Abstract
Reaction monitoring mass spectrometry has emerged as a powerful tool for targeted detection and quantification of proteins in clinical samples. Here, we report the use of gel electrophoresis for protein fractionation and liquid chromatography coupled to multiple reaction monitoring mass spectrometry (LC-MRM) screening for quantitative analysis of components from the Wnt/β-catenin signaling pathway, which contributes to colon tumor formation and progression. In silico tools are used to design LC-MRM screens for each target protein. Following successful peptide detection, stable isotope labeled peptides are synthesized and developed as internal standards. Then, the assays are implemented in colon cancer cell lines to achieve detection in minimal amounts of cells, compatible with direct translation to clinical specimens. Selected assays are compared with qualitative results from immunoblotting (Westerns) and translated to individual frozen colon tissue sections and laser capture microdissected tumor cells. This LC-MRM platform has been translated from in vitro models to clinical specimens, forming the basis for future experiments in patient assessment.
Keywords: Wnt/β-catenin signaling, colon cancer, frozen tissue, laser capture microdissection, liquid chromatography coupled to multiple reaction monitoring, biomarker assessment
Introduction
The Wnt signaling pathway is dysregulated in most colon cancer tumors primarily due to mutations occurring in either the Adenomatous Polyposis Coli (APC) protein or in β-catenin that disrupt the degradation of β-catenin.,1–6 Evaluation of 27 APC wild type colon cell lines for mutations in other canonical Wnt pathway genes, showed that 13 of the cell lines contained mutations in the β-catenin gene,7 and that mutations in the β-catenin regulatory domain and APC are mutually exclusive. However, this pathway can also dysregulated by upregulation or downregulation of protein expression.8 In normal cells, excess β-catenin is phosphorylated and targeted for proteasome degradation through the destruction complex, which includes the interacting proteins: APC, Axin, Glycogen Synthase Kinase 3β (GSK3β), and Casein Kinase I.9,10 Axin is required to recruit β-catenin to the destruction complex,11 but once APC is phosphorylated by GSK3β, β-catenin has a higher affinity for APC.12–14 β-Catenin is then ubiquitinated and targeted for degradation by the proteasome.15–18 Truncation of the APC protein within the mutational cluster region (MCR) results in the loss of the β-catenin binding site, thus preventing the phosphorylation of β-catenin.19 When β-catenin reaches sufficient levels in the cytoplasm, β-catenin is recruited to the nucleus where it binds to the TCF/LEF transcription factors that are bound to Wnt response elements,20,21 displacing the co-repressors bound to TCF/LEF.22–24 The destruction complex is reset when protein phosphatase 2A (PP2A) dephosphorylates APC.25,26 While post-translational modification are important molecular switches for driving these processes, the overall outcome can be measured by the expression of β-catenin. Because the accumulation of β-catenin mediates its tumorigenic activity, this pathway presents an optimal model system to illustrate the value of monitoring protein expression.
The expression levels of additional proteins in this pathway, particularly those that transcriptionally regulated by β-catenin, also have biological relevance and potential clinical significance. For example, transcriptional dysregulation causing increased expression of Cyclin D127 and Myc28,29 produces many downstream effects required for tumor development and progression. Increase expression of matrix metalloproteases-7 driven by β-catenin also has implications in invasion and metastasis.30 Elevated levels of cortactin are correlated with more aggressive colorectal cancer and poor survival.31 In contrast, reduced levels of Axin are correlated with poor prognosis in esophageal and lung cancer.32,33 Modulation of Src occurs early in colon cancer development; increases in expression34,35 and activation36,37 both correlate with tumor progression and poor prognosis. Thus, important gains in patient assessment could be made by simultaneous quantitative measurements of the expression of β-catenin interacting proteins, particularly because several of these proteins do not have prognostic value as individual biomarkers.38
While Wnt signaling has known biological and clinical relevance, its significance in patient treatment is increasing as inhibitors are developed for Wnt pathway proteins. In addition to existing and emerging targeted therapeutics for Cyclin D1, Myc, and Src, Ionomycin and Quercetin have both reported to downregulate β-catenin/Tcf signaling in colon cancer cell lines.39,40 Isoreserpine treatment enhances degradation of β-catenin.41 Nonsteroidal anti-inflammatory drugs42 and mesalazine43 have been proposed as a long term chemopreventive agent for patients with inflammatory bowel disease, as they effect inhibition of PP2A activity and decreases nuclear β-catenin as well as transcription of downstream proteins. Additional small molecule inhibitors have been shown to block the interaction between β-catenin and TCF/LEF both in vitro and in gene expression assays44 Taken together, these observations strongly support in situ analysis of Wnt pathway proteins as a relevant approach for examining colon cancer biology, determining relevant protein targets for pharmacological intervention, and evaluating the prognostic value of Wnt pathway proteins as prognostic biomarkers.
