Abstract
Aldehyde dehydrogenase (ALDH) enzymes are critical in the detoxification of endogenous and exogenous aldehydes. Our previous findings indicate that the ALDH3B1 enzyme is expressed in several mouse tissues and is catalytically active toward aldehydes derived from lipid peroxidation, suggesting a potential role against oxidative stress. The aim of this study was to elucidate by immunohistochemistry the tissue, cellular, and subcellular distribution of ALDH3B1 in normal human tissues and in tumors of human lung, colon, breast, and ovary. Our results indicate that ALDH3B1 is expressed in a tissue-specific manner and in a limited number of cell types, including hepatocytes, proximal convoluted tubule cells, cerebellar astrocytes, bronchiole ciliated cells, testis efferent ductule ciliated cells, and histiocytes. ALDH3B1 expression was upregulated in a high percentage of human tumors (lung > breast = ovarian > colon). Increased ALDH3B1 expression in tumor cells may confer a growth advantage or be the result of an induction mechanism mediated by increased oxidative stress. Subcellular localization of ALDH3B1 was predominantly cytosolic in tissues, with the exception of normal human lung and testis, in which localization appeared membrane-bound or membrane-associated. The specificity of ALDH3B1 distribution may prove to be directly related to the functional role of this enzyme in human tissues. (J Histochem Cytochem 58:765–783, 2010)
Keywords: aldehyde dehydrogenase, ALDH3B1, immunohistochemistry, cancer, human, liver, lung, colon, breast, ovary
Aldehyde dehydrogenases (ALDHs) comprise a superfamily of 19 human enzymes within 11 distinct families that catalyze the oxidation of aldehydes to their respective carboxylic acids (Marchitti et al. 2008). Aldehydes are generated from a variety of precursors, including alcohols, lipids, neurotransmitters, drugs, and environmental agents, and are present in high levels in pollutants such as cigarette smoke and smog. Aldehydes are highly electrophilic and relatively long-lived. Consequently, they can react with biomolecules, such as proteins and nucleic acids, leading them to possess cytotoxic, mutagenic, and even carcinogenic activity (Lindahl 1992). Aldehydes have been implicated in many pathological states, including neurodegenerative diseases (Yoritaka et al. 1996), alcoholic liver disease (Koch et al. 2004), and cancer (Yokoyama et al. 2001).
Oxidative stress is characterized by an imbalance between the production and the removal of reactive oxygen species (ROS). It is a process that can occur in virtually all organs and is believed to contribute to the etiology and progression of a number of human pathologies, including liver disease, cancer, neurodegeneration, and male infertility (Brooks and Theruvathu 2005; Marchitti et al. 2007a; Seitz and Becker 2007; Shiraishi and Naito 2007; Cederbaum et al. 2009). Lipid peroxidation (LPO), the oxidative degradation of cellular membrane lipids, results from oxidative stress and leads to the generation of more than 200 reactive aldehyde species (Esterbauer et al. 1991). ALDH enzymes play a critical role in maintaining cellular homeostasis through the metabolism of reactive aldehydes, which can themselves initiate oxidative stress. The importance of ALDHs in health and disease is underscored by the observation that mutations in ALDH genes are the molecular basis of many disease states, such as Sjögren–Larsson syndrome, type II hyperprolinemia, γ-hydroxybutyric aciduria, and pyridoxine-dependent epilepsy (Vasiliou et al. 2004; Mills et al. 2006). In addition, ALDH gene mutations contribute to other pathophysiological conditions, including cancer (Lindahl 1992; Yokoyama et al. 2001). Aside from aldehyde detoxification, ALDHs mayserve to mitigate cellular oxidative stress by (a) scavenging hydroxyl radicals via the thiol groups of their Cys and Met residues (Estey et al. 2007), (b) generating NAD(P)H, which is critical for the regeneration of GSH, and (c) acting as direct antioxidants by reducing glutathiyl (GS•) and tyrosyl radicals (Kirsch and De Groot 2001). Differential expression of ALDH isozymes occurs in tissues and is related to functional significance. Indeed, ALDH enzymes believed to play a role in ethanol metabolism (ALDH2 and ALDH1A1) are highly expressed in liver (Maeda et al. 1988; Sladek 2003a), whereas others, including ALDH3A1, have negligible expression (Vasiliou et al. 1992).
ALDH proteins appear to have diverse roles in cancer, although they are not completely understood (Marchitti et al. 2008). ALDH expression is upregulated in a variety of human cancer tissues and cell lines (Kim et al. 2007; Patel et al. 2008), which can cause resistance to anticancer drugs (Sreerama and Sladek 1993; Wang et al. 2001). ALDH1A1 is a tumor stem cell–associated marker in lung cancer (Jiang et al. 2009) and colon cancer (Huang et al. 2009), and it has been identified as a flavopiridol-binding protein, which correlates with flavopiridol resistance (Schnier et al. 1999). ALDH3A1 activity may be a functional marker in lung cancer (Ucar et al. 2009). In breast cancer stem cells, ALDH activity is a factor in drug resistance, cell differentiation, and oxidative stress response (Moreb 2008). In ovarian cancer, ALDH1A1 expression correlates with a complete response to cisplatin-based chemotherapy and favorable patient prognosis (Chang et al. 2009). Aside from aldehyde metabolism, ALDH isozymes have been proposed to have multiple additional roles in tumors. ALDH3A1 not only detoxifies LPO-derived aldehydes in tumor cells (Canuto et al. 1999) but also appears to be involved in maintaining tumor cell growth, motility, and gene expression (Moreb et al. 2008).
The gene for human ALDH3B1, a member of the ALDH3 family (ALDH3A1, ALDH3A2, ALDH3B1, and ALDH3B2), is located on chromosome 11q13.2 and encodes a protein with subunits of ∼52 kDa. Human ALDH3B1 is 83% identical to the human ALDH3B2 protein and 53% and 55% identical to human ALDH3A1 and ALDH3A2 proteins, respectively. Relatively little is known about the properties and physiological significance of ALDH3B1. We have previously shown that ALDH3B1 is a metabolically active enzyme with distinct specificity for various aldehyde substrates, particularly medium- and long-chain aliphatic aldehydes. These substrates include many products that are formed during LPO, such as hexanal, 4-hydroxy-2-nonenal (4-HNE), octanal, and trans-2-nonenal (Marchitti et al. 2007b). In addition, stable expression of ALDH3B1 protects cultured cells from cytotoxicity induced by LPO-derived aldehydes. As such, we postulate that ALDH3B1, similar to other members of the ALDH3 family (Pappa et al. 2003), plays an important physiological role against cellular oxidative stress by detoxifying aldehydes derived from oxidative processes, such as ethanol metabolism and LPO (Marchitti et al. 2007b).
Using Western blot analyses, we previously have shown that an ALDH3B1 85% similar to the human enzyme is expressed in various mouse tissues, including kidney, liver, lung, and regions of the brain (Marchitti et al. 2007b). However, IHC permits cellular (and subcellular) protein expression to be identified; therefore, it can be used to further elucidate the potential physiological roles and significance of proteins in human tissues. Using IHC, we describe for the first time the tissue, cellular, and subcellular distribution of ALDH3B1 in a number of normal (or healthy) human tissues and in cancerous tissues of the human colon, lung, breast, and ovary.
Materials and Methods
Rabbit Anti-human ALDH3B1 Antibody
We previously described the production and characteristics of a rabbit polyclonal antibody against human ALDH3B1 that is specific to human ALDH3B1 and does not crossreact with other ALDH isoforms (Marchitti et al. 2007b). Briefly, a conserved amino acid sequence strictly unique to human ALDH3B1 (MDPLGDTLRRLREAFHAG, aa 1–18, N-terminal) was used to produce a synthetic peptide (Alpha Diagnostics International; San Antonio, TX). Following conjugation to keyhole limpet hemocyanin, a two-rabbit antibody production was initiated. Sera were collected and characterized for reactivity, and those that were optimal in specificity were affinity purified. Immunoblots demonstrating the efficacy of this antibody have been published (Marchitti et al. 2007b).
Western Blot Analyses
Western blot analyses of human spleen and testis were performed using prerun Western blotting strips (Dip-N-Blots; Protein Biotechnologies, Ramona, CA). Briefly, flash-frozen (within 5–10 min of collection) human tissue specimens (Integrated Laboratory Services – Biotech; Chestertown, MD) were homogenized in modified radioimmunoprecipitation assay buffer [62.5 mM Tris (pH 6.8), 25 mM TCEP, 12.5% glycerol, 1% SDS, 0.005% bromophenol blue], centrifuged, and subjected to Western blot analyses (20 μg of total protein loaded onto 4–20% precast 1D-PAGE Tris–HCl gradient gel) using anti-human ALDH3B1 antibody (1:200 dilution). Antibody binding was detected using peroxidase-conjugated goat anti-rabbit IgG (1:5000 dilution; Calbiochem, San Diego, CA), and protein bands were visualized using chemiluminescence (NEN Life Science Products; Boston, MA) and hyperfilm (GE Healthcare; Piscataway, NJ).
Human Tissues
Archival, formalin-fixed, paraffin-embedded, normal human lung, testis, liver, kidney, ovary, and cerebellum sections were procured by IHCtech (Aurora, CO) from the Control Tissue Bank through membership in the National Society for Histotechnology and Immunohistochemistry Resource Group in accordance with relevant approvals and guidelines for IHC staining.
Normal human tissues, including placenta, pancreas, salivary gland, bone marrow, spleen, thyroid, endometrium, and colon, and abnormal human tumor tissues of the ovary, lung, colon, and breast were purchased as tissue microarray slides (T-MTA-6A) from the Cooperative Human Tissue Network and the Tissue Array Research Program of the National Cancer Institute (NCI), National Institutes of Health (NIH), Bethesda, MD. Tissue microarrays provided by the NCI are produced in a standardized and extremely well-controlled manner in order that comparative studies of tumor tissue samples can be performed and that the samples retain proper antigenicity. Their diagnostic application and usefulness in the discovery and validation of tumor markers have been well documented (Hewitt 2006,2009). Patient diagnosis, gender, age, and cancer characteristics were provided for most but not all human tumor tissue samples and were used in our evaluation of factors that may be involved in ALDH3B1 expression.
