar]
- 99.Kobayashi-Sakamoto M, Hirose K, Nishikata M, Isogai E, Chiba I. Osteoprotegerin protects endothelial cells against apoptotic cell death induced by Porphyromonas gingivalis cysteine proteinases. FEMS Microbiol Lett. 2006;264:238–245. doi: 10.1111/j.1574-6968.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 100.Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 101.Kontani M, Kimura S, Nakagawa I, Hamada S. Adherence of Porphyromonas gingivalis to matrix proteins via a fimbrial cryptic receptor exposed by its own arginine-specific protease. Mol Microbiol. 1997;24:1179–1187. doi: 10.1046/j.1365-2958.1997.4321788.x. [DOI] [PubMed] [Google Scholar]
- 102.Koziel J, Karim AY, Przybyszewska K, Ksiazek M, Rapala-Kozik M, Nguyen KA, Potempa J. Proteolytic inactivation of LL-37 by Karilysin, a novel virulence mechanism of Tannerella forsythia. J Innate Immun. 2010;2:288–293. doi: 10.1159/000281881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Krauss JL, Potempa J, Lambris JD, Hajishengallis G. Complementary Tolls in the periodontium: how periodontal bacteria modify complement and Toll-like receptor responses to prevail in the host. Periodontol 2000. 2010;52:141–162. doi: 10.1111/j.1600-0757.2009.00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kuboniwa M, Amano A, Shizukuishi S, Nakagawa I, Hamada S. Specific antibodies to Porphyromonas gingivalis Lys-gingipain by DNA vaccination inhibit bacterial binding to hemoglobin and protect mice from infection. Infect Immun. 2001;69:2972–2979. doi: 10.1128/IAI.69.5.2972-2979.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kuboniwa M, Hasegawa Y, Mao S, Shizukuishi S, Amano A, Lamont RJ, Yilmaz O. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10:122–128. doi: 10.1016/j.micinf.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuula H, Salo T, Pirila E, Hagstrom J, Luomanen M, Gutierrez-Fernandez A, Romanos GE, Sorsa T. Human beta-defensin-1 and -2 and matrix metalloproteinase-25 and -26 expression in chronic and aggressive periodontitis and in peri-implantitis. Arch Oral Biol. 2008;53:175–186. doi: 10.1016/j.archoralbio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 107.Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Levi M, van der Poll T. Inflammation and coagulation. Crit Care Med. 2010;38:S26–34. doi: 10.1097/CCM.0b013e3181c98d21. [DOI] [PubMed] [Google Scholar]
- 109.Lewis JP, Dawson JA, Hannis JC, Muddiman D, Macrina FL. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J Bacteriol. 1999;181:4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li N, Yun P, Nadkarni MA, Ghadikolaee NB, Nguyen KA, Lee M, Hunter N, Collyer CA. Structure determination and analysis of a hemolytic gingipain adhesin domain from Porphyromonas gingivalis. Mol Microbiol. 2010 Mar 10; doi: 10.1111/j.1365-2958.2010.07123.x. [Epub ahead of print] PMID: 20233299. [DOI] [PubMed] [Google Scholar]
- 111.Liu X, Sroka A, Potempa J, Genco CA. Coordinate expression of the Porphyromonas gingivalis lysine-specific gingipain proteinase, Kgp, arginine-specific gingipain proteinase, RgpA, and the heme/hemoglobin receptor, HmuR. Biol Chem. 2004;385:1049–1057. doi: 10.1515/BC.2004.136. [DOI] [PubMed] [Google Scholar]
- 112.Lourbakos A, Chinni C, Thompson P, Potempa J, Travis J, Mackie EJ, Pike RN. Cleavage and activation of proteinase-activated receptor-2 on human neutrophils by gingipain-R from Porphyromonas gingivalis. FEBS Lett. 1998;435:45–48. doi: 10.1016/s0014-5793(98)01036-9. [DOI] [PubMed] [Google Scholar]
- 113.Lourbakos A, Potempa J, Travis J, D'Andrea MR, Andrade-Gordon P, Santulli R, Mackie EJ, Pike RN. Arginine-specific protease from Porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion. Infect Immun. 2001;69:5121–5130. doi: 10.1128/IAI.69.8.5121-5130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lourbakos A, Yuan YP, Jenkins AL, Travis J, Andrade-Gordon P, Santulli R, Potempa J, Pike RN. Activation of protease-activated receptors by gingipains from Porphyromonas gingivalis leads to platelet aggregation: a new trait in microbial pathogenicity. Blood. 2001;97:3790–3797. doi: 10.1182/blood.v97.12.3790. [DOI] [PubMed] [Google Scholar]
- 115.Madianos PN, Papapanou PN, Sandros J. Porphyromonas gingivalis infection of oral epithelium inhibits neutrophil transepithelial migration. Infect Immun. 1997;65:3983–3990. doi: 10.1128/iai.65.10.3983-3990.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mahtout H, Chandad F, Rojo JM, Grenier D. Porphyromonas gingivalis mediates the shedding and proteolysis of complement regulatory protein CD46 expressed by oral epithelial cells. Oral Microbiol Immunol. 2009;24:396–400. doi: 10.1111/j.1399-302X.2009.00532.x. [DOI] [PubMed] [Google Scholar]
