Abstract
Emerging porcine models of cystic fibrosis (CF) are expected to mimic the human disease more closely than current mouse models do. However, little is known of the tissue and cellular expression patterns of the porcine CF transmembrane conductance regulator (pCFTR) and possible differences from human CFTR (hCFTR). Here, the expression pattern of pCFTR was systematically established on the mRNA and protein levels. Using specific anti-pCFTR antibodies, the majority of the protein was immunohistochemically detected on paraffin-embedded sections and on cryostate sections in the apical cytosol of intestinal crypt epithelial cells, nasal, tracheal, and bronchial epithelial cells, and other select, mostly glandular epithelial cells. Confocal laser scanning microscopy with co-localization of the Golgi marker 58K localized the protein in the cytosol between the Golgi apparatus and the apical cell membrane with occasional punctate or diffuse staining of the apical membrane. The tissue and cellular distribution patterns were confirmed by RT-PCR from whole tissue lysates or select cells after laser capture microdissection. Thus, expression of pCFTR was found to largely resemble that of hCFTR except for the kidney, brain, and cutaneous glands, which lack expression in pigs. Species-specific differences between pCFTR and hCFTR may become relevant for future interpretations of the CF phenotype in pig models. (J Histochem Cytochem 58:785–797, 2010)
Keywords: CFTR, expression pattern, cystic fibrosis, pig, animal model
Eleven different mouse models have been generated to date to investigate the pathology of cystic fibrosis (CF) (Guilbault et al. 2007). Although several relevant biochemical, electrophysiological, and cellular aspects of the disease have been studied in these models, they all fail to develop the complex human CF pathology, especially the typical CF lung disease including secondary bacterial and viral infections (Scholte et al. 2004). Instead, the murine CF phenotype is mostly dominated by intestinal disease. Moreover, the pancreatic lesions that shaped the name of the disease are not observed in CF mice (Davidson and Rolfe 2001). The mouse models available are thus unsuitable to elucidate the mechanisms of the pulmonary changes, which dominate human CF and other aspects of the disease. In addition, differences in size and physiology make CF mice less than optimal models to test new therapeutic approaches.
Several lines of evidence suggest that the pig may serve as a more suitable animal model for CF. The anatomy, biochemistry, physiology, size, and genetics of pigs resemble those of humans more closely than those of mice do (Rogers et al. 2008a). This is particularly true for CF-relevant tissues, including the lungs, intestine, and pancreas. In fact, first observations on CF pigs seem to support this idea in that newborn pigs not only develop meconium ileus but also pancreatic insufficiency and biliary cirrhosis, similar to human CF patients (Rogers et al. 2008b; Meyerholz et al. 2010). Importantly, the microanatomy of the porcine airways including the distribution of submucosal glands, which are thought to be critically involved in the pathogenesis of CF, closely matches that of humans (Rogers et al. 2008a; Elferink and Beuers 2009). Moreover, the longevity of pigs allows investigating the pathogenesis and development of the disease with respect to changes that might occur only after a longer period of time, including chronic airway infections. Also, the size of pigs enables testing of new therapeutic strategies developed for humans (Rogers et al. 2008a). Because of their many similarities to humans, pigs have been used successfully as organ donors in several xenotransplantation efforts (Cooper et al. 2008). Emerging pig models of CF are, therefore, expected to help elucidate the mechanisms of the disease and establish new therapeutic approaches (Rogers et al. 2008a,b).
To interpret the phenotype of new animal models and to draw appropriate conclusions, species-specific differences have to be carefully addressed. First, differences between the human CF transmembrane conductance regulator (hCFTR) and porcine CFTR (pCFTR) have been reported in terms of their intracellular protein processing (Ostedgaard et al. 2007). Furthermore, a comparative characterization of functional properties of chloride channels in the respiratory epithelium has revealed substantial, species-specific differences between pigs and humans (Liu et al. 2008).
Nevertheless, there is convincing evidence that the majority of chloride conductance in the porcine intestine is based on pCFTR (Leonhard-Marek et al. 2009).
A sound understanding of the tissue and cellular expression patterns of pCFTR, in addition to its molecular processing and function, will be relevant for characterizing CF in pig models. However, no systematic characterization of the pCFTR expression pattern and no specific anti-pCFTR antibodies have been reported to date. Two previous studies have used anti-hCFTR antibodies to localize the pCFTR protein in the lung and intestine of piglets. The anti-hCFTR antibodies labeled a protein in the apical membranes of respiratory and intestinal crypt epithelial cells (Rogers et al. 2008b) or goblet cells but not non-goblet cell enterocytes in the intestine of piglets (Hayden and Carey 1996). Tissues other than the lung and intestine have not been studied yet.
The aim of this study was, thus, to localize the mRNA and protein distribution patterns of pCFTR in all CF-relevant tissues using pig-specific molecular probes and antibodies, respectively. The results were compared with previous reports on the expression of the hCFTR mRNA and protein in humans.
Materials and Methods
Bioinformatic Analyses
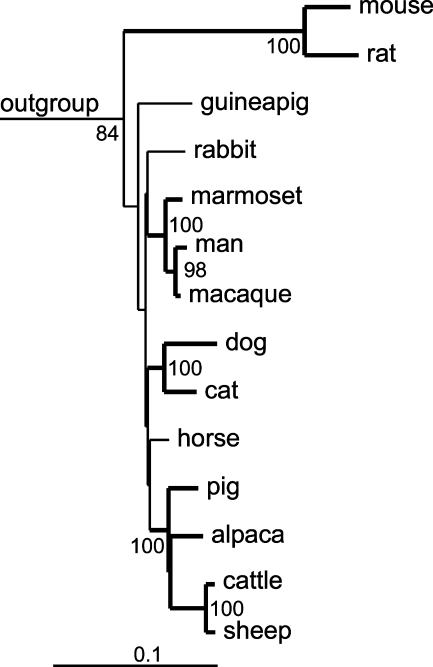
CFTR genomic and protein sequences were taken from the Ensembl genome database (http://www.ensembl.org) and from the GenBank database (http://www.ncbi.nlm.nih.gov). Regulatory regions in mammalian species were identified by Basic Local Alignment Search Tool (BLAST) search using the 2.5-kb upstream region of CFTR and the published human sequences of the −20.9- and +15.6-kb insulators (Blackledge et al. 2007) as query sequences. Phylogenetic analysis was performed as described (Klymiuk et al. 2006). In brief, protein or nucleotide sequences were aligned by ClustalW (Jeanmougin et al. 1998) and manipulated by hand in BioEdit (Hall 1999). The resulting alignments were used to generate single genetic distance or most parsimony trees and consensus trees of 100 bootstrapped alignments by the PHYLIP software package (http://evolution.genetics.washington.edu/phylip.html; Figure 1). The resulting trees were prepared by TreeView (http://taxonomy.zoology.gla.ac.uk/rod/rod.html). Identities or similarities were calculated by pairwise alignment using BioEdit.
Figure 1.
Phylogeny of cystic fibrosis transmembrane conductance regulator (CFTR) orthologs in closely related mammals. The tree was based on a genetic distance tree with branches confirmed by the most parsimony method shown in bold and branch nodes occurring more than 60 times in 100 neighbor joining trees indicated. Ten species of fish, amphibians, birds, and non-eutherian mammals were defined as outgroup.
Animals and Tissue Processing
Tissues from three 6-week-old male pigs (EUROC x Pietrain), two female pigs (2 and 3 months old, mixed breed), and one 7-month-old male pig (mixed breed) were used. The pigs were originally used by others for experiments unrelated to this study, with approval from local ethics authorities. The following tissues were immersion-fixed in 4% neutral buffered formaldehyde or shock-frozen in liquid nitrogen: nasal cavity, larynx, trachea, tracheal bronchus, left principal bronchus, lung (cranial left lobe, left main lobe, accessory lobe), esophagus, stomach (glandular and non-glandular parts), duodenum, jejunum, ileum, cecum, colon, rectum, parotid salivary gland, pancreas, liver, gall bladder, kidney, urinary bladder, mandibular lymph node, spleen, heart, aorta, brain (cortex, cerebellum, medulla), pituitary gland, thoracic ganglion, eyes, skin (perineum, rooting disc, prepuce), testicles, epididymides, spermatic cord, prostate gland, uterus, and ovaries.