Antibody Titration and Optimization
Antibody titration and optimization of IHC procedures followed standard methods and were performed on formalin–fixed, paraffin-embedded tissue sections as described (Coskran et al. 2006). Briefly, antibody titration involved first testing a series of antibody dilutions (1:50, 1:100, 1:500, and 1:1000) and incubation times. This was followed by the evaluation of a series of antigen retrieval pretreatments, including heat-induced epitope retrieval (HIER) using citrate buffer (pH 6), HIER using EDTA buffer (pH 9), and enzyme-induced epitope retrieval using proteinase K digestion. Antigen retrieval for formalin-fixed, paraffin-embedded tissue sections is a standard IHC practice and serves to unmask antigens or proteins crosslinked by formalin fixation. Optimal results were achieved with HIER pretreatment using citrate buffer (pH 6) and 1:50 dilution of anti-human ALDH3B1 antibody with 1-hr incubation at room temperature. During optimization procedures, multiple negative controls of identical tissue sections were prepared and tested using identical staining protocols with the exception that the primary antibody was either replaced with antibody diluent buffer or preadsorbed with the antigenic blocking peptide.
Immunohistochemistry
IHC was performed using standard avidin–biotin immunoperoxidase methods (Coskran et al. 2006) and an automated immunostainer (Dako; Carpenteria, CA). Tissues imbedded in paraffin were cut into 5-μm sections and mounted onto slides. After deparaffinization in xylene and rehydration in graded ethanol, slides were pretreated for antigen retrieval using HIER and citrate buffer (pH 6) in HpH Reveal (BioCare Medical; Concord, CA) for 25 min. Sections were then incubated with Dual Endogenous Enzyme Block (Dako) for 10 min (to block endogenous peroxidase activity) and then with Protein Blocker (Open Biosystems; Huntsville, AL) for 5 min (to block nonspecific protein binding). Human ALDH3B1 was detected by incubating the slides with rabbit anti-human ALDH3B1 antibody at a dilution of 1:50 in Protein Blocker for 60 min at room temperature. After washing, the slides were treated with MACH 2 Rabbit HRP Polymer secondary antibody (BioCare Medical) for 30 min and then with Stable DAB (Open Biosystems) for 10 min. Slides were counterstained with Auto Hematoxylin (Open Biosystems) for 1 min, followed by dehydration and sealing with Permount Mounting Media (Sigma; St Louis, MO). Negative control slides were processed at the same time as experimental slides, with the exception that the primary anti-human ALDH3B1 antibody was preadsorbed with the conjugated ALDH3B1 antigenic blocking peptide (Alpha Diagnostics International) before application to the tissue slides or replaced with antibody diluent buffer. Negative control slides were included to ensure that any observed positive staining was specific to the primary anti-human ALDH3B1 antibody and not due to endogenous peroxidase or nonspecific binding of the detection reagents or antibody to the tissue.
Two hundred individual tumor samples of colon, breast, lung, and ovary (50 samples each) were evaluated for IHC staining intensity and extensiveness of tumor cell immunoreactivity, as previously described and verified by others (Luo et al. 2002; Sladek et al. 2002; Patel et al. 2008). Briefly, antibody binding was identified microscopically as brown cytoplasmic staining, and intensity was rated on a scale of 0–3, with 0 indicating no staining and 3 indicating strong staining. The extensiveness was rated as the approximate percentage (0–100%) of visible tumor cells immunoreactive for ALDH3B1. Samples were scored by two independent blinded observers (SAM and DJO) and compared for bias and consistency. Those samples lacking enough tissue for evaluation were excluded. Cellular localization, tumor differentiation, tumor invasiveness, and any notable characteristics of the cancer samples were also identified. To compare and analyze the relative ALDH3B1 immunoreactivity of different types of tumors, an overall immunoreactivity score (mean ± SEM) was assigned to each case by multiplying the intensity score by the extensiveness score.
Results
Detection of Human ALDH3B1 in Normal Human Tissues by Western Blot Analysis
Western blot analyses of human testis and spleen using the anti-human ALDH3B1 antibody demonstrate a protein band at ∼52 kDa, which is consistent with the expected molecular mass of human ALDH3B1 (Figure 1). No cross-reactivity or extraneous band staining was seen, verifying the specificity of this antibody for use in human tissues for the recognition of human ALDH3B1. Similar results have been found in fixed ALDH3B1-transfected cultured human embryonic kidney (HEK293) cells, verifying the validity of this antibody for the recognition of formalin-fixed human ALDH3B1 (Marchitti SA, et al., unpublished data).
Figure 1.
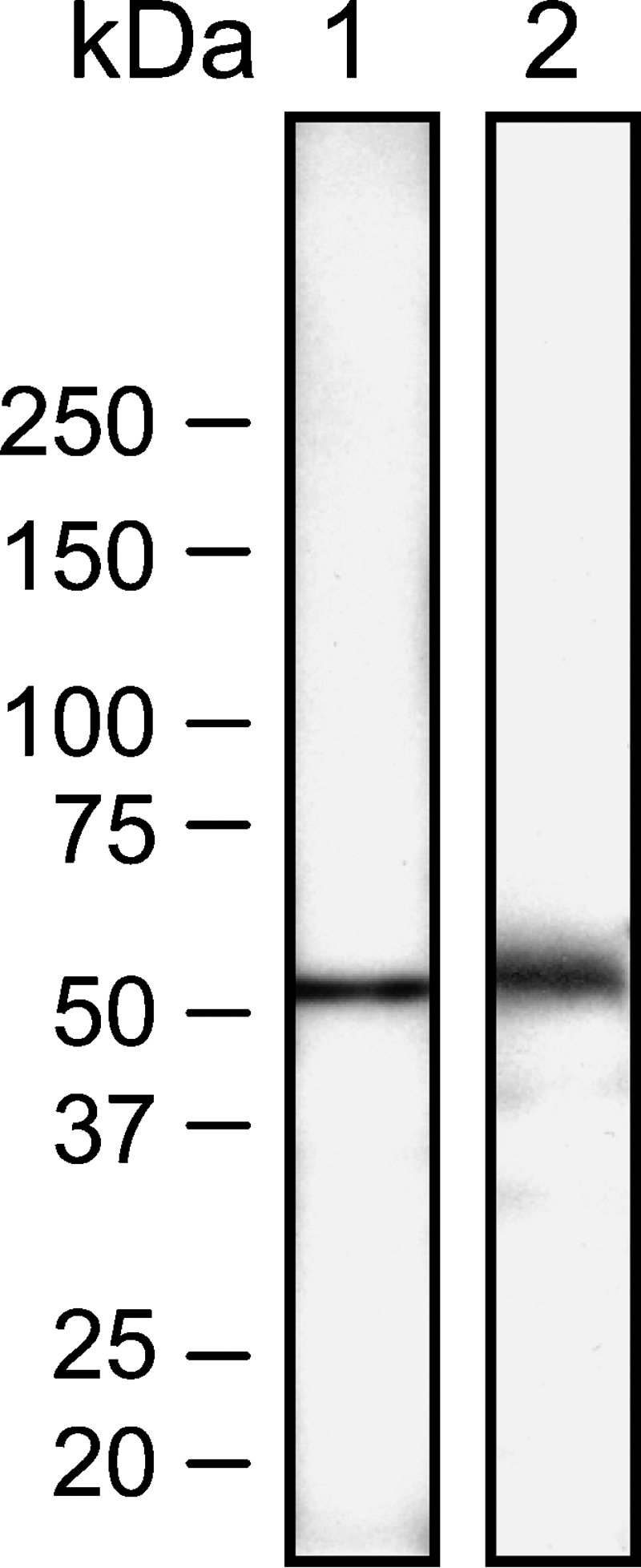
Expression of aldehyde dehydrogenase 3B1 (ADH3B1) in human tissues. Western blot analysis of human testis (Lane 1) and spleen (Lane 2) using anti-human ALDH3B1 antibody. Molecular mass markers are shown to the left. A protein band at ∼52 kDa was found in both human testis and spleen, which is consistent with the expected molecular mass of human ALDH3B1. No cross-reactivity or extraneous band staining from the anti-human ALDH3B1 antibody was found.
Detection of Human ALDH3B1 in Normal Human Tissues by IHC
ALDH3B1 immunoreactivity was localized to specific cell types in a wide variety of normal human tissues, including lung, testis, liver, and kidney (Table 1). Immunoreactivity was predominantly cytoplasmic, with the exception of lung and testis. No immunoreactivity was seen in negative controls from any tissue examined.
Table 1.
Summary of ALDH3B1 immunoreactivity in normal human tissues
| Human tissuea | ALDH3B1 immunoreactivity | Cell type | Localization |
|---|---|---|---|
| Bone marrow | + | Histiocytes | Cytoplasm |
| Cerebellum | + | Astrocytes, molecular layer of gray matter | ND |
| Colon | − | NA | NA |
| Endometrium | + | Few histiocytes | Cytoplasm |
| Kidney cortex | + | Proximal convoluted tubule cells | Cytoplasm |
| Kidney medulla | + | Loop of Henle cells | Cytoplasm |
| + | Collecting duct cells | Membrane-associatedb | |
| Liver | + | Hepatocytes | Cytoplasm |
| Lung | + | Ciliated brionchiole epithelial cells | Membrane-bound/associatedc |
| Ovary | − | NA | NA |
| Placenta | + | Intermediate trophoblasts | Cytoplasm |
| Pancreas | + | Acinar cells | Cytoplasm |
| Salivary gland | + | Striated duct cells | Membrane-associatedb |
| Spleen | + | Sinusoidal endothelial cells of red pulp | Cytoplasm |
| Testis | + | Ciliated columnar efferent ductule cells | Membrane-bound/associatedc |
| Thyroid | + | Few histiocytes | cytoplasm |
Human tissues immunoreactive (+) and non-immunoreactive (−) for ALDH3B1 are listed.
In kidney medulla and salivary gland, staining of duct cells appeared cytoplasmic but was stronger on the luminal surface of ducts, indicating a potential membrane association.
In lung and testis, staining was limited to cilia on the apical surface of cells; it was unclear whether localization was membrane-bound or cytoplasmic but membrane-associated.
NA, not applicable; ND, not determined.
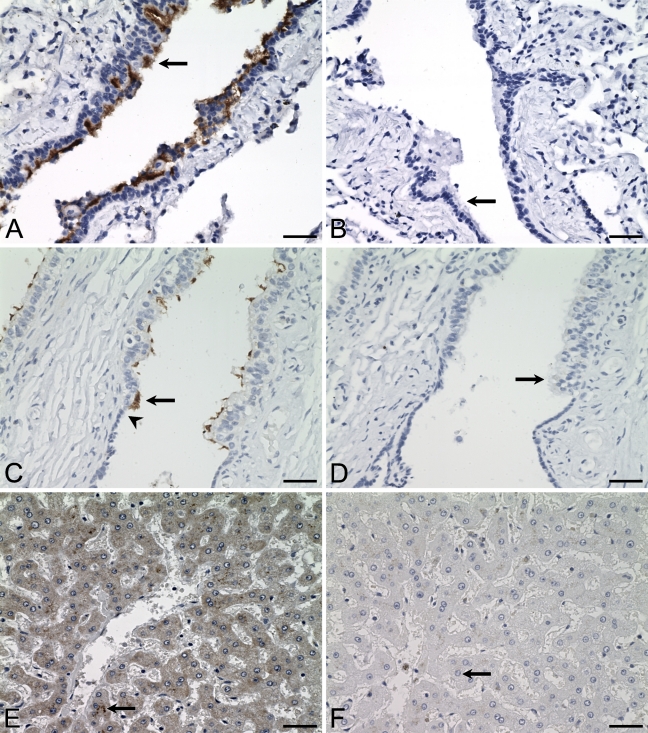
Human Lung
ALDH3B1 immunoreactivity was seen in the small airways (terminal bronchioles) and was specific to the apical surface of ciliated epithelial cells of bronchioles (Figure 2A, arrow). No other immunoreactivity was seen, including in the surrounding alveolar parenchyma, connective tissue, respiratory bronchioles, or vessels. Large airways, including the bronchi, larynx, and trachea, were not evaluated.