- 117.Mailhot JM, Potempa J, Stein SH, Travis J, Sterrett JD, Hanes PJ, Russell CM. A relationship between proteinase activity and clinical parameters in the treatment of periodontal disease. J Clin Periodontol. 1998;25:578–584. doi: 10.1111/j.1600-051x.1998.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 118.Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, Stavropoulos MF, Yilmaz O, Lamont RJ. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9:1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Matsushita K, Imamura T, Tomikawa M, Tancharoen S, Tatsuyama S, Maruyama I. DX-9065a inhibits proinflammatory events induced by gingipains and factor Xa. J Periodontal Res. 2006;41:148–156. doi: 10.1111/j.1600-0765.2005.00853.x. [DOI] [PubMed] [Google Scholar]
- 120.McAlister AD, Sroka A, Fitzpatrick RE, Quinsey NS, Travis J, Potempa J, Pike RN. Gingipain enzymes from Porphyromonas gingivalis preferentially bind immobilized extracellular proteins: a mechanism favouring colonization? J Periodontal Res. 2009;44:348–353. doi: 10.1111/j.1600-0765.2008.01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mezyk-Kopec R, Bzowska M, Potempa J, Jura N, Sroka A, Black RA, Bereta J. Inactivation of membrane tumor necrosis factor alpha by gingipains from Porphyromonas gingivalis. Infect Immun. 2005;73:1506–1514. doi: 10.1128/IAI.73.3.1506-1514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mikolajczyk-Pawlinska J, Travis J, Potempa J. Modulation of interleukin-8 activity by gingipains from Porphyromonas gingivalis: implications for pathogenicity of periodontal disease. FEBS Lett. 1998;440:282–286. doi: 10.1016/s0014-5793(98)01461-6. [DOI] [PubMed] [Google Scholar]
- 123.Mikolajczyk J, Boatright KM, Stennicke HR, Nazif T, Potempa J, Bogyo M, Salvesen GS. Sequential autolytic processing activates the zymogen of Arg-gingipain. J Biol Chem. 2003;278:10458–10464. doi: 10.1074/jbc.M210564200. [DOI] [PubMed] [Google Scholar]
- 124.Milner P, Batten JE, Curtis MA. Development of a simple chemically defined medium for Porphyromonas gingivalis: requirement for alpha-ketoglutarate. FEMS Microbiol Lett. 1996;140:125–130. doi: 10.1016/0378-1097(96)00159-0. [DOI] [PubMed] [Google Scholar]
- 125.Miyachi K, Ishihara K, Kimizuka R, Okuda K. Arg-gingipain A DNA vaccine prevents alveolar bone loss in mice. J Dent Res. 2007;86:446–450. doi: 10.1177/154405910708600511. [DOI] [PubMed] [Google Scholar]
- 126.Monteiro AC, Scovino A, Raposo S, Gaze VM, Cruz C, Svensjo E, Narciso MS, Colombo AP, Pesquero JB, Feres-Filho E, Nguyen KA, Sroka A, Potempa J, Scharfstein J. Kinin danger signals proteolytically released by gingipain induce fimbriae-specific IFN-gamma- and IL-17-producing T cells in mice infected intramucosally with Porphyromonas gingivalis. J Immunol. 2009;183:3700–3711. doi: 10.4049/jimmunol.0900895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Morgan MJ, Kim YS, Liu ZG. TNFalpha and reactive oxygen species in necrotic cell death. Cell Res. 2008;18:343–349. doi: 10.1038/cr.2008.31. [DOI] [PubMed] [Google Scholar]
- 128.Mydel P, Takahashi Y, Yumoto H, Sztukowska M, Kubica M, Gibson FC, III, Kurtz DM, Jr, Travis J, Collins LV, Nguyen KA, Genco CA, Potempa J. Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection. PLOS Pathogens. 2006;2:e76. doi: 10.1371/journal.ppat.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Naito M, Sakai E, Shi Y, Ideguchi H, Shoji M, Ohara N, Yamamoto K, Nakayama K. Porphyromonas gingivalis-induced platelet aggregation in plasma depends on Hgp44 adhesin but not Rgp proteinase. Mol Microbiol. 2006;59:152–167. doi: 10.1111/j.1365-2958.2005.04942.x. [DOI] [PubMed] [Google Scholar]
- 130.Nakagawa I, Inaba H, Yamamura T, Kato T, Kawai S, Ooshima T, Amano A. Invasion of epithelial cells and proteolysis of cellular focal adhesion components by distinct types of Porphyromonas gingivalis fimbriae. Infect Immun. 2006;74:3773–3782. doi: 10.1128/IAI.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nakayama K. Molecular genetics of Porphyromonas gingivalis: gingipains and other virulence factors. Curr Protein Pept Sci. 2003;4:389–395. doi: 10.2174/1389203033486983. [DOI] [PubMed] [Google Scholar]
- 132.Nakayama K, Ratnayake DB, Tsukuba T, Kadowaki T, Yamamoto K, Fujimura S. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol Microbiol. 1998;27:51–61. doi: 10.1046/j.1365-2958.1998.00656.x. [DOI] [PubMed] [Google Scholar]
- 133.Nakayama K, Yoshimura F, Kadowaki T, Yamamoto K. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol. 1996;178:2818–2824. doi: 10.1128/jb.178.10.2818-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nakhjiri SF, Park Y, Yilmaz O, Chung WO, Watanabe K, El-Sabaeny A, Park K, Lamont RJ. Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol Lett. 2001;200:145–149. doi: 10.1111/j.1574-6968.2001.tb10706.x. [DOI] [PubMed] [Google Scholar]
- 135.Nelson D, Potempa J, Kordula T, Travis J. Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human alpha1-proteinase inhibitor. J Biol Chem. 1999;274:12245–12251. doi: 10.1074/jbc.274.18.12245. [DOI] [PubMed] [Google Scholar]