Analysis of the pCFTR mRNA Tissue Expression Pattern
Total RNA was isolated from tissues as described previously (Plog et al. 2009). PCR primers were designed using the Beacon Designer 3.0 software (Premier Biosoft International; Palo Alto, CA) and are listed in Table 1. PCR amplification using the DreamTaq DNA Polymerase (Fermentas; St Leon-Rot, Germany) included 34 cycles at 95C for 2 min, 95C for 30 sec, 64.8C for 30 sec, and 72C for 1 sec with a time increment of 1 sec per cycle and a final extension at 72C for 10 min. To clarify their specificity, amplicons from spleen, lung, heart, and gall bladder were completely sequenced. All experiments were repeated at least two times in each animal. To control the mRNA quality and efficacy of reverse transcription, a 68-bp fragment of the housekeeping gene EF-1a was RT-PCR-amplified from each sample using primers 5′-CAAAAACGACCCACCAATGG-3′ (sense) and 5′-GGCCTGGATGGTTCAGGATA-3′ (antisense; Gruber and Levine 1997).
Table 1.
Primers used for specific pCFTR amplification
| Primer sequences | Position in pCFTR sequence (GenBank accession number NM_001104950) | Amplicon length (bp) |
|---|---|---|
| 5′-CCTGGGAAGCTTGATTTTG-3′ (upstream) | nt 4000–4018 | 450 |
| 5′-CTAAAGTCTTGTTTCTTGC-3′ (downstream) | nt 4431–4449 | |
| 5′-AACCTGAACAAGTTTGATGAAG-3′ (upstream) | nt 559–580 | 128 |
| 5′-AAGGCAAGTCCACAGAAGGC-3′ (downstream) | nt 667–686 |
pCFTR, porcine cystic fibrosis transmembrane conductance regulator.
Laser Capture Microdissection
Frozen bronchial tissue samples from three pigs were cut into multiple consecutive sections of 6 μm thickness and mounted onto glass slides coated with a polyethylene naphthalate membrane (PALM membrane slides; P.A.L.M. Microlaser Technologies, Martinsried, Germany). Sections were fixed for 1.5 min in 95% ethanol at −20C and air-dried, followed by staining with hematoxylin dissolved in diethylpyrocarbonate (DEPC)-treated water for 45 sec, rinsing in DEPC-treated water for 45 sec, and dipping three times in 1% eosin dissolved in DEPC-treated water. Sections were dehydrated in ascending graded ethanol and air-dried at room temperature for 5 min. A total area of 3.0 × 106 μm2 from the acinar cells of the submucosal glands or the bronchial epithelium was separated by laser microdissection and laser-pressure catapulted into the reaction caps of 0.5-ml tubes containing 30 μl RLT buffer (RNeasy Mini Kit; Qiagen, Hilden, Germany). Tubes were filled to 350 μl with RLT buffer, and total RNA was extracted and purified according to the manufacturer's protocol, including digestion with RNase-free DNase I (Qiagen). Approximately 30 ng of total RNA was reverse-transcribed in a 21-μl reaction volume (SuperScript III First-Strand Synthesis System; Invitrogen, Karlsruhe, Germany) using random hexamer primers in the presence of RNase inhibitor (RNase Out, Qiagen).
Generation of Antibodies
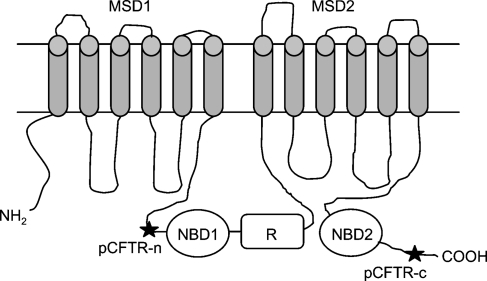
Two oligopeptides were synthesized based on immunogenicity prediction using the amino acid sequence of the pCFTR protein (GenBank accession no NP_001098420), one located N-terminally of the nucleotide-binding domain 1 (pCFTR-N, corresponding to aa 410–423, EKAKQNNNSRKISN) and the other located close to the carboxy-terminus of pCFTR (pCFTR-C, corresponding to aa 1451–1464, LLPHRNSSKQRSRS; Figure 2). NCBI BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of both antibodies revealed no significant homologies to porcine proteins other than CFTR. Oligopeptides were coupled to keyhole limpet hemocyanin and used for standard immunization of two rabbits each. Preimmune sera were collected before immunization and used as controls in the immunodetection experiments. The four antisera were designated pCFTR-N-1, pCFTR-N-2, pCFTR-C-1, and pCFTR-C-2. The antisera pCFTR-C-1 and pCFTR-C-2 were pooled and affinity-immunopurified with the oligopeptide used for immunization. The resulting immunopurified antibodies were designated pCFTR-C-1/2-p.
Figure 2.
Protein structure of CFTR and target regions of anti-pCFTR antibodies. Two different epitopes (stars) were selected for generating anti-pCFTR antibodies pCFTR-N (corresponding to aa 410–423, EKAKQNNNSRKISN) and pCFTR-C (corresponding to aa 1451–1464, LLPHRNSSKQRSRS). pCFTR, porcine cystic fibrosis transmembrane conductance regulator; NBD, nucleotide-binding domain; R, regulatory domain; MSD, membrane-spanning domain. Drawn using a description from the CFTR database.
Transient Transfection of HEK 293 Cells
HEK 293 cells were grown on 6-well plates in DMEM containing 10% heat-inactivated fetal calf serum, 2% glutamate, and 1% penicillin/streptomycin (culture medium) at 37C in the presence of 5% CO2. If not mentioned otherwise, cells were washed between medium changes with prewarmed PBS. HEK 293 cells were transfected with pcDNA3.1 plasmid containing the pCFTR open reading frame (kindly donated by Dr. Welsh; Ostedgaard et al. 2007) using Turbofect (Fermentas), according to the manufacturer's protocol. Briefly, cells grown on six-well plates were transfected with 2 μg plasmid DNA and 8 μl Turbofect suspended in 400 μl DMEM. Cells transfected with pcDNA3.1 vector alone served as negative controls (mock-transfected).
Immunoblot Analyses
At 48 hr after transfection, HEK 293 cells were lysed on ice in 150 μl standard lysis buffer [50 mM Tris–HCl (pH 7.4), 50 mM NaCl, 1.0% Triton X-100], supplemented with a mixture of protease inhibitors (1 mM PMSF, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 5 μg/ml antipain, 5 μg/ml leupeptin, 100 μg/ml trypsin–chymotrypsin inhibitor) for 30 min. To remove any cellular debris, the samples were spun at 13,000 × g for 15 min at 4C. Because of the known rapid degradability of pCFTR, samples were heated either at 95C for 7 min, 56C for 7 min, 37C for 7 min, or left at room temperature for 30 min in 5-fold SDS-PAGE Laemmli buffer containing 150 mM dithiothreitol. No specific bands were detected in samples boiled at 95C for 7 min, whereas specific bands were detected in all other samples independent of the pretreatment (data not shown). For future experiments, heating at 37C for 7 min was used as pretreatment. Samples were separated by 5% SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes (Carl Roth; Karlsruhe, Germany). The membranes were blocked for 1 hr with Roti-Block (Carl Roth) according to the manufacturer's protocol and subsequently probed overnight at 4C with pCFTR-C-1, pCFTR-C-1/2-p, or pCFTR-N-1 as primary antibodies (1:1000 dilution each) diluted in PBS + 0.1% Tween-20. Preabsorption of the antibodies was performed using 10 μg/ml of the respective specific peptides or an irrelevant peptide for 30 min at room temperature under vigorous shaking. Membranes were then incubated for 2 hr at room temperature with secondary antibody diluted 1:10,000 in PBS + 0.1% Tween-20. Protein labeling was developed using enhanced chemiluminescence.