Figure 2.
Distribution of ALDH3B1 in human lung, testis, and liver. IHC was performed using antibody against human ALDH3B1 on human lung (A), testis (C), and liver (E). In lung, ciliated cells of small bronchioles are immunoreactive (A, arrow). In testis, ciliated cells of efferent ductules are immunoreactive (C, arrow). Arrowhead in C indicates the transition from the low cuboidal cells of the rete to the tall columnar cells of the efferent ductule. In liver, zone 3 hepatocytes surrounding the central vein are immunoreactive (E, arrow). (B,D,F) Corresponding cells from negative controls in which IHC of human lung, testis, and liver was performed using antibody against human ALDH3B1 preadsorbed with peptide show no immunoreactivity (arrows). Bar = 50 μm.
Human Testis
ALDH3B1 immunoreactivity was limited to the ciliated columnar epithelial cells of the efferent ductules (Figure 2C, arrow). Of particular interest is the transition in the testis from the simple low cuboidal cells of the rete, which show no immunoreactivity, to the tall columnar ciliated cells of the efferent ductules, where immunoreactivity begins (Figure 2C, arrowhead). In addition, although the efferent ductules are composed of a mixture of true ciliated columnar cells and shorter cuboidal epithelial cells, only the ciliated columnar cells were ALDH3B1-immunoreactive. Similar to the lung, immunoreactivity was specific to the apical surface of these cells and predominantly localized to the cilia themselves. No other testicular cell type displayed immunoreactivity, including cells of the seminiferous tubules and epididymis. However, in the connective tissue of one sample, several histiocytes were immunoreactive (data not shown).
Human Liver
ALDH3B1 immunoreactivity in normal human liver was specific to the cytoplasm of hepatocytes and strongest in those of zone 3 surrounding the central vein (Figure 2E). Moderate to weak immunoreactivity was seen in hepatocytes of zones 2 and 1. No other liver cell type was immunoreactive.
Human Kidney
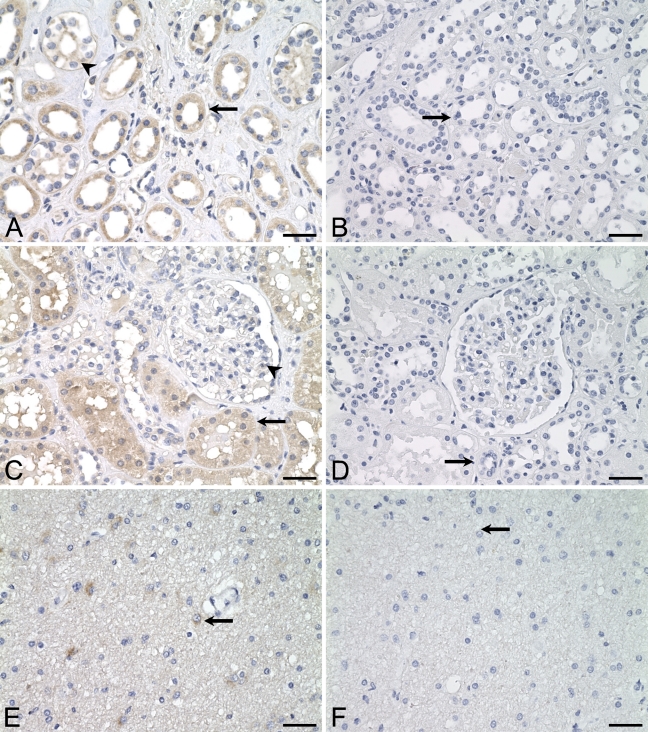
ALDH3B1 immunoreactivity in human kidney was seen in both medulla and cortex. In the medulla, loop of Henle cells (Figure 3A, arrow) and collecting duct cells (Figure 3A, arrowhead) were immunoreactive. Immunoreactivity was uniformly cytoplasmic in loop of Henle cells, whereas in the collecting ducts, it appeared more membrane-associated and was localized more to the microvilli along the luminal edge of the cells. In kidney cortex, cytoplasmic immunoreactivity was observed in cells of the proximal convoluted tubules, including all S1, S2, and S3 sections (Figure 3C, arrow). No other immunoreactivity was seen, including in distal convoluted tubules, collecting ducts, renal corpuscle (Figure 3C, arrowhead), and Bowman's capsule.
Figure 3.
Distribution of ALDH3B1 in human kidney cortex, kidney medulla, and cerebellum. IHC was performed using antibody against human ALDH3B1 on human kidney medulla (A), kidney cortex (C), and cerebellum (E). In kidney medulla, loop of Henle cells (A, arrow) and cells of the collecting ducts (A, arrowhead) are immunoreactive. In kidney cortex, proximal convoluted tubule cells are immunoreactive (C, arrow), whereas endothelial cells of the renal corpuscle lacked immunoreactivity (C, arrowhead). In cerebellum, astrocytes in the molecular layer of the gray matter are immunoreactive (E, arrow). (B,D,F) Corresponding cells from negative controls in which IHC of human kidney cortex, kidney medulla, and cerebellum was performed using antibody against human ALDH3B1 preadsorbed with peptide show no immunoreactivity (arrows). Bar = 50 μm.
Human Cerebellum
Human ALDH3B1 immunoreactivity in normal human cerebellum was limited to astrocytes of the molecular layer of the gray matter (Figure 3E, arrow). The subcellular localization of astrocyte immunoreactivity was difficult to ascertain but appeared cytoplasmic. Cells of the white matter and granular layer did not show ALDH3B1 immunoreactivity (data not shown).
Human Placenta
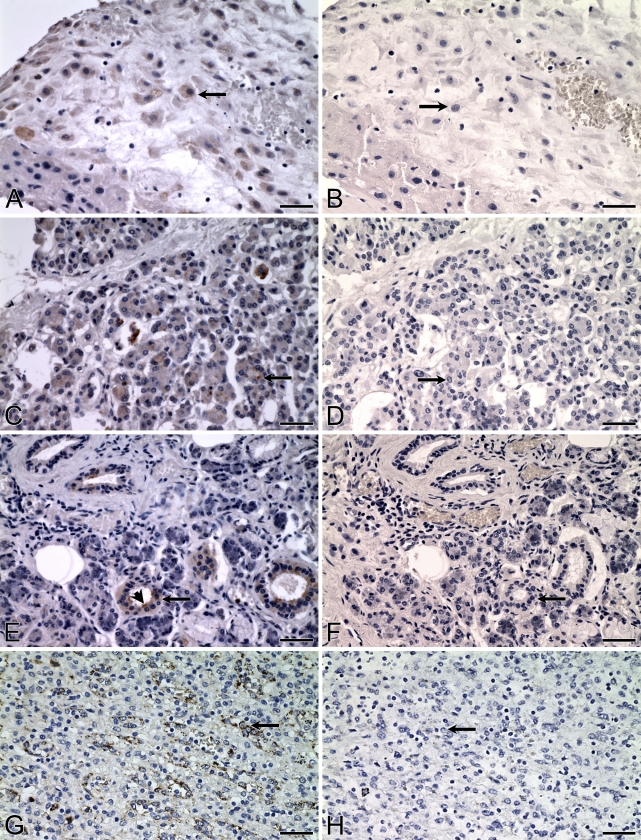
In placenta, cytoplasmic ALDH3B1 immunoreactivity was seen in intermediate trophoblasts (Figure 4A, arrow). No other placental cell type was immunoreactive.
Figure 4.
Distribution of ALDH3B1 in human placenta, pancreas, salivary gland, and spleen. IHC was performed using antibody against human ALDH3B1 on human placenta (A), pancreas (C), salivary gland (E), and spleen (G). In placenta, intermediate trophoblast cells are immunoreactive (A, arrow). In pancreas, acinar cells are immunoreactive (C, arrow). In salivary gland, epithelial cells of the salivary ducts are immunoreactive (E, arrow) with greater reactivity on the apical or luminal surface of the cells (E, arrowhead). In spleen, splenic endothelial cells lining the sinusoids of the red pulp are ALDH3B1-immunoreactive (G, arrow). (B,D,F,H) Corresponding cells from negative controls in which IHC of human placenta, pancreas, salivary gland, and spleen was performed using antibody against human ALDH3B1 preadsorbed with peptide show no immunoreactivity (arrows). Bar = 50 μm.
Human Pancreas
ALDH3B1 immunoreactivity was limited to pancreatic acinar epithelial cells (Figure 4C, arrow). Immunoreactivity was uniformly cytoplasmic; however, some acinar cells displayed intense granules of immunoreactivity, and there were others that did not appear immunoreactive at all.
Human Salivary Gland
ALDH3B1 immunoreactivity in normal human salivary gland was limited to cells of the striated salivary ducts (Figure 4E, arrow). Immunoreactivity appeared to be stronger on the luminal surface of some ducts (Figure 4E, arrowhead). Salivary gland acinar cells and connective tissue cells (including adipocytes, arteries, and veins) lacked immunoreactivity.
Human Spleen
In normal human spleen, the cytoplasm of endothelial cells lining the sinusoids of the red pulp displayed strong ALDH3B1-immunoreactivity (Figure 4G, arrow). Splenic white pulp, cells of the periarterial lymphatic sheath, and splenic cord cells of the red pulp were not immunoreactive.