- 136.Nguyen KA, Travis J, Potempa J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-negative bacteria? J Bacteriol. 2007;189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nhien NT, Huy NT, Naito M, Oida T, Uyen DT, Huang M, Kikuchi M, Harada S, Nakayama K, Hirayama K, Kamei K. Neutralization of toxic haem by Porphyromonas gingivalis haemoglobin receptor. J Biochem. 2010;147:317–325. doi: 10.1093/jb/mvp164. [DOI] [PubMed] [Google Scholar]
- 138.Nisapakultorn K, Ross KF, Herzberg MC. Calprotectin expression in vitro by oral epithelial cells confers resistance to infection by Porphyromonas gingivalis. Infect Immun. 2001;69:4242–4247. doi: 10.1128/IAI.69.7.4242-4247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nylander M, Lindahl TL, Bengtsson T, Grenegard M. The periodontal pathogen Porphyromonas gingivalis sensitises human blood platelets to epinephrine. Platelets. 2008;19:352–358. doi: 10.1080/09537100802056102. [DOI] [PubMed] [Google Scholar]
- 140.O'Brien-Simpson NM, Paolini RA, Hoffmann B, Slakeski N, Dashper SG, Reynolds EC. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect Immun. 2001;69:7527–7534. doi: 10.1128/IAI.69.12.7527-7534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.O'Brien-Simpson NM, Paolini RA, Reynolds EC. RgpA-Kgp peptide-based immunogens provide protection against Porphyromonas gingivalis challenge in a murine lesion model. Infect Immun. 2000;68:4055–4063. doi: 10.1128/iai.68.7.4055-4063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.O'Brien-Simpson NM, Pathirana RD, Paolini RA, Chen YY, Veith PD, Tam V, Ally N, Pike RN, Reynolds EC. An immune response directed to proteinase and adhesin functional epitopes protects against Porphyromonas gingivalis-induced periodontal bone loss. J Immunol. 2005;175:3980–3989. doi: 10.4049/jimmunol.175.6.3980. [DOI] [PubMed] [Google Scholar]
- 143.O'Brien-Simpson NM, Pathirana RD, Walker GD, Reynolds EC. Porphyromonas gingivalis RgpA-Kgp proteinase-adhesin complexes penetrate gingival tissue and induce proinflammatory cytokines or apoptosis in a concentration-dependent manner. Infect Immun. 2009;77:1246–1261. doi: 10.1128/IAI.01038-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.O'Brien-Simpson NM, Veith PD, Dashper SG, Reynolds EC. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr Protein Pept Sci. 2003;4:409–426. doi: 10.2174/1389203033487009. [DOI] [PubMed] [Google Scholar]
- 145.Oda H, Saiki K, Numabe Y, Konishi K. Effect of gamma-immunoglobulin on the asaccharolytic growth of Porphyromonas gingivalis. J Periodontal Res. 2007;42:438–442. doi: 10.1111/j.1600-0765.2006.00966.x. [DOI] [PubMed] [Google Scholar]
- 146.Oda H, Saiki K, Tonosaki M, Yajima A, Konishi K. Participation of the secreted dipeptidyl and tripeptidyl aminopeptidases in asaccharolytic growth of Porphyromonas gingivalis. J Periodontal Res. 2009;44:362–367. doi: 10.1111/j.1600-0765.2008.01117.x. [DOI] [PubMed] [Google Scholar]
- 147.Odell EW, Wu PJ. Susceptibility of Porphyromonas gingivalis and P. asaccharolytica to the non-oxidative killing mechanisms of human neutrophils. Arch Oral Biol. 1992;37:597–601. doi: 10.1016/0003-9969(92)90121-n. [DOI] [PubMed] [Google Scholar]
- 148.Ohno T, Okahashi N, Morisaki I, Amano A. Signaling pathways in osteoblast proinflammatory responses to infection by Porphyromonas gingivalis. Oral Microbiol Immunol. 2008;23:96–104. doi: 10.1111/j.1399-302X.2007.00393.x. [DOI] [PubMed] [Google Scholar]
- 149.Okahashi N, Inaba H, Nakagawa I, Yamamura T, Kuboniwa M, Nakayama K, Hamada S, Amano A. Porphyromonas gingivalis induces receptor activator of NF-kappaB ligand expression in osteoblasts through the activator protein 1 pathway. Infect Immun. 2004;72:1706–1714. doi: 10.1128/IAI.72.3.1706-1714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Olczak T, Dixon DW, Genco CA. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J Bacteriol. 2001;183:5599–5608. doi: 10.1128/JB.183.19.5599-5608.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Olczak T, Simpson W, Liu X, Genco CA. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol Rev. 2005;29:119–144. doi: 10.1016/j.femsre.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 152.Olczak T, Sroka A, Potempa J, Olczak M. Porphyromonas gingivalis HmuY and HmuR: further characterization of a novel mechanism of heme utilization. Arch Microbiol. 2008;189:197–210. doi: 10.1007/s00203-007-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oleksy A, Banbula A, Bugno M, Travis J, Potempa J. Proteolysis of interleukin-6 receptor (IL-6R) by Porphyromonas gingivalis cysteine proteinases (gingipains) inhibits interleukin-6-mediated cell activation. Microb Pathog. 2002;32:173–181. doi: 10.1006/mpat.2002.0491. [DOI] [PubMed] [Google Scholar]
- 154.Oliver RC, Brown LJ, Loe H. Periodontal diseases in the United States population. J Periodontol. 1998;69:269–278. doi: 10.1902/jop.1998.69.2.269. [DOI] [PubMed] [Google Scholar]