To detect pCFTR protein also in whole tissue lysates, 100 mg of all tissue samples was homogenized in 1 ml of standard lysis buffer with 1% protease inhibitors using a Precellys 24 homogenizer (peqlab Biotechnologie; Erlangen, Germany). Thirty μg of tissue lysates was subjected to immunoblot analyses. Lysate samples were also either boiled at 96C or incubated at 56C or 37C in standard 3-fold SDS-PAGE Laemmli buffer for 7 min and separated by SDS-PAGE. Immunoblot analysis was performed as described earlier. In addition, cell lysates from cultured Caco-2 and T84 cells were analyzed as described earlier.
Immunohistochemistry
Paraffin-embedded tissues were cut at 3 μm thickness, mounted on adhesive glass slides, and prepared as described previously (Leverkoehne and Gruber 2002). Frozen tissues were cut at 5 μm thickness, mounted on adhesive glass slides, and fixed in ice-cold acetone for 10 min, followed by blocking of endogenous avidin-binding sites (Dako; Carpinteria, CA). The slides were rinsed in PBS containing 0.05% Triton X-100 and blocked with PBS containing 2% BSA and 20% normal goat serum for 30 min. Anti-pCFTR antibodies, preimmune sera, three different antibodies from rabbits immunized with non-CFTR-related antigen or an irrelevant purified rabbit antibody (ecr10; Anton et al. 2005) were incubated overnight at 4C. Antibody dilutions ranged from 1:500 to 1:40,000 with optimal results for 1:10,000. The non-purified antibodies were preabsorbed using 0.25 mg/ml of the respective specific peptides or an irrelevant peptide for 1 hr at room temperature under vigorous shaking. In the preabsorption experiments, pCFTR-C-1 was diluted 1:15,000 in PBS + 2% BSA and pCFTR-N-1 was diluted 1:1500 in PBS. Sections were rinsed repeatedly in PBS containing 0.05% Triton X-100 and incubated for 1 hr with biotinylated secondary goat anti-rabbit antibody diluted 1:200. Color was developed by incubating the slides with ABC solution (VECTASTAIN Elite ABC Kit; Vector Laboratories, Burlingame, CA), followed by repeated washes and exposure to diaminobenzidine (Merck; Darmstadt, Germany). Sections were counterstained with hematoxylin, dehydrated in ascending graded ethanol, cleared in xylene, and coverslipped. Consecutive sections from each tissue sample were stained with hematoxylin and eosin for histological examination. All experiments were repeated at least two times in each animal.
Immunofluorescence and Confocal Laser Scanning Microscopy
For immunofluorescent analyses, slides were pretreated as described earlier, incubated with an antibody against the 58-kDa Golgi membrane protein (58K-9; 1:30 dilution, ACRIS antibodies, Herford, Germany) overnight, washed repeatedly in PBS containing 0.05% Triton X-100, and incubated with an FITC-conjugated secondary anti-mouse antibody (Dianova; Hamburg, Germany) for 60 min. After repeated washes, the slides were incubated with the anti-pCFTR antibodies diluted 1:100 overnight, followed by incubation with Dylight 549–conjugated secondary anti-rabbit antibody (Dianova) diluted 1:200 in PBS for 60 min. Sections were examined using a confocal microscope (TCS SP2; Leica Microsystems, Wetzlar, Germany), with FITC excited at 495 nm and emission of green fluorescence detected at 517 nm. Dylight 549 was excited at 555 nm and emission of red fluorescence was detected at 568 nm. Single optical scans were repeated several times and recorded separately to be subsequently combined as an average of three images.
Results
Bioinformatic Analyses
Species-specific variations in genomic structure may give rise to limited comparability of expression data observed for a specific molecule in different species. For example, entire gene duplications or silencing of genes have been observed in the pig but not in humans for the chloride channels, calcium-activated (CLCA) family of genes, which have been proposed as potential modulators of the CF phenotype (Plog et al. 2009). Our computational genomic analyses revealed no such differences in the pCFTR gene locus, including duplications, premature stop codons within the coding region, or other structural differences that could account for principally different expressions when compared with hCFTR (not shown).
To establish the relative evolutionary relationship between pCFTR and hCFTR, sequence data were compared with related sequences in the databases, and rudimentary phylogenetic analyses were conducted for 45 species based on the Ensembl genome database (http://www.ensembl.org/Homo_sapiens/Gene/Compara_Tree?db=core;g=ENSG00000001626;r=7:117127791-117127983;t=ENST00000454343). However, these data are either largely based on fragmentary sequences or deduced from bioinformatically defined exons, which turned out to be imperfect on closer inspection and resulted in unclear or artificial branching of the resulting phylogenetic trees (data not shown). We, therefore, recruited additional sequence information from the GenBank database and generated phylogenetic trees from the entire amino acid alignment of all mammals available. The resolution of trees was still unclear in certain branches, but was elucidated when we removed six hypervariable regions from the 1539-aa alignment and used the resulting 1199-aa alignment instead. The resulting trees reflect the prevalent evolutionary tree of species, with some minor discrepancies for the mammalian species (Figure 1). Sequence ambiguities within rodents and lagomorpha resulted in unexpected and inconsistent branch pattern, whereas primates, carnivores, and ungulates branch together. In this context, the large genetic distance of mouse and rat compared with the other eutherians is conspicuous. Whereas the 12 non-murine eutherians had at least 88.0% identity or 94.0% similarity (based on the BLOSUM62 matrix), the homology of mouse and rat revealed only 80.2% identity or 89.3% similarity to this species. The homology of rabbit and guinea pig to mouse and rat is below 80.2% identity or 89.3% similarity, whereas their homology to the other eutherians is higher than 88.0% identity or 94.0% similarity. Mouse and rat separate from the other eutherian species similarly to the birds examined, whereas the distance of all eutherians compared with analyzed fish species is consistently 55–59% identity or 73–77% similarity. Taken together, the data indicate a close evolutionary relationship between the pCFTR and hCFTR genes but a clear divergence in the mouse and rat when compared with other eutherians, indicating some diverging functional adaptation of the CFTR genes of the murinae branch during evolution but not in the pig.
Specificity of the pCFTR Antibodies
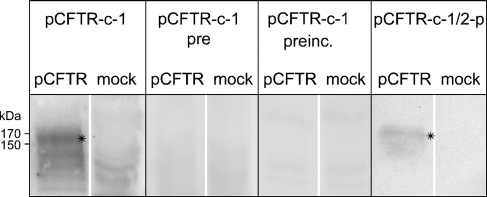
Cell lysates of transiently pCFTR-transfected HEK 293 cells were immunoblotted with antibodies pCFTR-C-1 or pCFTR-N-1. Antibody pCFTR-C-1 detected a specific band between 150 and 170 kDa (Figure 3), which was not detected with the preimmune serum. Preincubation with the peptide used for immunization abolished this band. In contrast, preincubation with an irrelevant peptide at the same concentration showed no reduction of the band (data not shown). The pooled and affinity-immunopurified antibodies pCFTR-C-1 and pCFTR-C-2, named pCFTR-C-1/2-p, only detected a specific band between 150 and 170 kDa, without any unspecific background.
Figure 3.
Anti-pCFTR antibodies detected an ∼170-kDa protein on immunoblots of transiently transfected HEK 293 cells. Antibody pCFTR-C-1 detected a specific band between 150 and 170 kDa (asterisks). This band was not detected in lysates from vector-alone-transfected cells (mock-transfected) or when the respective preimmune serum was used (pCFTR-C-1 pre), and the band disappeared when the antibody was preincubated with the respective peptide (pCFTR-C-1 preinc.). Immunopurified antibody pCFTR-C-1/2-p detected a similar protein between 150 and 170 kDa. All immune sera were diluted 1:1000.
Antibody pCFTR-N-1 failed to detect any specific bands in immunoblot experiments despite various technical variations and attempts (data not shown).