Human Colon
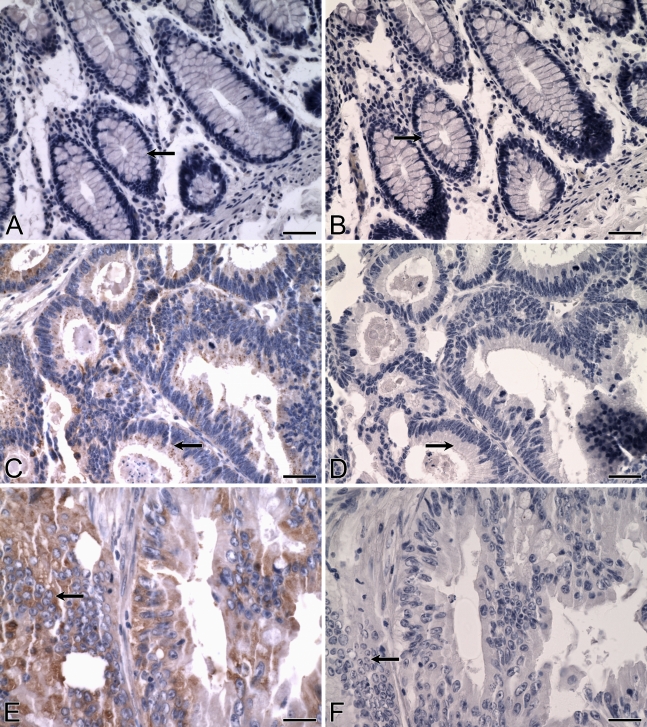
No immunoreactivity was detected in any cell type or structure of normal human colon, including in the glandular epithelium (Figure 5A, arrow), connective tissue, adipose tissue, blood vessels, and lymphatics.
Figure 5.
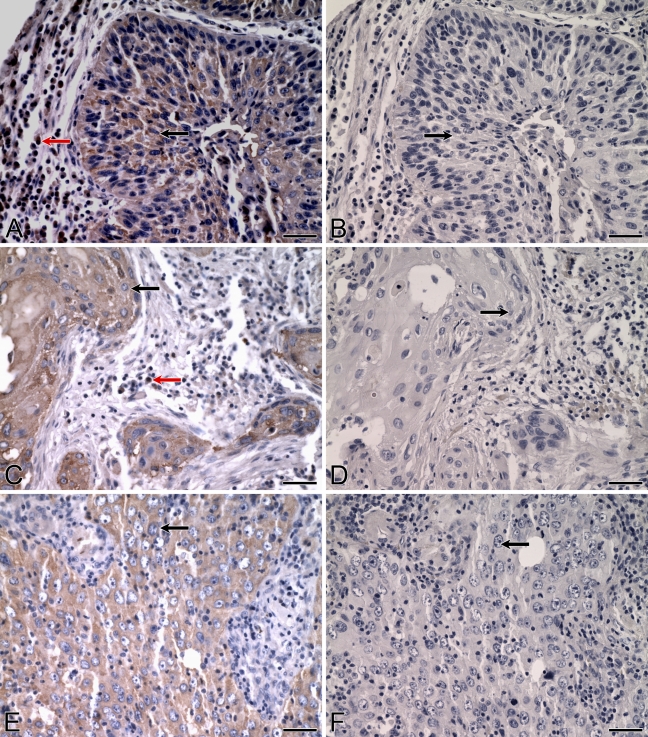
Expression of ALDH3B1 in normal and cancerous colon. IHC was performed using antibody against human ALDH3B1 on normal human colon (A) and colonic adenocarcinoma (C,E). (A) In normal human colon, glandular epithelial cells (arrow) and cells of the connective tissue, adipose tissue, arteries, and lymphatics lack immunoreactivity. In colonic adenocarcinoma, glandular epithelial cancer cells are ALDH3B1-immunoreactive, including well-differentiated (C, arrow) and moderately differentiated (E, arrow), invasive tumors. (B,D,F) Corresponding cells from negative controls in which IHC of normal and cancerous human colon was performed using antibody against human ALDH3B1 preadsorbed with peptide show no immunoreactivity (arrows). Bar = 50 μm.
Human Thyroid
Thyroid endocrine cells were not immunoreactive (data not shown). However, some histiocytes located in the connective tissue displayed immunoreactivity.
Human Endometrium
Endometrium epithelial glands were not ALDH3B1-immunoreactive (data not shown). However, similar to several other normal human tissues, some histiocytes located in the connective tissue were immunoreactive.
Human Ovary
No ALDH3B1 immunoreactivity was detected in human ovary (data not shown).
Human Bone Marrow
Cytoplasmic immunoreactivity was detected in histiocytes of normal human bone marrow (data not shown). No other cell type was immunoreactive.
Detection of Human ALDH3B1 in Abnormal and Cancerous Tissues
ALDH3B1 expression was examined in tumors of the human colon, lung, breast, and ovary. In cancerous epithelial cells, immunoreactivity was limited to the cytoplasm and appeared granular in nature. A small number of colon, lung, and breast cancer samples displayed immunoreactive histiocytes in the connective tissue. No immunoreactivity was seen in negative controls from any tumor sample examined.
Human Colon Cancer
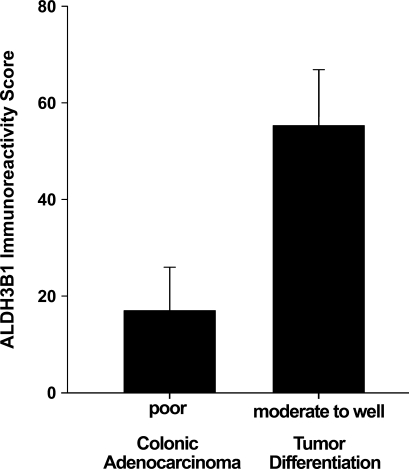
An overall ALDH3B1 immunoreactivity score of 47 ± 9 was determined for the colonic adenocarcinomas evaluated (Table 2). Of the 44 colon tumors, 20 (45%) were ALDH3B1-immunoreactive. These included poorly, moderately (Figure 5E), and well-differentiated tumors (Figure 5C) and those that were both invasive and non-invasive. ALDH3B1 expression was higher in moderately to well-differentiated colon tumors when compared to poorly differentiated colon tumors (Figure 6).
Table 2.
Patient and tumor characteristics
| Tumor type | Colon | Lung | Breast | Ovarian |
|---|---|---|---|---|
| Patient, n | 44 | 45 | 44 | 43 |
| Age, median (range) | 71 (38–90) | 66 (38–84) | 57 (36–76) | 59 (23–78) |
| Gender F/M, n | 19/17 (8 ND) | 17/17 (11 ND) | 44/0 | 43/0 |
| Tumor characteristics | ||||
| Invasive/non-invasive, n | 28/8 (8 ND) | 35/2 (8 ND) | 38/3 (3 ND) | 26/9 (8 ND) |
| Poorly/moderately/well-differentiated, n | 10/14/2020 | 13/29/3 | 25/16/3 | 15/21/7 |
| ALDH3B1-immunoreactive, n | 20 | 37 | 35 | 30 |
| Overall immunoreactivity score | 47 ± 9 | 131 ± 9 | 97 ± 11 | 89 ± 13 |
ND, not disclosed or not determined.
Figure 6.
ALDH3B1 expression in poorly differentiated vs moderately to well-differentiated colonic adenocarcinoma tumors. Moderately to well-differentiated colon tumors displayed higher ALDH3B1 expression than poorly differentiated colon tumors. Bars reflect mean overall immunoreactivity score ± SEM as evaluated by IHC.
Human Lung Hyperplasia and Cancer
In hyperplastic respiratory epithelium, human ALDH3B1 immunoreactivity was cytoplasmic with a granular appearance (Figure 7A, black arrow). Histiocytes located in the connective tissue were also immunoreactive (Figure 7A, red arrow).
Figure 7.
Expression of ALDH3B1 in human lung hyperplasia and lung cancer. IHC was performed using antibody against human ALDH3B1 on hyperplastic lung (A), squamous cell carcinoma of the lung (C), and adenocarcinoma of the lung (E). In lung hyperplasia, hyperplastic epithelial cells are immunoreactive (A, black arrow). In invasive squamous cell carcinoma of the lung, moderately differentiated squamous epithelial cancer cells are immunoreactive (C, black arrow). Histiocytes are also ALDH3B1-immunoreactive in lung hyperplasia (A, red arrow) and squamous cell carcinoma of the lung (C, red arrow). In invasive adenocarcinoma of the lung, poorly differentiated mucin-producing glandular epithelial cancer cells are immunoreactive (E, arrow). (B,D,F) Corresponding cells from negative controls in which IHC of human lung was performed using antibody against human ALDH3B1 preadsorbed with peptide show no immunoreactivity (arrows). Bar = 50 μm.
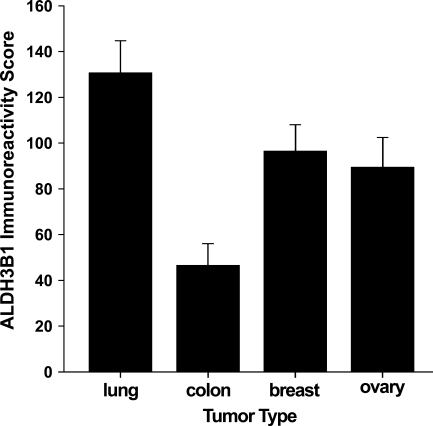
Of the 45 lung tumors evaluated for ALDH3B1 expression, 37 (82%) were ALDH3B1-immunoreactive, and an overall lung tumor immunoreactivity score of 131 ± 14 was determined (Table 2). ALDH3B1-immunoreactive tumors included 86% of non-small cell carcinomas (score 148 ± 39, n=7), 84% of squamous cell carcinomas (score 144 ± 23, n=19; Figure 7C), 76% of lung adenocarcinomas (score 112 ± 21, n=17; Figure 7E), and 100% of bronchoalveolar carcinomas (score 100 ± 50, n=2). No difference in ALDH3B1 immunoreactivity was seen among the different lung cancer diagnoses. However, ALDH3B1 expression in lung tumors was higher than that in colon tumors (Figure 8). Higher ALDH3B1 expression in lung tumors was also seen in comparison with breast and ovarian tumors.
Figure 8.
Summary of ALDH3B1 expression in tumors of the human lung, colon, breast, and ovary. Bars reflect the mean overall immunoreactivity score ± SEM for each tumor type as evaluated by IHC. Lung, ovarian, and breast tumors demonstrated higher ALDH3B1 expression than colon tumors.
Human Breast Cancer
Forty-four human breast cancer tumors were evaluated for ALDH3B1 expression; an overall immunoreactivity score of 97 ± 11 was determined (Table 2). Thirty-six (82%) of the forty-four tumors were ALDH3B1-immunoreactive. These included 81% of ductal adenocarcinomas (score 99 ± 13, n=36), 80% of lobular adenocarcinomas (score 102 ± 29, n=5), 50% of adenocarcinomas (score 75 ± 75, n=2), and 100% of ductal carcinomas in situ (score 15, n=1). No difference in immunoreactivity was observed between the different breast tumor diagnoses. However, ALDH3B1 expression in breast tumors was higher than that in the colon tumors evaluated in this study (Figure 8). Of the breast tumors, ALDH3B1 immunoreactivity was seen in a variety of tumors, including those that were poorly differentiated and highly invasive (Figure 9A), well-differentiated and mucin-producing (Figure 9C), non-mucin-producing (Figure 9G), and non-invasive (Figure 9E). However, the majority of immunoreactive breast tumors were invasive (86%) and poorly differentiated (57%) (Figure 9G).