- 155.Ouhara K, Komatsuzawa H, Yamada S, Shiba H, Fujiwara T, Ohara M, Sayama K, Hashimoto K, Kurihara H, Sugai M. Susceptibilities of periodontopathogenic and cariogenic bacteria to antibacterial peptides, {beta}-defensins and LL37, produced by human epithelial cells. J Antimicrob Chemother. 2005;55:888–896. doi: 10.1093/jac/dki103. [DOI] [PubMed] [Google Scholar]
- 156.Papapanou PN, Baelum V, Luan WM, Madianos PN, Chen X, Fejerskov O, Dahlen G. Subgingival microbiota in adult Chinese: prevalence and relation to periodontal disease progression. J Periodontol. 1997;68:651–666. doi: 10.1902/jop.1997.68.7.651. [DOI] [PubMed] [Google Scholar]
- 157.Paramaesvaran M, Nguyen KA, Caldon E, McDonald JA, Najdi S, Gonzaga G, Langley DB, DeCarlo A, Crossley MJ, Hunter N, Collyer CA. Porphyrin-mediated cell surface heme capture from hemoglobin by Porphyromonas gingivalis. J Bacteriol. 2003;185:2528–2537. doi: 10.1128/JB.185.8.2528-2537.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Park Y, Lamont RJ. Contact-dependent protein secretion in Porphyromonas gingivalis. Infect Immun. 1998;66:4777–4782. doi: 10.1128/iai.66.10.4777-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, Hackett M, Yoshimura F, Demuth DR, Lamont RJ. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 161.Pathirana RD, O'Brien-Simpson NM, Brammar GC, Slakeski N, Reynolds EC. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect Immun. 2007;75:1436–1442. doi: 10.1128/IAI.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pathirana RD, O'Brien-Simpson NM, Reynolds EC. Host immune responses to Porphyromonas gingivalis antigens. Periodontol 2000. 2010;52:218–237. doi: 10.1111/j.1600-0757.2009.00330.x. [DOI] [PubMed] [Google Scholar]
- 163.Pathirana RD, O'Brien-Simpson NM, Veith PD, Riley PF, Reynolds EC. Characterization of proteinase-adhesin complexes of Porphyromonas gingivalis. Microbiology. 2006;152:2381–2394. doi: 10.1099/mic.0.28787-0. [DOI] [PubMed] [Google Scholar]
- 164.Pathirana RD, O'Brien-Simpson NM, Visvanathan K, Hamilton JA, Reynolds EC. Flow cytometric analysis of the adherence of Porphyromonas gingivalis to oral epithelial cells. Infect Immun. 2007 doi: 10.1128/IAI.02004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pathirana RD, O'Brien-Simpson NM, Visvanathan K, Hamilton JA, Reynolds EC. The role of the RgpA-Kgp proteinase-adhesin complexes in the adherence of Porphyromonas gingivalis to fibroblasts. Microbiology. 2008;154:2904–2911. doi: 10.1099/mic.0.2008/019943-0. [DOI] [PubMed] [Google Scholar]
- 166.Pham K, Feik D, Hammond BF, Rams TE, Whitaker EJ. Aggregation of human platelets by gingipain-R from Porphyromonas gingivalis cells and membrane vesicles. Platelets. 2002;13:21–30. doi: 10.1080/09537100120104863. [DOI] [PubMed] [Google Scholar]
- 167.Pike R, McGraw W, Potempa J, Travis J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J Biol Chem. 1994;269:406–411. [PubMed] [Google Scholar]
- 168.Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. 2007;178:7242–7250. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- 169.Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol 2000. 2000;24:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. [DOI] [PubMed] [Google Scholar]
- 170.Potempa J, Mikolajczyk-Pawlinska J, Brassell D, Nelson D, Thogersen IB, Enghild JJ, Travis J. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J Biol Chem. 1998;273:21648–21657. doi: 10.1074/jbc.273.34.21648. [DOI] [PubMed] [Google Scholar]
- 171.Potempa J, Pike R, Travis J. Titration and mapping of the active site of cysteine proteinases from Porphyromonas gingivalis (gingipains) using peptidyl chloromethanes. Biol Chem. 1997;378:223–230. doi: 10.1515/bchm.1997.378.3-4.223. [DOI] [PubMed] [Google Scholar]
- 172.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr Protein Pept Sci. 2003;4:397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 173.Potempa M, Potempa J, Kantyka T, Nguyen KA, Wawrzonek K, Manandhar SP, Popadiak K, Riesbeck K, Eick S, Blom AM. Interpain A, a cysteine proteinase from Prevotella intermedia, inhibits complement by degrading complement factor C3. PLoS Pathog. 2009;5:e1000316. doi: 10.1371/journal.ppat.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Potempa M, Potempa J, Okroj M, Popadiak K, Eick S, Nguyen KA, Riesbeck K, Blom AM. Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J Immunol. 2008;181:5537–5544. doi: 10.4049/jimmunol.181.8.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Puklo M, Guentsch A, Hiemstra PS, Eick S, Potempa J. Analysis of neutrophil-derived antimicrobial peptides in gingival crevicular fluid suggests importance of cathelicidin LL-37 in the innate immune response against periodontogenic bacteria. Oral Microbiol Immunol. 2008;23:328–335. doi: 10.1111/j.1399-302X.2008.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Rajapakse PS, O'Brien-Simpson NM, Slakeski N, Hoffmann B, Reynolds EC. Immunization with the RgpA-Kgp proteinase-adhesin complexes of Porphyromonas gingivalis protects against periodontal bone loss in the rat periodontitis model. Infect Immun. 