Expression Pattern of pCFTR mRNA
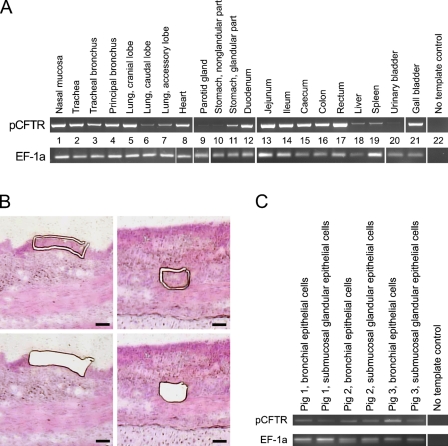
Detection of the pCFTR mRNA yielded strong signals in several tissues that were consistent in all animals examined (Figure 4A). Two repeated amplifications with each of the two primer pairs gave virtually identical results. Most prominent signals were observed in the upper respiratory tract including the nasal mucosa, trachea, and bronchi, with minor expression in the periphery of the lung. pCFTR mRNA was also consistently found in the gastrointestinal tract, including minor expression in the glandular part of the stomach. In addition, mRNA was detected in the parotid gland, pituitary gland, heart, gall bladder, liver, spleen, testicle, epididymis, uterus, and, weakly but consistently, in the esophagus, pancreas, urinary bladder, and non-glandular part of the stomach. Amplicons from the spleen, lung, heart, and gall bladder were sequenced, confirming specific detection of pCFTR. All other organs tested were negative, including the brain, the kidney, and all three regions from the skin. Parallel successful RT-PCR amplification of a fragment of the housekeeping gene EF-1a confirmed integrity of each cDNA sample used.
Figure 4.
Broad tissue expression of pCFTR mRNA detected by RT-PCR. (A) Strong expression of pCFTR mRNA was detected in the upper airways (Lanes 1–7), the heart (Lane 8), the glandular part of the stomach (Lane 11), all segments of the intestine (Lanes 12–17), and the gall bladder (Lane 21). pCFTR mRNA expression was also strongly detected in the uterus, testicle, and epididymis (not shown) and weakly detected in the parotid gland (Lane 9), the esophagus (not shown), the non-glandular part of the stomach (Lane 10), the urinary bladder (Lane 20), and the pancreas (not shown). All other organs were negative, including the brain, the kidney, and all three regions of the skin. A fragment of the mRNA coding for the housekeeping gene EF-1a was RT-PCR–amplified to control for RNA integrity and efficacy of reverse transcription. The RT-PCR results shown are representative for all animals examined. Lane 22: No template control. (B) Laser capture microdissection of bronchial epithelial cells (left panel) and submucosal glands (right panel). Cells are shown before (upper panel) and after (lower panel) dissection. Bar = 50 μm. (C) pCFTR mRNA was consistently detected by RT-PCR in bronchial epithelial cells and submucosal gland cells.
To establish whether the submucosal glands or respiratory epithelium contained pCFTR mRNA in the airways, we performed laser capture microdissection of bronchial tissue samples from three animals and tested each of these compartments separately (Figure 4B). The results show that the pCFTR mRNA was clearly present in both the respiratory epithelial cells and acini of submucosal glands, with a slightly lower expression in the latter (Figure 4C).
Tissue and Cellular Expression Patterns of the pCFTR Protein
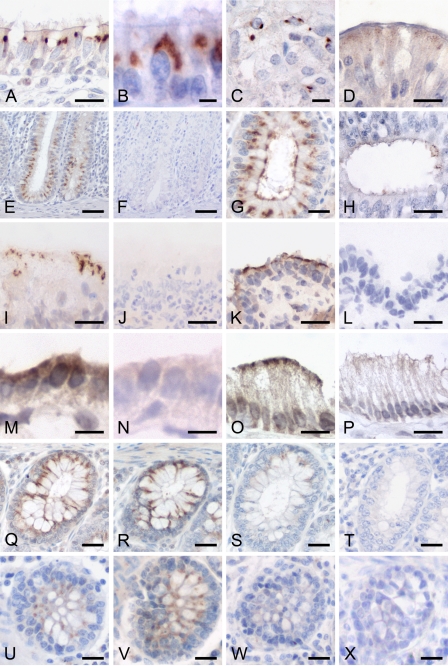
Immunohistochemical analyses using two different specific antibodies (Figure 2) performed on paraffin-embedded sections or on cryostate sections (Figure 5) yielded virtually identical results for both antibodies and for all animals tested. Signals obtained from the N-terminal antibody pCFTR-N-1 (Figure 5U) were similar to those obtained from pCFTR-C-1 and pCFTR-C-2 but weaker, and the antibody had to be used in a higher concentration. The non-purified antibodies were used in all immunohistochemical experiments because of substantial reduction in sensitivity after immunopurification. In the respiratory tract, strong and consistent signals were identified within ciliated epithelial cells and goblet cells in the nasal, tracheal, and bronchial mucosa (Figures 5A and 5B). The pCFTR protein was mostly detected intracellularly between the nucleus and the apical cell membrane. However, we consistently failed to detect the protein in any submucosal glands throughout the entire respiratory tract. No signals were observed within alveolar or bronchiolar epithelia.
Figure 5.
The pCFTR protein is expressed in several epithelial and glandular tissues. The pCFTR protein was detected on formalin-fixed, paraffin-embedded sections (A–H) or on cryostate sections (I–P, antibody pCFTR-C-1, 1:10,000 dilution) with a mainly intracytoplasmic staining pattern. In addition, especially, cryostate sections also had clear apical membrane staining, but cellular morphology and signal-to-noise ratio were clearly superior on formalin-fixed, paraffin-embedded tissues. Strong signals were found throughout the airways, including trachea (not shown) and large bronchi (A,B) and throughout the gastrointestinal tract from the cardiac glands of the glandular part of the stomach (C) and the duodenum (D) to all segments of the large intestine (E, colon; G, rectum). pCFTR protein was only weakly detected in the excretory ducts of the prostate gland (H). On cryostate sections, pCFTR was detected more apically, as seen in the nasal cavity (I) and the large bronchi (K). In addition, pCFTR was also found on cryostate sections within the excretory ducts of the parotid gland (M) and on the apical surface of some gall bladder epithelial cells (O). After preabsorption with the respective peptides used for immunization, signal intensities were abolished or markedly reduced (pCFTR-C-1, 1:15,000 dilution, S; pCFTR-N-1, 1:1500 dilution, W) when compared with non-absorbed antibodies (Q,U) or antibodies absorbed with an irrelevant peptide at the same concentration (R,V). Sections incubated with preimmune sera or four purified or non-purified immune sera from rabbits immunized with non-CFTR-related antigens at the same dilutions failed to develop any staining (F,J,L,N,P,T,X). Bars: E,F = 50 μm; Q–X = 25 μm; A,D,G–P = 10 μm; B,C = 5 μm.
In the intestine, the pCFTR protein was consistently detected in the cytosol between the nucleus and the apical membrane and less frequently associated with the apical cell membrane in the crypt epithelial cells of all intestinal segments with a distinct increase in signal intensities from small to large intestine and a decrease along the crypt–villus axis (Figures 5D, 5E, and 5G). Strong signals were observed in nearly all crypt epithelial cells of the colon and rectum, whereas crypt cells in the small intestine were only weakly and inconsistently stained. Both goblet cells and non-goblet cells of the intestinal crypts stained positive. In contrast to all other intestinal segments, the duodenum had only very faint and granular signals within non-goblet epithelial cells (Figure 5D). Brunner's glands were consistently negative. Signals were also observed within the cardiac glands of the glandular part of the stomach, which displayed a less intense and spottier appearance than the signals within the intestinal crypts (Figure 5C).