Figure 9.
Expression of ALDH3B1 in human breast cancer. IHC was performed using antibody against human ALDH3B1 on ductal adenocarcinoma of the breast (A,C,E,G). Cancerous glandular epithelial cells of the human breast are strongly immunoreactive, including poorly differentiated, highly invasive mucin-producing pleimorphic epithelial tumor cells (A, arrow), poorly differentiated, highly invasive non-mucin-producing epithelial tumor cells (G, arrow), well-differentiated, invasive, mucin-producing epithelial tumor cells (C, arrow), and well-differentiated, non-invasive, mucin-producing epithelial tumor cells (E, arrow). (B,D,F,H) Corresponding cells from negative controls in which IHC of human breast was performed using antibody against human ALDH3B1 preadsorbed with peptide show no immunoreactivity (arrows). Bar = 50 μm.
Human Ovarian Cancer
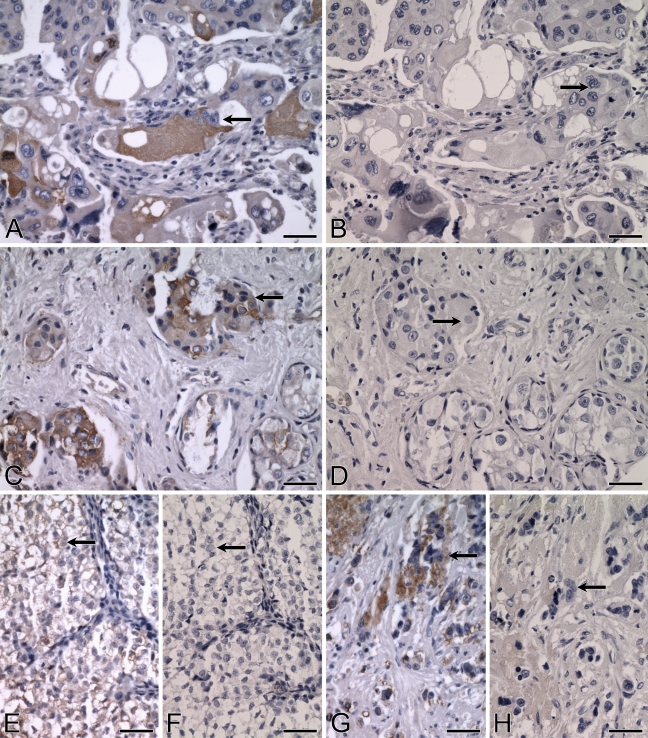
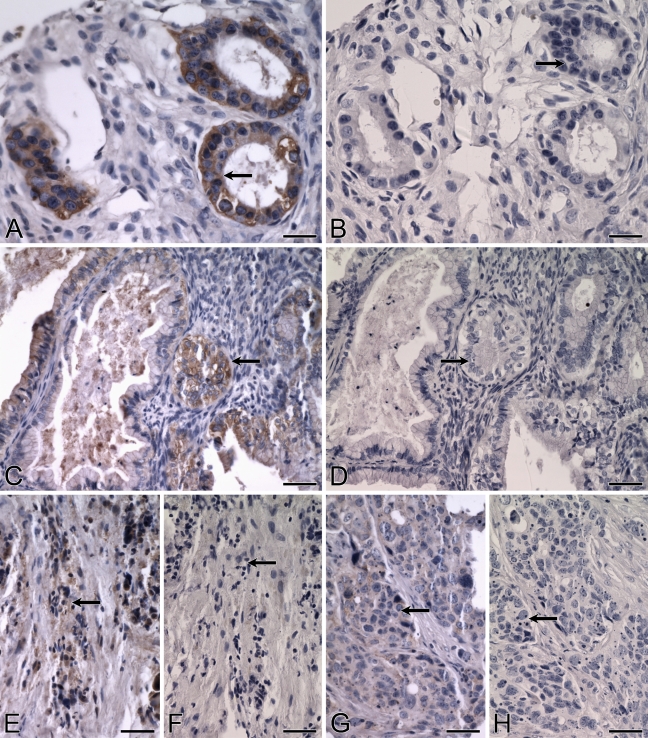
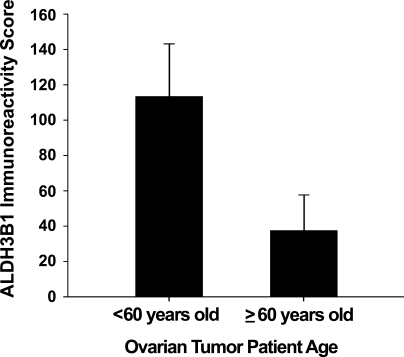
An overall ALDH3B1 immunoreactivity score of 89 ± 13 was determined for the ovarian tumors evaluated (Table 2). Of the 43 ovarian tumors, 30 (70%) were ALDH3B1-immunoreactive. Immunoreactivity was seen in nearly all diagnoses, including in 67% of serous papillary adenocarcinomas (score 79 ± 14, n=27; Figures 10E and 10G), 86% of clear cell adenocarcinomas (score 106 ± 34, n=7), 75% of endometroid adenocarcinomas (score 108 ± 66, n=4; Figure 10A), and 100% of mucinous adenocarcinomas (score 183 ± 44, n=3; Figure 10C). The majority of ALDH3B1-immunoreactive ovarian tumors were invasive (63%; Figures 10E and 10G) and moderately differentiated (53%; Figure 10A) or poorly differentiated (34%; Figures 10E and 10G). No difference in ALDH3B1 expression was observed between the different ovarian cancer diagnoses. However, ovarian tumor patients less than 60 years of age had higher ALDH3B1 tumor expression when compared with patients 60 years of age or older (Figure 11).
Figure 10.
Expression of ALDH3B1 in human ovarian cancer. (A–H) IHC was performed using antibody against human ALDH3B1 on ovarian cancer. Cancerous glandular epithelial cells of the human ovary are strongly immunoreactive, including moderately differentiated endometroid adenocarcinoma cells (A, arrow), well-differentiated, non-invasive, mucinous adenocarcinoma cells (C, arrow), and poorly differentiated, invasive, serous papillary adenocarcinoma cells (E, arrow; G, arrow). (B,D,F,H) Corresponding cells from negative controls in which IHC of human ovary was performed using antibody against human ALDH3B1 preadsorbed with peptide show no immunoreactivity (arrows). Bar = 50 μm.
Figure 11.
ALDH3B1 expression in ovarian tumors from patients less than 60 years of age vs tumors from patients 60 years of age or older. Tumors from ovarian cancer patients less than 60 years of age demonstrated higher ALDH3B1 tumor expression than tumors from patients 60 years of age or older. Bars reflect mean immunoreactivity score ± SEM as evaluated by IHC.
Discussion
ALDH-mediated detoxification of aldehydes represents a critical cellular defense. Increasing evidence suggests that the ALDH3 family plays important aldehyde-detoxifying roles in a variety of cell types. Genetic mutations leading to the catalytic inactivity of ALDH3A2, which detoxifies fatty aldehydes in human skin fibroblasts, result in Sjögren–Larsson syndrome, a neurocutaneous disorder (Rizzo and Carney 2005). In corneal epithelial and stromal cells, ALDH3A1 protects ocular structures against oxidative damage and cataract formation through the metabolism of LPO-derived aldehydes (Pappa et al. 2003). Similarly, our previous findings indicate that ALDH3B1 may be involved in the protection of cells against oxidative stress through the detoxification of LPO-derived aldehydes (Marchitti et al. 2007b). Delineation of tissue and cellular distribution of ALDH3B1 in healthy and diseased human tissues is essential for advancing our understanding of the physiological role and pathophysiological significance of ALDH3B1 in humans.
The cell-specific expression of ALDH3B1 detailed in this study is supportive of a physiological role of ALDH3B1 against aldehydes and oxidative stress. In the lung and testis, ALDH3B1 localized to the apical surface of ciliated cells, suggesting that the enzyme may play a role in the maintenance of ciliary function and serve as a first line of defense against the penetration of aldehydes further into the cell. Acetaldehyde and aldehyde–protein adducts have been implicated to play a role in the impairment of lung ciliary motion (Sisson et al. 1991; Elliott et al. 2007). Additionally, the lung is exposed to reactive, carcinogenic aldehydes, such as acetaldehyde, present in cigarette smoke and other pollutants (Patel et al. 2008). In the testis, oxidative stress and the production of 4-HNE impair spermatogenesis (Shiraishi and Naito 2007), and increased aldehydes, protein adducts, and DNA damage occur from environmental pollutants, including cigarette smoke, pesticides, and gasoline exhaust (Che et al. 2009; Vaithinathan et al. 2009). In the liver, zone 3 hepatocytes have high levels of cytochrome P450 but low levels of GSH, which leads to increased oxidative stress and LPO, particularly during ethanol metabolism (Cederbaum et al. 2009). Oxidative stress has been implicated in renal diseases, including diabetic nephropathy, which is associated with increased production of ROS and cell death (Han et al. 2005; Brezniceanu et al. 2007). Kidney proximal convoluted tubule and loop of Henle cells are susceptible to direct damage by toxins, including those that initiate oxidative stress and LPO (Hughes et al. 1998; de Castro et al. 2004; Schwerdt et al. 2007). In the placenta, fetal antioxidant defense mechanisms are critical in the prevention of placental toxicity, trophoblast damage, and early pregnancy loss (Guilbert et al. 1993; Lunghi et al. 2007). In the pancreas, acinar cells are the main site of ethanol metabolism, and alcohol abuse is a major cause of acinar cell death, believed to occur as a result of acetaldehyde and production of LPO-derived aldehydes (Gukovskaya et al. 2002; Apte et al. 2007; Palmieri et al. 2007). Cigarette smoke also causes increased oxidative stress and aldehyde production in acinar cells (Chowdhury and Walker 2008), which express multiple ALDH isozymes, including ALDH1A1, ALDH2, and ALDH3A1 (Chiang et al. 2009). In salivary gland striated duct cells, increased DNA adducts and 4-HNE accumulation have been reported with certain disease states (Kurimoto et al. 2007). The spleen is also sensitive to aldehyde toxicity (Warholm et al. 1984), and ALDH induction occurs following exposure to toxins, including dioxin (Germolec et al. 1996) and methylcholanthrene (Vasiliou and Marselos 1989).