2002;70:2480–2486. doi: 10.1128/IAI.70.5.2480-2486.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ramachandran R, Hollenberg MD. Proteinases and signalling: pathophysiological and therapeutic implications via PARs and more. Br J Pharmacol. 2008;153 1:S263–282. doi: 10.1038/sj.bjp.0707507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ramseier CA, Kinney JS, Herr AE, Braun T, Sugai JV, Shelburne CA, Rayburn LA, Tran HM, Singh AK, Giannobile WV. Identification of pathogen and host-response markers correlated with periodontal disease. J Periodontol. 2009;80:436–446. doi: 10.1902/jop.2009.080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rautemaa R, Jarvensivu A, Kari K, Wahlgren J, DeCarlo A, Richardson M, Sorsa T. Intracellular localization of Porphyromonas gingivalis thiol proteinase in periodontal tissues of chronic periodontitis patients. Oral Dis. 2004;10:298–305. doi: 10.1111/j.1601-0825.2004.01021.x. [DOI] [PubMed] [Google Scholar]
- 180.Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 182.Rudney JD, Chen R, Sedgewick GJ. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect Immun. 2001;69:2700–2707. doi: 10.1128/IAI.69.4.2700-2707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rudney JD, Chen R, Sedgewick GJ. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J Dent Res. 2005;84:59–63. doi: 10.1177/154405910508400110. [DOI] [PubMed] [Google Scholar]
- 184.Sakai E, Naito M, Sato K, Hotokezaka H, Kadowaki T, Kamaguchi A, Yamamoto K, Okamoto K, Nakayama K. Construction of recombinant hemagglutinin derived from the gingipain-encoding gene of Porphyromonas gingivalis, identification of its target protein on erythrocytes, and inhibition of hemagglutination by an interdomain regional peptide. J Bacteriol. 2007;189:3977–3986. doi: 10.1128/JB.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sato Y, Kishi J, Suzuki K, Nakamura H, Hayakawa T. Sonic extracts from a bacterium related to periapical disease activate gelatinase A and inactivate tissue inhibitor of metalloproteinases TIMP-1 and TIMP-2. Int Endod J. 2009;42:1104–1111. doi: 10.1111/j.1365-2591.2009.01640.x. [DOI] [PubMed] [Google Scholar]
- 187.Schaffner A, Rhyn P, Schoedon G, Schaer DJ. Regulated expression of platelet factor 4 in human monocytes--role of PARs as a quantitatively important monocyte activation pathway. J Leukoc Biol. 2005;78:202–209. doi: 10.1189/jlb.0105024. [DOI] [PubMed] [Google Scholar]
- 188.Scragg MA, Alsam A, Rangarajan M, Slaney JM, Shepherd P, Williams DM, Curtis MA. Nuclear targeting of Porphyromonas gingivalis W50 protease in epithelial cells. Infect Immun. 2002;70:5740–5750. doi: 10.1128/IAI.70.10.5740-5750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Seers CA, Slakeski N, Veith PD, Nikolof T, Chen YY, Dashper SG, Reynolds EC. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J Bacteriol. 2006;188:6376–6386. doi: 10.1128/JB.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sharp L, Poole S, Reddi K, Fletcher J, Nair S, Wilson M, Curtis M, Henderson B, Tabona P. A lipid A-associated protein of Porphyromonas gingivalis, derived from the haemagglutinating domain of the RI protease gene family, is a potent stimulator of interleukin 6 synthesis. Microbiology. 1998;144:3019–3026. doi: 10.1099/00221287-144-11-3019. [DOI] [PubMed] [Google Scholar]
- 191.Sheets SM, Potempa J, Travis J, Casiano CA, Fletcher HM. Gingipains from Porphyromonas gingivalis W83 induce cell adhesion molecule cleavage and apoptosis in endothelial cells. Infect Immun. 2005;73:1543–1552. doi: 10.1128/IAI.73.3.1543-1552.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Sheets SM, Potempa J, Travis J, Fletcher HM, Casiano CA. Gingipains from Porphyromonas gingivalis W83 synergistically disrupt endothelial cell adhesion and can induce caspase-independent apoptosis. Infect Immun. 2006;74:5667–5678. doi: 10.1128/IAI.01140-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sheets SM, Robles-Price AG, McKenzie RM, Casiano CA, Fletcher HM. Gingipain-dependent interactions with the host are important for survival of Porphyromonas gingivalis. Front Biosci. 2008;13:3215–3238. doi: 10.2741/2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shelburne CE, Coulter WA, Olguin D, Lantz MS, Lopatin DE. Induction of {beta}-defensin resistance in the oral anaerobe Porphyromonas gingivalis. Antimicrob Agents Chemother. 2005;49:183–187. doi: 10.1128/AAC.49.1.183-187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Shelburne CE, Gleason RM, Coulter WA, Lantz MS, Lopatin DE. Differential display analysis of Porphyromonas gingivalis gene activation response to heat and oxidative stress. Oral Microbiol Immunol. 2005;20:233–238. doi: 10.1111/j.1399-302X.2005.00219.x. [DOI] [PubMed] [Google Scholar]