None of the reproductive organs had specific staining except for rare faint signals in epithelial cells of solitary excretory duct epithelia within the prostate gland (Figure 5H). All other organs tested, including the brain and the kidney, all three skin regions, and other CF-relevant organs such as the bile ducts, the pancreas, and the gall bladder were consistently negative for pCFTR protein on formalin-fixed, paraffin-embedded sections. Staining with preimmune sera or purified and non-purified immune sera from rabbits immunized with four different non-CFTR-related antigens confirmed specificity of the pCFTR signals. Moreover, after preincubation of the antibodies with their respective peptides, the signals disappeared or their intensities were strongly reduced (Figures 5S and 5W) compared with the non-preincubated antibodies (Figures 5Q and 5U) or with antibodies preincubated with an irrelevant peptide at the same concentration (Figures 5R and 5V).
Immunohistochemical pCFTR protein detection on cryostate sections yielded results very similar to paraffin-embedded tissues. However, tissue and cellular morphologies were much more poorly preserved. Of note, in addition to the supranuclear patterns seen in formalin-fixed, paraffin-embedded sections, the protein was more prominently detected in the apical cell membranes, ranging from spotty to diffuse membrane staining (Figures 5I, 5K, 5M, and 5O). Moreover, the protein was detected in few parotid duct cells and in the apical membranes of few gall bladder epithelial cells (Figures 5M and 5O). As in formalin-fixed, paraffin-embedded tissues, staining with preimmune sera or purified and non-purified immune sera from rabbits immunized with four different non-CFTR-related antigens confirmed specificity of the pCFTR signals.
Immunoblot Analyses on Tissue Samples
Whole tissue lysates from several organs, including the trachea, bronchi, lung, and segments of the small and large intestine, were subjected to immunoblot analyses, but none of the antibodies tested detected a specific pCFTR protein band in any of the tissues, despite strong and specific reactivity of the same antibodies in the immunohistochemical experiments and detection of specific bands in lysates of pCFTR-transfected HEK 293 cells. To test for crossreactivity of the antibodies with the hCFTR protein (GenBank accession number P13569), lysates from the human cell lines Caco-2 and T84 that express hCFTR (Denning et al. 1992) were also tested by immunoblotting. Again, none of our anti-pCFTR antibodies tested detected any specific protein band in the cell lysates of these cells (data not shown).
Confocal Laser Scanning Microscopy
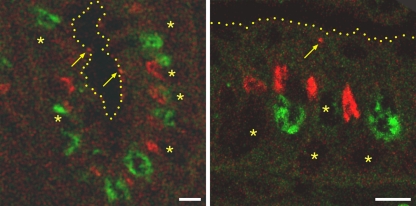
Subcellular immunohistochemical detection localized the majority of pCFTR to the apical cytosol with only rare signals in cell surface membranes (Figure 5). To better define this intracytoplasmic compartment, a confocal laser scan co-localization analysis was performed using the Golgi marker 58K. As shown exemplarily for the colon in Figure 6, significantly more crypt epithelial cells were labeled for the Golgi apparatus than for pCFTR (Figure 6, left panel), and pCFTR was found above the Golgi apparatus within the crypt epithelial cytosol (Figure 6, right panel).
Figure 6.
pCFTR is located apically to the Golgi apparatus and occasionally in the apical cell membrane. Using immunofluorescent detection on formalin-fixed, paraffin-embedded tissues, the pCFTR protein was detected in the cytosol of crypt epithelial cells, including goblet cells and non-goblet cells. In the majority of cells, the pCFTR protein (red) was detected apically to the Golgi marker 58K (green). Punctate signals were occasionally detected in or directly beneath the apical plasma membrane (arrows). Left panel, crypt epithelial cells in the colon; right panel, higher magnification of single cells; yellow dots, apical cell membranes; asterisks, locations of the nuclei. Antibody pCFTR-C-1, 1:100 dilution. Bar = 20 μm.
Discussion
Phenotyping and interpretation of the CF pig models that have recently been generated (Rogers et al. 2008b; Welsh et al. 2009; Meyerholz et al. 2010) and those that are currently being established by other groups will depend on a sound understanding of the function and cellular expression pattern of the pCFTR, which is the target molecule in all of these models. Our preparatory genetic studies failed to identify any evidence that the pCFTR genomic structure may be altered such that its expression would be fundamentally different from that of humans and other mammals. The goal of this study was, therefore, to systematically characterize the cellular and tissue distribution patterns of pCFTR and to compare them with those of hCFTR.
Despite its molecular cloning 20 years ago (Riordan et al. 1989; Rommens et al. 1989), the tissue and cellular expression patterns of the hCFTR protein are still far from being fully understood. Several studies using different antibodies, tissue sources, and processing techniques have resulted in partially contradictory results (see list of references in Table 2). It is generally accepted that among the most important reasons for this is the overall very low and varying expression level of the protein with only a few to a few hundreds of protein copies per cell (Crawford et al. 1991; Claass et al. 2000; Farinha et al. 2004). The expression data observed here for the pCFTR ortholog mirror these difficulties in that several tissues and cell types had borderline or no detectable protein expression, especially in immunoblot experiments with whole tissue lysates, whereas pCFTR mRNA was clearly detectable by RT-PCR. To corroborate the immunohistochemical staining patterns and to exclude the possibility of unspecific antibody interactions, two different sets of antibodies were generated here against distinct epitopes of the protein. Both sets of antibodies yielded virtually identical staining patterns on tissues, which, together with the other controls included here, argue for highly specific detection of the pCFTR protein in the experiments described. Rather low protein expression levels may be the cause for consistent lack of pCFTR protein detection on Western blots of whole tissue lysates performed in this study.
Table 2.
Comparison of porcine and human CFTR expression patterns
+, CFTR detection; (+), weak CFTR detection; −, no CFTR detection; ND, not determined.
Overall comparisons between the distribution patterns of the pCFTR and hCFTR proteins identified wide overlaps and similarities with only few obvious differences (Table 2). Concordant pCFTR protein and mRNA expression was observed only in the respiratory and intestinal tracts, which are of prime significance for the CF pathology. Virtually all respiratory epithelial cells, including goblet cells in the nasal mucosa, trachea, and bronchi, and crypt epithelial cells throughout the intestine contained pCFTR protein. Strong intestinal expression of pCFTR protein with emphasis on the ileum and colon explains the meconium ileus that newborn CF piglets develop, similar to infants (Rogers et al. 2008b). However, the cellular expression pattern of pCFTR in the intestine is at some variance with the reports by Hayden and Carey (1996), who detected the protein only in intestinal goblet cells but not in non-goblet cell enterocytes. This difference may again be attributed to different sensitivities of the antibodies used or different expression levels in the specific cell types. Decreasing expression levels along the crypt–villus axis are reminiscent of what has been observed for hCFTR (Strong et al. 1994), but increasing staining intensities along the oral–aboral axis are contrary to previous reports on hCFTR (Kälin et al. 1999). Importantly, both the intracellular, putatively vesicle-associated and the apical membrane pCFTR protein clearly had increasing staining intensities in the large intestine when compared with the small intestine (compare Figure 5D with Figure 5G). This seems paradoxical because in pigs, in contrast to humans, the majority of intestinal fluid is absorbed in the colon and the small intestine plays a dominant role in fluid secretion (Fordtran and Locklear 1966; Hamilton and Roe 1977). Our findings, thus, suggest that the regulation of secretion and absorption of intestinal fluids may be more complex in the pig than previously thought, and porcine CF models will have to be interpreted with caution in that respect.
Staining patterns in the apical cytosol between the Golgi apparatus and cell surface membrane, indicative of the protein residing in the trans-Golgi network or clathrin-coated vesicles, are consistent with the reports on hCFTR (Bradbury 1999; Kälin et al. 1999). Furthermore, the pCFTR signals obtained from goblet cells are clearly located between the nucleus and porcine goblet cell granule marker pCLCA1 (Plog et al. 2009). Unfortunately, no other antibodies were identified that could serve for co-localization studies of pCFTR with specific subcellular structures, mostly due to their lack of crossreactivity on pig tissues (data not shown). hCFTR is known to be stored in an intracellular vesicular pool (Guggino and Stanton 2006), and taking into account the location of the pCFTR protein apically to the Golgi and below the goblet cell granules, we have at present no reason to assume any differences between pCFTR and hCFTR in subcellular location. Importantly, the pCFTR protein was also occasionally associated directly with or just beneath the apical cell surface, albeit more prominently on cryostate sections, which apparently allowed for a slightly more sensitive detection. In contrast, however, we failed to identify a subpopulation with high amounts of pCFTR protein (CFTR high expressor cells) that has been described in rats (Ameen et al. 1999,2000).