Although ALDH3B1 appears to be expressed in an epithelium-specific manner, the enzyme was also localized to cerebellar astrocytes and histiocytes (macrophages), where it likely also plays a role in aldehyde and oxidative stress defense. In the central nervous system, including the cerebellum, alcohol exposure leads to increased aldehyde production (Smith et al. 2005), and astrocytes are a critical cell type involved in homeostasis, oxidative stress defense (Belanger and Magistretti 2009), and the detoxification of LPO-derived aldehydes (Kubatova et al. 2006). Macrophages are also susceptible to oxidative stress (Kirkham 2007), and they themselves generate ROS as part of the immune response (Nakamura et al. 1999). As such, the ALDH-mediated detoxification of LPO-derived aldehydes is an important macrophage defense system in various tissues, including lung, liver, and spleen (Germolec et al. 1995; Luckey and Petersen 2001).
It is of note that certain epithelial tissues lacked ALDH3B1 expression. Surface epithelium from human colon, lung type II pneumocytes, kidney distal convoluted tubule cells, thyroid follicle epithelial cells, germinal epithelium of the ovary, and glandular epithelium of the endometrium all lacked immunoreactivity. In contrast to a generalized expression throughout all tissues, which would be indicative of a less-specific “house-keeping” function, this selectivity in tissue and cellular distribution is more consistent, with ALDH3B1 serving specific functions in the cells in which it is expressed. Aside from aldehyde metabolism, ALDH3B1 may prove to have additional, but not yet determined, cellular roles. Indeed, many ALDH enzymes possess multiple catalytic functions, including esterase and nitrate reductase activity (Sladek 2003a; Vasiliou and Nebert 2005), and several of these act as binding proteins for various endogenous (e.g., androgen, cholesterol, and thyroid hormone) and exogenous (e.g., acetaminophen) compounds (Graves et al. 2002; Vasiliou et al. 2004). ALDH3A1, in addition to aldehyde detoxification, catalyzes ester hydrolysis, filters UV light as a corneal crystallin, scavenges ROS, and may have a role in cell cycle regulation and DNA repair (Pappa et al. 2005; Lassen et al. 2006,2007; Estey et al. 2007). Further investigation of the putative additional roles of ALDH3B1 in tissues is warranted.
Similar to other ALDH enzymes (Marselos and Michalopoulos 1987; Krupenko and Oleinik 2002; Kim et al. 2007), ALDH3B1 was found to be highly expressed in a large percentage of colon, lung, breast, and ovarian tumors. Consistent with other reports (Wroczynski et al. 2005; Chang et al. 2009), high variability of ALDH3B1 expression among patients with tumor was seen, and it remains unclear why ALDH3B1 was expressed in many but not all tumors. In colon tumors, ALDH3B1 expression was higher in patients with moderate to well-differentiated tumors when compared with those with poorly differentiated tumors. In this study, among patients with ovarian tumors, ALDH3B1 expression was higher in patients less than 60 years of age when compared with patients 60 years of age or older, who represent the majority of patients with epithelial derived ovarian tumor. Age-related changes in tumor protein expression have been reported and may correlate with patient outcome and survival (Yan et al. 2009). Moreover, differential expression of ALDH isozymes appears to be a factor in patient diagnosis, treatment, and survival. In colon cancer, ALDH1A1 expression is a marker for malignancy and tumor-initiating ability, and ALDH-expressing colon stem cells contribute to tumorigenesis (Carpentino et al. 2009; Huang et al. 2009). In human colon carcinoma cell lines, ALDH3A1 mediates cellular resistance to oxazaphosphorine anticancer drugs through the detoxification of aldehyde metabolites (Rekha et al. 1994). Similarly, high ALDH expression in multidrug-resistant colonic adenocarcinoma cells eliminates the toxicity of the anticancer bovine serum amine oxidase, which acts through hydrogen peroxide and aldehyde metabolites (Agostinelli et al. 2006). In lung tumors, expression of ALDH3B1 was consistent with that reported for ALDH1A1 and ALDH3A1 (Patel et al. 2008), which have been suggested to be biomarkers for early detection and clinical prognosis (Jiang et al. 2009; Rubporn et al. 2009; Ucar et al. 2009). ALDH1A1 and ALDH3A1 are also highly expressed in breast cancer, which may be of value in predicting the therapeutic potential of cyclophosphamide (Sreerama and Sladek 1997; Sladek et al. 2002). Indeed, custom treatment plans involving the downregulation of ALDH1A1 and ALDH3A1 lead to increased sensitivity to anticancer drugs and suppression of tumor cell growth (Rekha et al. 1998; Sladek 1999; Moreb et al. 2000; Muzio et al. 2006). In breast cancer, high ALDH activity appears to be a factor in drug resistance, cell differentiation, metastasis, and oxidative stress response (Croker et al. 2008; Moreb 2008). ALDH1A1, a marker of breast cancer stem cells, mediates the resistance to paclitaxel and epirubicin-based chemotherapy and is a predictor of poor clinical outcome (Ginestier et al. 2007; Morimoto et al. 2009; Tanei et al. 2009). In contrast, ALDH1A1 expression in the ovary correlates with low-grade tumors, successful cisplatin-based chemotherapy, and a favorable patient prognosis (Tanner et al. 1997; Chang et al. 2009). The significance of ALDH3B1 expression in tumors and its potential role in treatment and patient outcome remains to be identified.
ALDH3B1 may be upregulated in tumors as a result of oxidative stress processes. Alcohol consumption or cigarette smoke exposure, known risk factors for many cancers, causes high levels of aldehyde production, hydroxyl radicals, ROS, and DNA damage (Wright et al. 1999; Seitz and Becker 2007). Much of the negative effects of ethanol and cigarette smoke are mediated by acetaldehyde, which causes oxidative stress and is a carcinogen itself (Seitz and Becker 2007; Minegishi et al. 2007; Eom et al. 2009). These processes could be of particular significance in patients with diminished cellular antioxidant defenses or in tissues with low antioxidant capacity. Indeed, low ALDH levels in the colon lead to the highest acetaldehyde exposure of any tissue during ethanol metabolism, which is believed to be a factor in alcohol-related colon cancer (Salaspuro 1996). ALDH2-mediated metabolism of acetaldehyde is a major detoxification route, and carriers of the ALDH2*2 polymorphism, which confers loss of ALDH2 enzymatic activity, have increased risk for alcohol-related colon cancer (Murata et al. 1999) and smoke- and alcohol-related lung cancer (Minegishi et al. 2007; Eom et al. 2009). Although acetaldehyde itself is poorly metabolized by ALDH3B1, downstream LPO-derived aldehydes, including the highly cytotoxic 4-HNE, appear to be good substrates (Marchitti et al. 2007b). 4-HNE can inhibit the ALDH-mediated metabolism of aldehydes, including ALDH2, thus preventing the detoxification of acetaldehyde (Mitchell and Petersen 1991; Florang et al. 2007). In this regard, ALDH3B1-mediated detoxification of 4-HNE would be an important cellular defense system during acetaldehyde metabolism. The precise molecular mechanism by which ALDH3B1 is upregulated in tumors and whether oxidative stress plays a role remain to be determined. However, in breast cancer cell lines, ALDH3A1 is induced from the transient exposure to electrophiles, which activate an electrophile-responsive element (EpRE), and from exposure to Ah-receptor agonists such as polycyclic aromatic hydrocarbons, which activate a xenobiotic-responsive element (XRE) (Sreerama and Sladek 2001; Sladek 2003b). The presence of an EpRE or an XRE in the ALDH3B1 gene has yet to be reported.
In summary, ALDH3B1 is expressed in a number of normal human tissues in a very cell-specific manner, indicating functional specificity of this enzyme. Aside from cerebellar astrocytes and histiocytes, the ALDH3B1 expression in tissues was limited to epithelial cells. Localization of ALDH3B1 was uniformly cytosolic in tissues, with the exception of normal human lung and testis, in which it appeared to be either membrane-bound or membrane-associated. The majority of ALDH3B1 expression appeared granular in nature, suggesting that the protein may be associated with vesicles, such as lysosomes or peroxisomes, or is localized to the lumen of the endoplasmic reticulum. The upregulation of ALDH3B1 expression in human tumors could indicate a conferring of a growth advantage or, alternatively, could be the result of an induction mechanism mediated by oxidative stress and related processes such as LPO and the production of reactive aldehydes. Similar to other ALDH enzymes, differential ALDH3B1 expression in tumors may prove to have important implications for patient treatment and prognosis. What remains to be clarified is what factors mediate ALDH3B1 expression in tumors and whether this expression could be used as a tumor marker. Given the diversity of the ALDH family, it is possible that ALDH3B1 has multiple functions aside from aldehyde metabolism in normal and diseased tissues. In conclusion, we believe this study represents the most comprehensive immunohistochemical investigation of any ALDH enzyme to date, and the results presented herein will lead to further understanding of the significance of ALDH3B1 under physiological and pathophysiological conditions.
Acknowledgments
The work was supported by NIH Grants EY11490 and EY17963 (to VV) and NIH/National Institute on Alcohol Abuse and Alcoholism Predoctoral Fellowship AA016875 (to SAM).
The authors thank Dr. David Thompson for critical reading and discussion of this manuscript and IHCtech for technical support.
This article is distributed under the terms of a License to Publish Agreement (http://www.jhc.org/misc/ltopub.shtml). JHC deposits all of its published articles into the U.S. National Institutes of Health (http://www.nih.gov/) and PubMed Central (http://www.pubmedcentral.nih.gov/) repositories for public release twelve months after publication.