- 196.Shi Y, Ratnayake DB, Okamoto K, Abe N, Yamamoto K, Nakayama K. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J Biol Chem. 1999;274:17955–17960. doi: 10.1074/jbc.274.25.17955. [DOI] [PubMed] [Google Scholar]
- 197.Shoji M, Naito M, Yukitake H, Sato K, Sakai E, Ohara N, Nakayama K. The major structural components of two cell surface filaments of Porphyromonas gingivalis are matured through lipoprotein precursors. Mol Microbiol. 2004;52:1513–1525. doi: 10.1111/j.1365-2958.2004.04105.x. [DOI] [PubMed] [Google Scholar]
- 198.Silva-Boghossian CM, Luiz RR, Colombo AP. Periodontal status, sociodemographic, and behavioral indicators in subjects attending a public dental school in Brazil: analysis of clinical attachment loss. J Periodontol. 2009;80:1945–1954. doi: 10.1902/jop.2009.090242. [DOI] [PubMed] [Google Scholar]
- 199.Simpson W, Olczak T, Genco CA. Lysine-specific gingipain K and heme/hemoglobin receptor HmuR are involved in heme utilization in Porphyromonas gingivalis. Acta Biochim Pol. 2004;51:253–262. [PubMed] [Google Scholar]
- 200.Slakeski N, Bhogal PS, O'Brien-Simpson NM, Reynolds EC. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin-binding motif involved in association of the prtR and prtK proteinases and adhesins into large complexes. Microbiology. 1998;144:1583–1592. doi: 10.1099/00221287-144-6-1583. [DOI] [PubMed] [Google Scholar]
- 201.Slaney JM, Curtis MA. Mechanisms of evasion of complement by Porphyromonas gingivalis. Front Biosci. 2008;13:188–196. doi: 10.2741/2669. [DOI] [PubMed] [Google Scholar]
- 202.Smalley JW, Birss AJ, Silver J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the mu-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol Lett. 2000;183:159–164. doi: 10.1111/j.1574-6968.2000.tb08951.x. [DOI] [PubMed] [Google Scholar]
- 203.Smalley JW, Birss AJ, Szmigielski B, Potempa J. The HA2 haemagglutinin domain of the lysine-specific gingipain (Kgp) of Porphyromonas gingivalis promotes micro-oxo bishaem formation from monomeric iron(III) protoporphyrin IX. Microbiology. 2006;152:1839–1845. doi: 10.1099/mic.0.28835-0. [DOI] [PubMed] [Google Scholar]
- 204.Smalley JW, Birss AJ, Szmigielski B, Potempa J. Sequential action of R- and K-specific gingipains of Porphyromonas gingivalis in the generation of the haem-containing pigment from oxyhaemoglobin. Arch Biochem Biophys. 2007;465:44–49. doi: 10.1016/j.abb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 205.Smalley JW, Birss AJ, Szmigielski B, Potempa J. Mechanism of methaemoglobin breakdown by the lysine-specific gingipain of the periodontal pathogen Porphyromonas gingivalis. Biol Chem. 2008;389:1235–1238. doi: 10.1515/BC.2008.XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Smalley JW, Thomas MF, Birss AJ, Withnall R, Silver J. A combination of both arginine- and lysine-specific gingipain activity of Porphyromonas gingivalis is necessary for the generation of the micro-oxo bishaem-containing pigment from haemoglobin. Biochem J. 2004;379:833–840. doi: 10.1042/BJ20031221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Smolik I, Robinson D, El-Gabalawy HS. Periodontitis and rheumatoid arthritis: epidemiologic, clinical, and immunologic associations. Compend Contin Educ Dent. 2009;30:188–190. 192, 194. passim; quiz 198, 210. [PubMed] [Google Scholar]
- 208.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 209.Soder B, Airila Mansson S, Soder PO, Kari K, Meurman J. Levels of matrix metalloproteinases-8 and -9 with simultaneous presence of periodontal pathogens in gingival crevicular fluid as well as matrix metalloproteinase-9 and cholesterol in blood. J Periodontal Res. 2006;41:411–417. doi: 10.1111/j.1600-0765.2006.00888.x. [DOI] [PubMed] [Google Scholar]
- 210.Sroka A, Sztukowska M, Potempa J, Travis J, Genco CA. Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J Bacteriol. 2001;183:5609–5616. doi: 10.1128/JB.183.19.5609-5616.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Stathopoulou PG, Galicia JC, Benakanakere MR, Garcia CA, Potempa J, Kinane DF. Porphyromonas gingivalis induce apoptosis in human gingival epithelial cells through a gingipain-dependent mechanism. BMC Microbiol. 2009;9:107. doi: 10.1186/1471-2180-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sugawara S, Nemoto E, Tada H, Miyake K, Imamura T, Takada H. Proteolysis of human monocyte CD14 by cysteine proteinases (gingipains) from Porphyromonas gingivalis leading to lipopolysaccharide hyporesponsiveness. J Immunol. 2000;165:411–418. doi: 10.4049/jimmunol.165.1.411. [DOI] [PubMed] [Google Scholar]
- 213.Tada H, Sugawara S, Nemoto E, Imamura T, Potempa J, Travis J, Shimauchi H, Takada H. Proteolysis of ICAM-1 on human oral epithelial cells by gingipains. J Dent Res. 2003;82:796–801. doi: 10.1177/154405910308201007. [DOI] [PubMed] [Google Scholar]