Significantly lower expression levels undetected on formalin-fixed, paraffin-embedded tissues but readily seen on cryostate sections were present in the apical membranes of parotid salivary glands and gall bladder epithelium, consistent with hCFTR expression in these tissues in humans (Kartner et al. 1992; Dray-Charier et al. 1995). In contrast, several organs that consistently had pCFTR mRNA amplified by RT-PCR failed to yield any pCFTR protein signals on both paraffin-embedded and cryostate sections. Given similar scenarios reported in numerous previous studies on hCFTR (Table 2), such discrepancies are not surprising in swine. These organs include the pancreas, heart muscle, pituitary gland, spleen, and testicle in which we were unable to localize the protein to a specific cell type or subcellular compartment. pCFTR expression in pancreas and bile duct epithelium is again compatible with the observation that newborn CF pigs develop pancreatic destruction and focal biliary cirrhosis (Rogers et al. 2008b). Nevertheless, pancreatic expression seems to be too low to be detected immunohistochemically, and the expressing cell type and subcellular compartment in the swine pancreas could not be established here. Human heart also contains hCFTR mRNA at low levels, but the expressing cell type and functional significance are similarly unknown to date (Warth et al. 1996; Yajima et al. 1997). The presence of a specific isoform of hCFTR mRNA in the heart has been reported (Levesque et al. 1992; Horowitz et al. 1993), but it is apparently not associated with functional protein expression (Lader et al. 2000).
Of high significance for the understanding of the homeostasis of the airway surface liquid (ASL) and CF pathology is the detection of pCFTR mRNA in laser microdissected submucosal glands from airway epithelia in which the protein was undetectable by IHC. CFTR expression in submucosal glands and its role in regulation of the ASL have been controversially discussed in the literature. Several studies indicate that submucosal glands are the major sites of CFTR expression in human and murine airways (Engelhardt et al. 1992; Kälin et al. 1999; Doucet et al. 2003), where it is thought to control the composition and depth of the ASL (Verkman et al. 2003). It is widely accepted that the airway pathology in CF patients is largely due to CFTR dysfunction in these glands, and the different anatomy between submucosal airway glands in humans and those in mice has been proposed to contribute to the lack of airway pathology in mice (Scholte et al. 2004).
In another study, Kreda et al. (2005) reported inconsistent and weak staining of hCFTR protein in submucosal glands compared with the expression in ciliated epithelial cells of the respiratory tract. The study suggested a minor role for CFTR in airway gland acinar fluid secretion, and the CFTR expression in ciliated epithelia was assumed to be more important for the regulation of the ASL. Clunes and Boucher (2007) also argue that the surface epithelium represents the largest surface area in close contact with the ASL and represents the main site for modification of the ASL volume. In this study, pCFTR protein was detected in surface ciliated epithelia, but was completely absent in acinar cells of submucosal glands. This may point toward an important role of CFTR in the regulation of the ASL by ciliated epithelia rather than by submucosal glands. The minor role of CFTR in porcine submucosal glands has recently been suggested by Lee and Foskett (2009), according to whom alternative chloride channels play a more important role in porcine submucosal chloride secretion than CFTR, at least in an in vitro cell system. In any case, detection of pCFTR mRNA in both microenvironments points toward a significant role in both systems, albeit with different protein expression levels. Functional in situ studies or cell-type–specific silencing or overexpression may shed further light on this debate in the future. Interestingly, pCFTR is not expressed in the porcine skin although hCFTR is strongly expressed in the human skin, specifically in the apical membrane of secretory coil cells and apical and basolateral membranes of absorptive duct cells (Kartner et al. 1992). Increased chloride concentrations (>60 mmol/liter) in their sweat have been used to identify CF patients clinically for more than 50 years (di Sant'Agnese et al. 1953). Absence of pCFTR expression from porcine skin is apparently due to anatomic differences. Pigs use different ways of controlling body temperature, and this specific kind of eccrine sweat gland is absent in most areas of the porcine skin. However, even skin from the routing disk that contains similar glands in pigs was consistently negative for pCFTR mRNA and protein in all three pigs examined. This difference should be considered when diagnosing CF in pigs and when comparing the CF phenotype in pigs with the human disease.
Detection of pCFTR mRNA but no protein in the spleen is also unlike previous studies on hCFTR, which has not been reported to be expressed in the spleen. However, endothelial cells are known to contain hCFTR mRNA and protein at low levels (Tousson et al. 1998), and the mRNA signal obtained here could possibly be explained by endothelial cells that are abundantly present in the spleen. Endothelial cells, however, were consistently negative in all of our immunohistochemical detections in all tissues investigated.
As a further discrepancy, pCFTR was not detected in the kidney on mRNA or protein levels despite detection of hCFTR protein in human kidney tubules (Crawford et al. 1991). This is surprising because a severe decrease in membrane stability and a low molecular weight proteinuria have been reported in CF mouse models and CF patients (Jouret et al. 2006). Our negative results in the kidney may, thus, be due to limited sensitivity of our assays. Alternatively, we cannot exclude differences in pCFTR expression in the porcine kidney that may limit the comparability of porcine CF models with the human disease regarding the renal lesions. In any case, this possible difference may be of little significance given the overall limited importance of the renal alterations in CF patients (Devuyst and Guggino 2002).
Furthermore, neuronal cell expression of CFTR as observed in rat and human brains (Johannesson et al. 1997; Mulberg et al. 1998) and in human ganglion cells (Niu et al. 2009) was not confirmed for pCFTR in this study despite inclusion of several brain regions and thoracic ganglion cells. It remains unresolved whether the mRNA signal obtained from the pituitary gland stems from endocrine or neuronal cells or may be due to its high content of endothelial cells, as hypothesized for the spleen.
Clearly, the overall tissue and cellular distribution patterns of pCFTR are similar or identical with those of hCFTR in humans, particularly in CF-relevant organs. Apart from anatomic differences in cutaneous sweat glands, the many similarities identified here support the suitability of the pig as a promising new model to study the CF pathology and novel therapeutic interventions.
Acknowledgments
This study was supported by the German Research Council (Deutsche Forschungsgemeinschaft MU 3015/1-1).
We thank Lynda Ostedgaard and Michael J. Welsh for the donation of the pCFTR clone, Jana Enders and Ursula Kobalz for excellent technical support, Astrid Bethe, Institute of Microbiology and Epizootics, Berlin, for helping with the confocal laser scanning microscopic analyses, and Christoph Hanski, Charité Medical School, Berlin, for donating the Caco-2 and T84 cell lines.
This article is distributed under the terms of a License to Publish Agreement (http://www.jhc.org/misc/ltopub.shtml). JHC deposits all of its published articles into the U.S. National Institutes of Health (http://www.nih.gov/) and PubMed Central (http://www.pubmedcentral.nih.gov/) repositories for public release twelve months after publication.