References
- Agostinelli E, Belli F, Dalla VL, Marra M, Crateri P, Arancia G (2006) Hyperthermia enhances cytotoxicity of amine oxidase and spermine on drug-resistant LoVo colon adenocarcinoma cells. Int J Oncol 28:1543–1553 [PubMed] [Google Scholar]
- Apte M, McCarroll J, Pirola R, Wilson J (2007) Pancreatic MAP kinase pathways and acetaldehyde. Novartis Found Symp 285:200–211 [DOI] [PubMed] [Google Scholar]
- Belanger M, Magistretti PJ (2009) The role of astroglia in neuroprotection. Dialogues Clin Neurosci 11:281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezniceanu ML, Liu F, Wei CC, Tran S, Sachetelli S, Zhang SL, Guo DF, et al. (2007) Catalase overexpression attenuates angiotensinogen expression and apoptosis in diabetic mice. Kidney Int 71:912–923 [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Theruvathu JA (2005) DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol 35:187–193 [DOI] [PubMed] [Google Scholar]
- Canuto RA, Muzio G, Ferro M, Maggiora M, Federa R, Bassi AM, Lindahl R, et al. (1999) Inhibition of class-3 aldehyde dehydrogenase and cell growth by restored lipid peroxidation in hepatoma cell lines. Free Radic Biol Med 26:333–340 [DOI] [PubMed] [Google Scholar]
- Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, Huang EH (2009) Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res 69:8208–8215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederbaum AI, Lu Y, Wu D (2009) Role of oxidative stress in alcohol-induced liver injury. Arch Toxicol 83:519–548 [DOI] [PubMed] [Google Scholar]
- Chang B, Liu G, Xue F, Rosen DG, Xiao L, Wang X, Liu J (2009) ALDH1 expression correlates with favorable prognosis in ovarian cancers. Mod Pathol 22:817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che W, Qiu H, Liu G, Ran Y, Zhang H, Zhang L, Wen W (2009) Oxidative damage of the extracts of condensate, particulate and semivolatile organic compounds from gasoline engine exhausts on testicles of rats. Bull Environ Contam Toxicol 83:42–47 [DOI] [PubMed] [Google Scholar]
- Chiang CP, Wu CW, Lee SP, Chung CC, Wang CW, Lee SL, Nieh S, et al. (2009) Expression pattern, ethanol-metabolizing activities, and cellular localization of alcohol and aldehyde dehydrogenases in human pancreas: implications for pathogenesis of alcohol-induced pancreatic injury. Alcohol Clin Exp Res 33:1059–1068 [DOI] [PubMed] [Google Scholar]
- Chowdhury P, Walker A (2008) A cell-based approach to study changes in the pancreas following nicotine exposure in an animal model of injury. Langenbecks Arch Surg 393:547–555 [DOI] [PubMed] [Google Scholar]
- Coskran TM, Morton D, Menniti FS, Adamowicz WO, Kleiman RJ, Ryan AM, Strick CA, et al. (2006) Immunohistochemical localization of phosphodiesterase 10A in multiple mammalian species. J Histochem Cytochem 54:1205–1213 [DOI] [PubMed] [Google Scholar]
- Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL (2008) High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med 13:2236–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro I, Burdmann EA, Seguro AC, Yu L (2004) Bothrops venom induces direct renal tubular injury: role for lipid peroxidation and prevention by antivenom. Toxicon 43:833–839 [DOI] [PubMed] [Google Scholar]
- Elliott MK, Sisson JH, Wyatt TA (2007) Effects of cigarette smoke and alcohol on ciliated tracheal epithelium and inflammatory cell recruitment. Am J Respir Cell Mol Biol 36:452–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eom SY, Zhang YW, Kim SH, Choe KH, Lee KY, Park JD, Hong YC, et al. (2009) Influence of NQO1, ALDH2, and CYP2E1 genetic polymorphisms, smoking, and alcohol drinking on the risk of lung cancer in Koreans. Cancer Causes Control 20:137–145 [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H (1991) Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med 11:81–128 [DOI] [PubMed] [Google Scholar]
- Estey T, Piatigorsky J, Lassen N, Vasiliou V (2007) ALDH3A1: a corneal crystallin with diverse functions. Exp Eye Res 84:3–12 [DOI] [PubMed] [Google Scholar]
- Florang VR, Rees JN, Brogden NK, Anderson DG, Hurley TD, Doorn JA (2007) Inhibition of the oxidative metabolism of 3,4-dihydroxyphenylacetaldehyde, a reactive intermediate of dopamine metabolism, by 4-hydroxy-2-nonenal. Neurotoxicology 28:76–82 [DOI] [PubMed] [Google Scholar]
- Germolec DR, Adams NH, Luster MI (1995) Comparative assessment of metabolic enzyme levels in macrophage populations of the F344 rat. Biochem Pharmacol 50:1495–1504 [DOI] [PubMed] [Google Scholar]
- Germolec DR, Henry EC, Maronpot R, Foley JF, Adams NH, Gasiewicz TA, Luster MI (1996) Induction of CYP1A1 and ALDH-3 in lymphoid tissues from Fisher 344 rats exposed to 2,3,7,8-tetrachlorodibenzodioxin (TCDD). Toxicol Appl Pharmacol 137:57–66 [DOI] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, et al. (2007) ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1:555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves PR, Kwiek JJ, Fadden P, Ray R, Hardeman K, Coley AM, Foley M, et al. (2002) Discovery of novel targets of quinoline drugs in the human purine binding proteome. Mol Pharmacol 62:1364–1372 [DOI] [PubMed] [Google Scholar]
- Guilbert L, Robertson SA, Wegmann TG (1993) The trophoblast as an integral component of a macrophage-cytokine network. Immunol Cell Biol 71(Pt 1):49–57 [DOI] [PubMed] [Google Scholar]
- Gukovskaya AS, Mouria M, Gukovsky I, Reyes CN, Kasho VN, Faller LD, Pandol SJ (2002) Ethanol metabolism and transcription factor activation in pancreatic acinar cells in rats. Gastroenterology 122:106–118 [DOI] [PubMed] [Google Scholar]
- Han HJ, Lee YJ, Park SH, Lee JH, Taub M (2005) High glucose-induced oxidative stress inhibits Na+/glucose cotransporter activity in renal proximal tubule cells. Am J Physiol Renal Physiol 288:F988–996 [DOI] [PubMed] [Google Scholar]
- Hewitt SM (2006) The application of tissue microarrays in the validation of microarray results. Methods Enzymol 410:400–415 [DOI] [PubMed] [Google Scholar]
- Hewitt SM (2009) Tissue microarrays as a tool in the discovery and validation of tumor markers. Methods Mol Biol 520:151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, et al. (2009) Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res 69:3382–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AK, Stricklett PK, Kohan DE (1998) Cytotoxic effect of Shiga toxin-1 on human proximal tubule cells. Kidney Int 54:426–437 [DOI] [PubMed] [Google Scholar]
- Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, et al. (2009) Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res 7:330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Lee HJ, Choi HY, Shin Y, Nam S, Seo G, Son DS, et al. (2007) Clinical validity of the lung cancer biomarkers identified by bioinformatics analysis of public expression data. Cancer Res 67:7431–7438 [DOI] [PubMed] [Google Scholar]
- Kirkham P (2007) Oxidative stress and macrophage function: a failure to resolve the inflammatory response. Biochem Soc Trans 35:284–287 [DOI] [PubMed] [Google Scholar]
- Kirsch M, De Groot H (2001) NAD(P)H, a directly operating antioxidant? FASEB J 15:1569–1574 [DOI] [PubMed] [Google Scholar]
- Koch OR, Pani G, Borrello S, Colavitti R, Cravero A, Farre S, Galeotti T (2004) Oxidative stress and antioxidant defenses in ethanol-induced cell injury. Mol Aspects Med 25:191–198 [DOI] [PubMed] [Google Scholar]
- Krupenko SA, Oleinik NV (2002) 10-formyltetrahydrofolate dehydrogenase, one of the major folate enzymes, is down-regulated in tumor tissues and possesses suppressor effects on cancer cells. Cell Growth Differ 13:227–236 [PubMed] [Google Scholar]
- Kubatova A, Murphy TC, Combs C, Picklo MJ Sr. (2006) Astrocytic biotransformation of trans-4-hydroxy-2-nonenal is dose-dependent. Chem Res Toxicol 19:844–851 [DOI] [PubMed] [Google Scholar]
- Kurimoto C, Kawano S, Tsuji G, Hatachi S, Jikimoto T, Sugiyama D, Kasagi S, et al. (2007) Thioredoxin may exert a protective effect against tissue damage caused by oxidative stress in salivary glands of patients with Sjogren's syndrome. J Rheumatol 34:2035–2043 [PubMed] [Google Scholar]
- Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, Duester G, et al. (2007) Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem 282:25668–25676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen N, Pappa A, Black WJ, Jester JV, Day BJ, Min E, Vasiliou V (2006) Antioxidant function of corneal ALDH3A1 in cultured stromal fibroblasts. Free Radic Biol Med 41:1459–1469 [DOI] [PubMed] [Google Scholar]
- Lindahl R (1992) Aldehyde dehydrogenases and their role in carcinogenesis. Crit Rev Biochem Mol Biol 27:283–335 [DOI] [PubMed] [Google Scholar]
- Luckey SW, Petersen DR (2001) Metabolism of 4-hydroxynonenal by rat Kupffer cells. Arch Biochem Biophys 389:77–83 [DOI] [PubMed] [Google Scholar]
- Lunghi L, Ferretti ME, Medici S, Biondi C, Vesce F (2007) Control of human trophoblast function. Reprod Biol Endocrinol 5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ, Ewing CM, et al. (2002) Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res 62:2220–2226 [PubMed] [Google Scholar]
- Maeda M, Hasumura Y, Takeuchi J (1988) Localization of cytoplasmic and mitochondrial aldehyde dehydrogenase isozymes in human liver. Lab Invest 59:75–81 [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, Vasiliou V (2008) Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol 4:697–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchitti SA, Deitrich RA, Vasiliou V (2007a) Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol Rev 59:125–150 [DOI] [PubMed] [Google Scholar]
- Marchitti SA, Orlicky DJ, Vasiliou V (2007b) Expression and initial characterization of human ALDH3B1. Biochem Biophys Res Commun 356:792–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marselos M, Michalopoulos G (1987) Changes in the pattern of aldehyde dehydrogenase activity in primary and metastatic adenocarcinomas of the human colon. Cancer Lett 34:27–37 [DOI] [PubMed] [Google Scholar]
- Mills PB, Struys E, Jakobs C, Plecko B, Baxter P, Baumgartner M, Willemsen MA, et al. (2006) Mutations in antiquitin in individuals with pyridoxine-dependent seizures. Nat Med 12:307–309 [DOI] [PubMed] [Google Scholar]
- Minegishi Y, Tsukino H, Muto M, Goto K, Gemma A, Tsugane S, Kudoh S, et al. (2007) Susceptibility to lung cancer and genetic polymorphisms in the alcohol metabolite-related enzymes alcohol dehydrogenase 3, aldehyde dehydrogenase 2, and cytochrome P450 2E1 in the Japanese population. Cancer 110:353–362 [DOI] [PubMed] [Google Scholar]