- 214.Tada H, Sugawara S, Nemoto E, Takahashi N, Imamura T, Potempa J, Travis J, Shimauchi H, Takada H. Proteolysis of CD14 on human gingival fibroblasts by arginine-specific cysteine proteinases from Porphyromonas gingivalis leading to down-regulation of lipopolysaccharide-induced interleukin-8 production. Infect Immun. 2002;70:3304–3307. doi: 10.1128/IAI.70.6.3304-3307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Takii R, Kadowaki T, Baba A, Tsukuba T, Yamamoto K. A functional virulence complex composed of gingipains, adhesins, and lipopolysaccharide shows high affinity to host cells and matrix proteins and escapes recognition by host immune systems. Infect Immun. 2005;73:883–893. doi: 10.1128/IAI.73.2.883-893.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tam V, O'Brien-Simpson NM, Chen YY, Sanderson CJ, Kinnear B, Reynolds EC. The RgpA-Kgp proteinase-adhesin complexes of Porphyromonas gingivalis inactivate the Th2 cytokines interleukin-4 and interleukin-5. Infect Immun. 2009;77:1451–1458. doi: 10.1128/IAI.01377-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tancharoen S, Sarker KP, Imamura T, Biswas KK, Matsushita K, Tatsuyama S, Travis J, Potempa J, Torii M, Maruyama I. Neuropeptide release from dental pulp cells by RgpB via proteinase-activated receptor-2 signaling. J Immunol. 2005;174:5796–5804. doi: 10.4049/jimmunol.174.9.5796. [DOI] [PubMed] [Google Scholar]
- 218.Turkoglu O, Emingil G, Kutukculer N, Atilla G. Gingival crevicular fluid levels of cathelicidin LL-37 and interleukin-18 in patients with chronic periodontitis. J Periodontol. 2009;80:969–976. doi: 10.1902/jop.2009.080532. [DOI] [PubMed] [Google Scholar]
- 219.Uehara A, Imamura T, Potempa J, Travis J, Takada H. Gingipains from Porphyromonas gingivalis synergistically induce the production of proinflammatory cytokines through protease-activated receptors with Toll-like receptor and NOD1/2 ligands in human monocytic cells. Cell Microbiol. 2008;10:1181–1189. doi: 10.1111/j.1462-5822.2008.01119.x. [DOI] [PubMed] [Google Scholar]
- 220.Uehara A, Muramoto K, Imamura T, Nakayama K, Potempa J, Travis J, Sugawara S, Takada H. Arginine-specific gingipains from Porphyromonas gingivalis stimulate production of hepatocyte growth factor (scatter factor) through protease-activated receptors in human gingival fibroblasts in culture. J Immunol. 2005;175:6076–6084. doi: 10.4049/jimmunol.175.9.6076. [DOI] [PubMed] [Google Scholar]
- 221.Uehara A, Naito M, Imamura T, Potempa J, Travis J, Nakayama K, Takada H. Dual regulation of interleukin-8 production in human oral epithelial cells upon stimulation with gingipains from Porphyromonas gingivalis. J Med Microbiol. 2008;57:500–507. doi: 10.1099/jmm.0.47679-0. [DOI] [PubMed] [Google Scholar]
- 222.Urnowey S, Ansai T, Bitko V, Nakayama K, Takehara T, Barik S. Temporal activation of anti- and pro-apoptotic factors in human gingival fibroblasts infected with the periodontal pathogen, Porphyromonas gingivalis: potential role of bacterial proteases in host signalling. BMC Microbiol. 2006;6:26. doi: 10.1186/1471-2180-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 223.Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, Triantafilou M, Triantafilou K, Lambris JD, Hajishengallis G. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010;3:ra11. doi: 10.1126/scisignal.2000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wegner N, Wait R, Sroka A, Eick S, Nguyen K, Lundberg K, Kinloch A, Culshaw S, Potempa J, Venables P. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: Implications for autoimmunity in rheumatoid arthritis. Arth Rheum. 2010 doi: 10.1002/art.27552. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wojtowicz H, Guevara T, Tallant C, Olczak M, Sroka A, Potempa J, Sola M, Olczak T, Gomis-Ruth FX. Unique structure and stability of HmuY, a novel heme-binding protein of Porphyromonas gingivalis. PLoS Pathog. 2009;5:e1000419. doi: 10.1371/journal.ppat.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wong DM, Tam V, Lam R, Walsh KA, Tatarczuch L, Pagel CN, Reynolds EC, O'Brien-Simpson NM, Mackie EJ, Pike RN. Protease-activated receptor 2 has pivotal roles in cellular mechanisms involved in experimental periodontitis. Infect Immun. 2010;78:629–638. doi: 10.1128/IAI.01019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Yamada M, Ikegami A, Kuramitsu HK. Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol Lett. 2005;250:271–277. doi: 10.1016/j.femsle.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 228.Yamatake K, Maeda M, Kadowaki T, Takii R, Tsukuba T, Ueno T, Kominami E, Yokota S, Yamamoto K. The role for gingipains in Porphyromonas gingivalis traffic to phagolysosomes and survival in human aortic endothelial cells. Infect Immun. 2007 doi: 10.1128/IAI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yasuhara R, Miyamoto Y, Takami M, Imamura T, Potempa J, Yoshimura K, Kamijo R. Lysine-specific gingipain promotes lipopolysaccharide- and active-vitamin D3-induced osteoclast differentiation by degrading osteoprotegerin. Biochem J. 2009;419:159–166. doi: 10.1042/BJ20081469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yokoyama K, Sugano N, Shimada T, Shofiqur RA, Ibrahim el SM, Isoda R, Umeda K, Sa NV, Kodama Y, Ito K. Effects of egg yolk antibody against Porphyromonas gingivalis gingipains in periodontitis patients. J Oral Sci. 2007;49:201–206. doi: 10.2334/josnusd.49.201. [DOI] [PubMed] [Google Scholar]