References
- Ameen NA, Martensson B, Bourguinon L, Marino C, Isenberg J, McLaughlin GE (1999) CFTR channel insertion to the apical surface in rat duodenal villus epithelial cells is upregulated by VIP in vivo. J Cell Sci 112(Pt 6):887–894 [DOI] [PubMed] [Google Scholar]
- Ameen NA, van Donselaar E, Posthuma G, de Jonge H, McLaughlin G, Geuze HJ, Marino C, et al. (2000) Subcellular distribution of CFTR in rat intestine supports a physiologic role for CFTR regulation by vesicle traffic. Histochem Cell Biol 114:219–228 [DOI] [PubMed] [Google Scholar]
- Anton F, Leverkoehne I, Mundhenk L, Thoreson WB, Gruber AD (2005) Overexpression of eCLCA1 in small airways of horses with recurrent airway obstruction. J Histochem Cytochem 53:1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Carter EJ, Evans JR, Lawson V, Rowntree RK, Harris A (2007) CTCF mediates insulator function at the CFTR locus. Biochem J 408:267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury NA (1999) Intracellular CFTR: localization and function. Physiol Rev 79:S175–191 [DOI] [PubMed] [Google Scholar]
- Brezillon S, Dupuit F, Hinnrasky J, Marchand V, Kälin N, Tümmler B, Puchelle E (1995) Decreased expression of the CFTR protein in remodeled human nasal epithelium from non-cystic fibrosis patients. Lab Invest 72:191–200 [PubMed] [Google Scholar]
- Chow YH, Plumb J, Wen Y, Steer BM, Lu Z, Buchwald M, Hu J (2000) Targeting transgene expression to airway epithelia and submucosal glands, prominent sites of human CFTR expression. Mol Ther 2:359–367 [DOI] [PubMed] [Google Scholar]
- Claass A, Sommer M, de Jonge H, Kälin N, Tümmler B (2000) Applicability of different antibodies for immunohistochemical localization of CFTR in sweat glands from healthy controls and from patients with cystic fibrosis. J Histochem Cytochem 48:831–837 [DOI] [PubMed] [Google Scholar]
- Clunes MT, Boucher RC (2007) Cystic fibrosis: the mechanisms of pathogenesis of an inherited lung disorder. Drug Discov Today Dis Mech 4:63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DK, Ezzelarab M, Hara H, Ayares D (2008) Recent advances in pig-to-human organ and cell transplantation. Expert Opin Biol Ther 8:1–4 [DOI] [PubMed] [Google Scholar]
- Crawford I, Maloney PC, Zeitlin PL, Guggino WB, Hyde SC, Turley H, Gatter KC, et al. (1991) Immunocytochemical localization of the cystic fibrosis gene product CFTR. Proc Natl Acad Sci USA 88:9262–9266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson DJ, Rolfe M (2001) Mouse models of cystic fibrosis. Trends Genet 17:S29–37 [DOI] [PubMed] [Google Scholar]
- Denning GM, Ostedgaard LS, Cheng SH, Smith AE, Welsh MJ (1992) Localization of cystic fibrosis transmembrane conductance regulator in chloride secretory epithelia. J Clin Invest 89:339–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devuyst O, Guggino WB (2002) Chloride channels in the kidney: lessons learned from knockout animals. J Am Soc Nephrol 18:707–718 [DOI] [PubMed] [Google Scholar]
- di Sant'Agnese PA, Darling RC, Perera GA, Shea E (1953) Abnormal electrolyte composition of sweat in cystic fibrosis of the pancreas. Pediatrics 12:549–563 [PubMed] [Google Scholar]
- Doucet L, Mendes F, Montier T, Delepine P, Penque D, Ferec C, Amaral MD (2003) Applicability of different antibodies for the immunohistochemical localization of CFTR in respiratory and intestinal tissues of human and murine origin. J Histochem Cytochem 51:1191–1199 [DOI] [PubMed] [Google Scholar]
- Dray-Charier N, Paul A, Veissiere D, Mergey M, Scoazec JY, Capeau J, Brahimi-Horn C, et al. (1995) Expression of cystic fibrosis transmembrane conductance regulator in human gallbladder epithelial cells. Lab Invest 73:828–836 [PubMed] [Google Scholar]
- Dupuit F, Kälin N, Brézillon S, Hinnrasky J, Tümmler B, Puchelle E (1995) CFTR and differentiation markers expression in non-CF and delta F 508 homozygous CF nasal epithelium. J Clin Invest 96:1601–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink RO, Beuers U (2009) Are pigs more human than mice? J Hepatol 50:838–841 [DOI] [PubMed] [Google Scholar]
- Engelhardt JF, Yankaskas JR, Ernst SA, Yang Y, Marino CR, Boucher RC, Cohn JA, et al. (1992) Submucosal glands are the predominant site of CFTR expression in the human bronchus. Nat Genet 2:240–248 [DOI] [PubMed] [Google Scholar]
- Engelhardt JF, Zepeda M, Cohn JA, Yankaskas JR, Wilson JM (1994) Expression of the cystic fibrosis gene in adult human lung. J Clin Invest 93:737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha CM, Penque D, Roxo-Rosa M, Lukacs G, Dormer R, McPherson M, Pereira M, et al. (2004) Biochemical methods to assess CFTR expression and membrane localization. J Cyst Fibros 3(suppl 2):73–77 [DOI] [PubMed] [Google Scholar]
- Fordtran JS, Locklear TW (1966) Ionic constituents and osmolality of gastric and small-intestinal fluids after eating. Am J Dig Dis 11:503–521 [DOI] [PubMed] [Google Scholar]
- Guggino WB, Stanton BA (2006) New insights into cystic fibrosis: molecular switches that regulate CFTR. Nat Rev Mol Cell Biol 7:426–436 [DOI] [PubMed] [Google Scholar]
- Guilbault C, Saeed Z, Downey GP, Radzioch D (2007) Cystic fibrosis mouse models. Am J Respir Cell Mol Biol 36:1–7 [DOI] [PubMed] [Google Scholar]
- Gruber AD, Levine RA (1997) In situ assessment of mRNA accessibility in heterogeneous tissue samples using elongation factor-1 alpha (EF-1 alpha). Histochem Cell Biol 107:411–416 [DOI] [PubMed] [Google Scholar]
- Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98 [Google Scholar]
- Hamilton DL, Roe WE (1977) Electrolyte levels and net fluid and electrolyte movements in the gastrointestinal tract of weanling swine. Can J Comp Med 41:241–250 [PMC free article] [PubMed] [Google Scholar]
- Hayden UL, Carey HV (1996) Cellular localization of cystic fibrosis transmembrane regulator protein in piglet and mouse intestine. Cell Tissue Res 283:209–213 [DOI] [PubMed] [Google Scholar]
- Hihnala S, Kujala M, Toppari J, Kere J, Holmberg C, Hoglund P (2006) Expression of SLC26A3, CFTR and NHE3 in the human male reproductive tract: role in male subfertility caused by congenital chloride diarrhoea. Mol Hum Reprod 12:107–111 [DOI] [PubMed] [Google Scholar]
- Horowitz B, Tsung SS, Hart P, Levesque PC, Hume JR (1993) Alternative splicing of CFTR Cl− channels in heart. Am J Physiol 264:H2214–2220 [DOI] [PubMed] [Google Scholar]
- Jacquot J, Puchelle E, Hinnrasky J, Fuchey C, Bettinger C, Spilmont C, Bonnet N, et al. (1993) Localization of the cystic fibrosis transmembrane conductance regulator in airway secretory glands. Eur Respir J 6:169–176 [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405 [DOI] [PubMed] [Google Scholar]
- Johannesson M, Bogdanovic N, Nordqvist AC, Hjelte L, Schalling M (1997) Cystic fibrosis mRNA expression in rat brain: cerebral cortex and medial preoptic area. Neuroreport 8:535–539 [DOI] [PubMed] [Google Scholar]
- Jouret F, Bernard A, Hermans C, Dom G, Terryn S, Leal T, Lebecque P, et al. (2006) Cystic fibrosis is associated with a defect in apical receptor-mediated endocytosis in mouse and human kidney. J Am Soc Nephrol 18:707–718 [DOI] [PubMed] [Google Scholar]