- Mitchell DY, Petersen DR (1991) Inhibition of rat hepatic mitochondrial aldehyde dehydrogenase-mediated acetaldehyde oxidation by trans-4-hydroxy-2-nonenal. Hepatology 13:728–734 [DOI] [PubMed] [Google Scholar]
- Moreb JS (2008) Aldehyde dehydrogenase as a marker for stem cells. Curr Stem Cell Res Ther 3:237–246 [DOI] [PubMed] [Google Scholar]
- Moreb JS, Baker HV, Chang LJ, Amaya M, Lopez MC, Ostmark B, Chou W (2008) ALDH isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol Cancer 7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreb JS, Maccow C, Schweder M, Hecomovich J (2000) Expression of antisense RNA to aldehyde dehydrogenase class-1 sensitizes tumor cells to 4-hydroperoxycyclophosphamide in vitro. J Pharmacol Exp Ther 293:390–396 [PubMed] [Google Scholar]
- Morimoto K, Kim SJ, Tanei T, Shimazu K, Tanji Y, Taguchi T, Tamaki Y, et al. (2009) Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci 100:1062–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Tagawa M, Watanabe S, Kimura H, Takeshita T, Morimoto K (1999) Genotype difference of aldehyde dehydrogenase 2 gene in alcohol drinkers influences the incidence of Japanese colorectal cancer patients. Jpn J Cancer Res 90:711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio G, Trombetta A, Maggiora M, Martinasso G, Vasiliou V, Lassen N, Canuto RA (2006) Arachidonic acid suppresses growth of human lung tumor A549 cells through down-regulation of ALDH3A1 expression. Free Radic Biol Med 40:1929–1938 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Yokoyama H, Okamura Y, Ohgo H, Fukuda M, Horie Y, Kato S, et al. (1999) Evidence for ethanol oxidation by Kupffer cells. Alcohol Clin Exp Res 23:92S–95S [DOI] [PubMed] [Google Scholar]
- Palmieri VO, Grattagliano I, Palasciano G (2007) Ethanol induces secretion of oxidized proteins by pancreatic acinar cells. Cell Biol Toxicol 23:459–464 [DOI] [PubMed] [Google Scholar]
- Pappa A, Brown D, Koutalos Y, DeGregori J, White C, Vasiliou V (2005) Human aldehyde dehydrogenase 3A1 inhibits proliferation and promotes survival of human corneal epithelial cells. J Biol Chem 280:27998–28006 [DOI] [PubMed] [Google Scholar]
- Pappa A, Chen C, Koutalos Y, Townsend AJ, Vasiliou V (2003) Aldh3a1 protects human corneal epithelial cells from ultraviolet- and 4-hydroxy-2-nonenal-induced oxidative damage. Free Radic Biol Med 34:1178–1189 [DOI] [PubMed] [Google Scholar]
- Patel M, Lu L, Zander DS, Sreerama L, Coco D, Moreb JS (2008) ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer 59:340–349 [DOI] [PubMed] [Google Scholar]
- Rekha GK, Devaraj VR, Sreerama L, Lee MJ, Nagasawa HT, Sladek NE (1998) Inhibition of human class 3 aldehyde dehydrogenase, and sensitization of tumor cells that express significant amounts of this enzyme to oxazaphosphorines, by chlorpropamide analogues. Biochem Pharmacol 55:465–474 [DOI] [PubMed] [Google Scholar]
- Rekha GK, Sreerama L, Sladek NE (1994) Intrinsic cellular resistance to oxazaphosphorines exhibited by a human colon carcinoma cell line expressing relatively large amounts of a class-3 aldehyde dehydrogenase. Biochem Pharmacol 48:1943–1952 [DOI] [PubMed] [Google Scholar]
- Rizzo WB, Carney G (2005) Sjögren–Larsson syndrome: diversity of mutations and polymorphisms in the fatty aldehyde dehydrogenase gene (ALDH3A2). Hum Mutat 26:1–10 [DOI] [PubMed] [Google Scholar]
- Rubporn A, Srisomsap C, Subhasitanont P, Chokchaichamnankit D, Chiablaem K, Svasti J, Sangvanich P (2009) Comparative proteomic analysis of lung cancer cell line and lung fibroblast cell line. Cancer Genomics Proteomics 6:229–237 [PubMed] [Google Scholar]
- Salaspuro M (1996) Bacteriocolonic pathway for ethanol oxidation: characteristics and implications. Ann Med 28:195–200 [DOI] [PubMed] [Google Scholar]
- Schnier JB, Kaur G, Kaiser A, Stinson SF, Sausville EA, Gardner J, Nishi K, et al. (1999) Identification of cytosolic aldehyde dehydrogenase 1 from non-small cell lung carcinomas as a flavopiridol-binding protein. FEBS Lett 454:100–104 [DOI] [PubMed] [Google Scholar]
- Schwerdt G, Holzinger H, Sauvant C, Konigs M, Humpf HU, Gekle M (2007) Long-term effects of ochratoxin A on fibrosis and cell death in human proximal tubule or fibroblast cells in primary culture. Toxicology 232:57–67 [DOI] [PubMed] [Google Scholar]
- Seitz HK, Becker P (2007) Alcohol metabolism and cancer risk. Alcohol Res Health 30:38–47 [PMC free article] [PubMed] [Google Scholar]
- Shiraishi K, Naito K (2007) Effects of 4-hydroxy-2-nonenal, a marker of oxidative stress, on spermatogenesis and expression of p53 protein in male infertility. J Urol 178:1012–1017 [DOI] [PubMed] [Google Scholar]
- Sisson JH, Tuma DJ, Rennard SI (1991) Acetaldehyde-mediated cilia dysfunction in bovine bronchial epithelial cells. Am J Physiol 260:L29–36 [DOI] [PubMed] [Google Scholar]
- Sladek NE (1999) Aldehyde dehydrogenase-mediated cellular relative insensitivity to the oxazaphosphorines. Curr Pharm Des 5:607–625 [PubMed] [Google Scholar]
- Sladek NE (2003a) Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol 17:7–23 [DOI] [PubMed] [Google Scholar]
- Sladek NE (2003b) Transient induction of increased aldehyde dehydrogenase 3A1 levels in cultured human breast (adeno)carcinoma cell lines via 5′-upstream xenobiotic, and electrophile, responsive elements is, respectively, estrogen receptor-dependent and -independent. Chem Biol Interact 143/144:63–74 [DOI] [PubMed] [Google Scholar]
- Sladek NE, Kollander R, Sreerama L, Kiang DT (2002) Cellular levels of aldehyde dehydrogenases (ALDH1A1 and ALDH3A1) as predictors of therapeutic responses to cyclophosphamide-based chemotherapy of breast cancer: a retrospective study. Rational individualization of oxazaphosphorine-based cancer chemotherapeutic regimens. Cancer Chemother Pharmacol 49:309–321 [DOI] [PubMed] [Google Scholar]
- Smith AM, Zeve DR, Grisel JJ, Chen WJ (2005) Neonatal alcohol exposure increases malondialdehyde (MDA) and glutathione (GSH) levels in the developing cerebellum. Brain Res Dev Brain Res 160:231–238 [DOI] [PubMed] [Google Scholar]
- Sreerama L, Sladek NE (1993) Identification and characterization of a novel class 3 aldehyde dehydrogenase overexpressed in a human breast adenocarcinoma cell line exhibiting oxazaphosphorine-specific acquired resistance. Biochem Pharmacol 45:2487–2505 [DOI] [PubMed] [Google Scholar]
- Sreerama L, Sladek NE (1997) Cellular levels of class 1 and class 3 aldehyde dehydrogenases and certain other drug-metabolizing enzymes in human breast malignancies. Clin Cancer Res 3:1901–1914 [PubMed] [Google Scholar]
- Sreerama L, Sladek NE (2001) Three different stable human breast adenocarcinoma sublines that overexpress ALDH3A1 and certain other enzymes, apparently as a consequence of constitutively upregulated gene transcription mediated by transactivated EpREs (electrophile responsive elements) present in the 5′-upstream regions of these genes. Chem Biol Interact 130/132:247–260 [DOI] [PubMed] [Google Scholar]
- Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, Tamaki Y, et al. (2009) Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res 15:4234–4241 [DOI] [PubMed] [Google Scholar]
- Tanner B, Hengstler JG, Dietrich B, Henrich M, Steinberg P, Weikel W, Meinert R, et al. (1997) Glutathione, glutathione S-transferase alpha and pi, and aldehyde dehydrogenase content in relationship to drug resistance in ovarian cancer. Gynecol Oncol 65:54–62 [DOI] [PubMed] [Google Scholar]
- Ucar D, Cogle CR, Zucali JR, Ostmark B, Scott EW, Zori R, Gray BA, et al. (2009) Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem Biol Interact 178:48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaithinathan S, Saradha B, Mathur PP (2009) Methoxychlor-induced alteration in the levels of HSP70 and clusterin is accompanied with oxidative stress in adult rat testis. J Biochem Mol Toxicol 23:29–35 [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Marselos M (1989) Tissue distribution of inducible aldehyde dehydrogenase activity in the rat after treatment with phenobarbital or methylcholanthrene. Pharmacol Toxicol 64:39–42 [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Nebert DW (2005) Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics 2:138–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliou V, Pappa A, Estey T (2004) Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev 36:279–299 [DOI] [PubMed] [Google Scholar]
- Vasiliou V, Puga A, Nebert DW (1992) Negative regulation of the murine cytosolic aldehyde dehydrogenase-3 (Aldh-3c) gene by functional CYP1A1 and CYP1A2 proteins. Biochem Biophys Res Commun 187:413–419 [DOI] [PubMed] [Google Scholar]
- Wang JS, Fang Q, Sun DJ, Chen J, Zhou XL, Lin GW, Lu HZ, et al. (2001) Genetic modification of hematopoietic progenitor cells for combined resistance to 4-hydroperoxycyclophosphamide, vincristine, and daunorubicin. Acta Pharmacol Sin 22:949–955 [PubMed] [Google Scholar]
- Warholm M, Holmberg B, Hogberg J, Kronevi T, Gotharson A (1984) The acute effects of single and repeated injections of acrolein and other aldehydes. Int J Tissue React 6:61–70 [PubMed] [Google Scholar]
- Wright RM, McManaman JL, Repine JE (1999) Alcohol-induced breast cancer: a proposed mechanism. Free Radic Biol Med 26:348–354 [DOI] [PubMed] [Google Scholar]
- Wroczynski P, Nowak M, Wierzchowski J, Szubert A, Polanski J (2005) Activities of cytosolic aldehyde dehydrogenase isozymes in colon cancer: determination using selective, fluorimetric assays. Acta Pol Pharm 62:427–433 [PubMed] [Google Scholar]
- Yan Y, Skliris GP, Penner C, Chooniedass-Kothari S, Cooper C, Nugent Z, Blanchard A, et al. (2009) Steroid Receptor RNA Activator Protein (SRAP): a potential new prognostic marker for estrogen receptor-positive/node-negative/younger breast cancer patients. Breast Cancer Res 11:R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Muramatsu T, Omori T, Yokoyama T, Matsushita S, Higuchi S, Maruyama K, et al. (2001) Alcohol and aldehyde dehydrogenase gene polymorphisms and oropharyngolaryngeal, esophageal and stomach cancers in Japanese alcoholics. Carcinogenesis 22:433–439 [DOI] [PubMed] [Google Scholar]
- Yoritaka A, Hattori N, Uchida K, Tanaka M, Stadtman ER, Mizuno Y (1996) Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc Natl Acad Sci USA 93:2696–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]