- 231.Yoneda M, Hirofuji T, Anan H, Matsumoto A, Hamachi T, Nakayama K, Maeda K. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: involvement of gingipains in a synergistic effect. J Periodontal Res. 2001;36:237–243. doi: 10.1034/j.1600-0765.2001.036004237.x. [DOI] [PubMed] [Google Scholar]
- 232.Yoneda M, Hirofuji T, Motooka N, Anan H, Hamachi T, Miura M, Ishihara Y, Maeda K. Antibody responses to Porphyromonas gingivalis infection in a murine abscess model--involvement of gingipains and responses to re-infection. J Periodontal Res. 2003;38:551–556. doi: 10.1034/j.1600-0765.2003.00685.x. [DOI] [PubMed] [Google Scholar]
- 233.Yoneda M, Yoshikane T, Motooka N, Yamada K, Hisama K, Naito T, Okada I, Yoshinaga M, Hidaka K, Imaizumi K, Maeda K, Hirofuji T. Stimulation of growth of Porphyromonas gingivalis by cell extracts from Tannerella forsythia. J Periodontal Res. 2005;40:105–109. doi: 10.1111/j.1600-0765.2005.00774.x. [DOI] [PubMed] [Google Scholar]
- 234.Yonezawa H, Ishihara K, Okuda K. Arg-gingipain A DNA vaccine induces protective immunity against infection by Porphyromonas gingivalis in a murine model. Infect Immun. 2001;69:2858–2864. doi: 10.1128/IAI.69.5.2858-2864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Yonezawa H, Kato T, Kuramitsu HK, Okuda K, Ishihara K. Immunization by Arg-gingipain A DNA vaccine protects mice against an invasive Porphyromonas gingivalis infection through regulation of interferon-gamma production. Oral Microbiol Immunol. 2005;20:259–266. doi: 10.1111/j.1399-302X.2005.00220.x. [DOI] [PubMed] [Google Scholar]
- 236.Yun LW, Decarlo AA, Collyer C, Hunter N. Enhancement of Th2 pathways and direct activation of B cells by the gingipains of Porphyromonas gingivalis. Clin Exp Immunol. 2003;134:295–302. doi: 10.1046/j.1365-2249.2003.02287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yun LW, Decarlo AA, Hunter N. Blockade of protease-activated receptors on T cells correlates with altered proteolysis of CD27 by gingipains of Porphyromonas gingivalis. Clin Exp Immunol. 2007;150:217–229. doi: 10.1111/j.1365-2249.2007.03488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yun PL, Decarlo AA, Chapple CC, Collyer CA, Hunter N. Binding of Porphyromonas gingivalis gingipains to human CD4(+) T cells preferentially down-regulates surface CD2 and CD4 with little affect on co-stimulatory molecule expression. Microb Pathog. 2005;38:85–96. doi: 10.1016/j.micpath.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 239.Yun PL, Decarlo AA, Chapple CC, Hunter N. Functional implication of the hydrolysis of platelet endothelial cell adhesion molecule 1 (CD31) by gingipains of Porphyromonas gingivalis for the pathology of periodontal disease. Infect Immun. 2005;73:1386–1398. doi: 10.1128/IAI.73.3.1386-1398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yun PL, Decarlo AA, Collyer C, Hunter N. Hydrolysis of interleukin-12 by Porphyromonas gingivalis major cysteine proteinases may affect local gamma interferon accumulation and the Th1 or Th2 T-cell phenotype in periodontitis. Infect Immun. 2001;69:5650–5660. doi: 10.1128/IAI.69.9.5650-5660.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yun PL, DeCarlo AA, Collyer C, Hunter N. Modulation of an interleukin-12 and gamma interferon synergistic feedback regulatory cycle of T-cell and monocyte cocultures by Porphyromonas gingivalis lipopolysaccharide in the absence or presence of cysteine proteinases. Infect Immun. 2002;70:5695–5705. doi: 10.1128/IAI.70.10.5695-5705.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Yun PL, DeCarlo AA, Hunter N. Modulation of major histocompatibility complex protein expression by human gamma interferon mediated by cysteine proteinase-adhesin polyproteins of Porphyromonas gingivalis. Infect Immun. 1999;67:2986–2995. doi: 10.1128/iai.67.6.2986-2995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Yun PL, Decarlo AA, Hunter N. Gingipains of Porphyromonas gingivalis modulate leukocyte adhesion molecule expression induced in human endothelial cells by ligation of CD99. Infect Immun. 2006;74:1661–1672. doi: 10.1128/IAI.74.3.1661-1672.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhou J, Windsor LJ. Porphyromonas gingivalis affects host collagen degradation by affecting expression, activation, and inhibition of matrix metalloproteinases. J Periodontal Res. 2006;41:47–54. doi: 10.1111/j.1600-0765.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- 245.Zipfel PF, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009;9:729–740. doi: 10.1038/nri2620. [DOI] [PubMed] [Google Scholar]