- Kälin N, Claass A, Sommer M, Puchelle E, Tümmler B (1999) DeltaF508 CFTR protein expression in tissues from patients with cystic fibrosis. J Clin Invest 103:1379–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartner N, Augustinas O, Jensen TJ, Naismith AL, Riordan JR (1992) Mislocalization of delta F508 CFTR in cystic fibrosis sweat gland. Nat Genet 1:321–327 [DOI] [PubMed] [Google Scholar]
- Klymiuk N, Müller M, Brem G, Aigner B (2006) Phylogeny, recombination and expression of porcine endogenous retrovirus gamma2 nucleotide sequences. J Gen Virol 87(Pt 4):977–986 [DOI] [PubMed] [Google Scholar]
- Kreda SM, Mall M, Mengos A, Rochelle L, Yankaskas J, Riordan JR, Boucher RC (2005) Characterization of wild-type and deltaF508 cystic fibrosis transmembrane regulator in human respiratory epithelia. Mol Biol Cell 16:2154–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lader AS, Wang Y, Jackson GR Jr, Borkan SC, Cantiello HF (2000) cAMP-activated anion conductance is associated with expression of CFTR in neonatal mouse cardiac myocytes. Am J Physiol Cell Physiol 278:C436–450 [DOI] [PubMed] [Google Scholar]
- Lee RJ, Foskett KJ (2009) Mechanisms of Ca2+-stimulated fluid secretion by porcine bronchial submucosal gland serous acinar cells. Am J Physiol Lung Cell Mol Physiol 298:L210–231 [DOI] [PubMed] [Google Scholar]
- Leonhard-Marek S, Hempe J, Schroeder B, Breves G (2009) Electrophysiological characterization of chloride secretion across the jejunum and colon of pigs as affected by age and weaning. J Comp Physiol B 179:883–896 [DOI] [PubMed] [Google Scholar]
- Leverkoehne I, Gruber AD (2002) The murine mCLCA3 (alias gob-5) protein is located in the mucin granule membranes of intestinal, respiratory and uterine goblet cells. J Histochem Cytochem 50:829–838 [DOI] [PubMed] [Google Scholar]
- Levesque PC, Hart PJ, Hume JR, Kenyon JL, Horowitz B (1992) Expression of cystic fibrosis transmembrane regulator Cl− channels in heart. Circ Res 71:1002–1007 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang Y, Jiang Y, Zhu N, Liang H, Xu L, Feng X, et al. (2008) Mild processing defect of porcine DeltaF508-CFTR suggests that DeltaF508 pigs may not develop cystic fibrosis disease. Biochem Biophys Res Commun 373:113–118 [DOI] [PubMed] [Google Scholar]
- Mendes F, Doucet L, Hinzpeter A, Ferec C, Lipecka J, Fritsch J, Edelman A, et al. (2004) Immunohistochemistry of CFTR in native tissues and primary epithelial cell cultures. J Cyst Fibros 3(suppl 2):37–41 [DOI] [PubMed] [Google Scholar]
- Meyerholz DK, Stoltz DA, Pezzulo AA, Welsh MJ (2010) Pathology of gastrointestinal organs in a porcine model of cystic fibrosis. Am J Pathol 176:1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulberg AE, Weyler RT, Altschuler SM, Hyde TM (1998) Cystic fibrosis transmembrane conductance regulator expression in human hypothalamus. Neuroreport 9:141–144 [DOI] [PubMed] [Google Scholar]
- Niu N, Zhang J, Guo Y, Yang C, Gu J (2009) Cystic fibrosis transmembrane conductance regulator expression in human spinal and sympathetic ganglia. Lab Invest 89:636–644 [DOI] [PubMed] [Google Scholar]
- Ostedgaard LS, Rogers CS, Dong Q, Randak CO, Vermeer DW, Rokhlina T, Karp PH, et al. (2007) Processing and function of CFTR-DeltaF508 are species-dependent. Proc Natl Acad Sci USA 104:15370–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrizio P, Salameh WA (1998) Expression of the cystic fibrosis transmembrane conductance regulator (CFTR) mRNA in normal and pathological adult human epididymis. J Reprod Fertil Suppl 53:261–270 [PubMed] [Google Scholar]
- Plog S, Mundhenk L, Klymiuk N, Gruber AD (2009) Genomic, tissue expression and protein characterization of pCLCA1, a putative modulator of cystic fibrosis in the pig. J Histochem Cytochem 57:1169–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier A, Dannhoffer L, Blouquit-Laye S, Bakari M, Naline E, Chinet T (2008) Expression of cystic fibrosis transmembrane conductance regulator in the human distal lung. Hum Pathol 39:368–376 [DOI] [PubMed] [Google Scholar]
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, et al. (1989) Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245:1066–1073 [DOI] [PubMed] [Google Scholar]
- Rogers CS, Abraham WM, Brogden KA, Engelhardt JF, Fisher JT, McCray PB Jr, McLennan G, et al. (2008a) The porcine lung as a potential model for cystic fibrosis. Am J Physiol Lung Cell Mol Physiol 295:L240–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, Rogan MP, et al. (2008b) Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science 321:1837–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommens JM, Zengerling-Lentes S, Kerem B, Melmer G, Buchwald M, Tsui LC (1989) Physical localization of two DNA markers closely linked to the cystic fibrosis locus by pulsed-field gel electrophoresis. Am J Hum Genet 45:932–941 [PMC free article] [PubMed] [Google Scholar]
- Scholte BJ, Davidson DJ, Wilke M, De Jonge HR (2004) Animal models of cystic fibrosis. J Cyst Fibros 3(suppl 2):183–190 [DOI] [PubMed] [Google Scholar]
- Strong TV, Boehm K, Collins FS (1994) Localization of cystic fibrosis transmembrane conductance regulator mRNA in the human gastrointestinal tract by in situ hybridization. J Clin Invest 93:347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XC, McCutheon C, Bertram P, Xie Q, Bonanno JA (2001) Studies on the expression of mRNA for anion transport related proteins in corneal endothelial cells. Curr Eye Res 22:1–7 [DOI] [PubMed] [Google Scholar]
- Tizzano EF, Chitayat D, Buchwald M (1993) Cell-specific localization of CFTR mRNA shows developmentally regulated expression in human fetal tissues. Hum Mol Genet 2:219–224 [DOI] [PubMed] [Google Scholar]
- Tizzano EF, Silver MM, Chitayat D, Benichou JC, Buchwald M (1994) Differential cellular expression of cystic fibrosis transmembrane regulator in human reproductive tissues. Clues for the infertility in patients with cystic fibrosis. Am J Pathol 144:906–914 [PMC free article] [PubMed] [Google Scholar]
- Tousson A, Van Tine BA, Naren AP, Shaw GM, Schwiebert LM (1998) Characterization of CFTR expression and chloride channel activity in human endothelia. Am J Physiol 275:C1555–1564 [DOI] [PubMed] [Google Scholar]
- Verkman AS, Song Y, Thiagarajah JR (2003) Role of airway surface liquid and submucosal glands in cystic fibrosis lung disease. Am J Physiol Cell Physiol 284:C2–15 [DOI] [PubMed] [Google Scholar]
- Warth JD, Collier ML, Hart P, Geary Y, Gelband CH, Chapman T, Horowitz B, et al. (1996) CFTR chloride channels in human and simian heart. Cardiovasc Res 31:615–624 [PubMed] [Google Scholar]
- Welsh MJ, Rogers CS, Stoltz DA, Meyerholz DK, Prather RS (2009) Development of a porcine model of cystic fibrosis. Trans Am Clin Climatol Assoc 120:149–162 [PMC free article] [PubMed] [Google Scholar]
- Wills NK, Weng T, Mo L, Hellmich HL, Yu A, Wang T, Buchheit S, et al. (2000) Chloride channel expression in cultured human fetal RPE cells: response to oxidative stress. Invest Ophthalmol Vis Sci 41:4247–4255 [PubMed] [Google Scholar]
- Yajima T, Nagashima H, Tsutsumi-Sakai R, Hagiwara N, Hosoda S, Quertermous T, Kasanuki H, et al. (1997) Functional activity of the CFTR Cl− channel in human myocardium. Heart Vessels 12:255–261 [DOI] [PubMed] [Google Scholar]
- Zheng XY, Chen GA, Wang HY (2004) Expression of cystic fibrosis transmembrane conductance regulator in human endometrium. Hum Reprod 19:2933–2941 [DOI] [PubMed] [Google Scholar]