Numerous options are currently available for quantitative analysis of protein expression. In prior studies, detection and quantification of β-catenin is typically achieved using antibody-based techniques.45–48 Traditional immunoblotting (Western analysis) enables qualitative assessment of protein expression with limitations in the number of targets and total protein required. Typically, 0.5–1×106 cells producing 50 to 100 μg of total protein are lysed for each Western blot and one protein is then detected within a given molecular weight range (without stripping and reprobing). Selected antibodies can be translated to tissue samples for immunohistochemical staining to localize the protein and assess expression in different samples. Antibody-based techniques that can provide quantification include: enzyme linked immunosorbent assays (ELISA), Western analysis with two color near-infrared fluorescence imagers (e.g. Li-Cor Odyssey),49,50 antibody arrays51,52,53, and protein arrays printed on nitrocellulose54 or reverse phase media.55,56 The first two methods are limited in the number of analytes that can be simultaneously assessed; however, ELISA is typically used for absolute quantification. Most commercially available antibody arrays currently assess less than 100 proteins, but antibody microarrays that can measure several hundred proteins are now available. Reverse phase protein arrays enable high throughput parallel sample analysis with individual antibodies.
Quantitative mass spectrometry, specifically reaction monitoring, provides a complementary methodology for protein quantification that does not require antibodies. The coupling of liquid chromatography and multiple reaction monitoring mass spectrometry (LC-MRM) enables selective detection of proteolytic peptides using their reverse phase retention time, precursor (intact) mass-to-charge ratio, and selected fragment ion mass-to-charge ratios. Quantification is achieved by integration of the peak areas; data can be normalized from run-to-run using internal standards for relative quantification or to stable isotope labeled synthetic peptides spiked at known amounts for absolute quantification (AQUA).57,58 The versatility of LC-MRM analysis has recently been demonstrated in the analysis of protein post-translational modifications such as phosphorylation 59,60 and oxidation,61 protein complexes,62 cellular extracts,63 and clinical samples.64,65 The combination of liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and LC-MRM has also recently been illustrated to enable comparison of abundant proteins and cancer-related targets. 66
LC-MRM holds promise for elucidation of biology and evaluation of clinical samples; therefore, a description of assay development and translation to patient specimens as well as comparison to currently available methods will be critical for its early evaluation as a research tool. Data are presented to illustrate the translation of LC-MRM analysis from cell lines to clinical specimens and enables comparisons to antibody-based techniques. To enable comparisons with Western blotting and build on the utility of SDS-PAGE coupled to LC-MS/MS or GeLC-MS/MS,67–70 the coupling of SDS-PAGE and LC-MRM is implemented for quantification of protein components of Wnt/β-catenin signaling pathway. Peptides were screened using total protein extracted from 50,000 cells and fractionated by SDS-PAGE. Selected assays are compared to Western Blots to examine concordance between the different measurement methods. The assays are evaluated for reproducibility using biological replicates, and the limits of detection and quantification are assessed using dilution series. Assays are translated to individual frozen sections of tumor and normal tissue as well as laser capture microdissected tumor cells;71–73 this process is illustrated with the data from four protein targets: β-catenin, protein phosphatase 2A catalytic subunit, c-Src, and c-Myc. These data are used to illustrate the utility of the platform and discuss its translation from cell lines to patient samples.
Experimental
Reagents
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO) at their highest available purity, unless otherwise noted. HPLC solvents (water and acetonitrile) were supplied by Burdick and Jackson (Honeywell, Muskegon, MI). Antibodies were purchased from BD Transduction Laboratories (β-catenin #610153, protein phosphatase 2A #610555, α-catenin #610193, cortactin #610049), Millipore (c-Src #05-184), Santa Cruz Biotechnologies (c-Myc #sc-40), Abcam (CD44 #ab51037) and Jackson Immunoresearch Laboratories, Inc (Donkey anti-Mouse #715-035-150, donkey anti-rabbit #711-035-152). These antibodies were selected based on availability and their reliable performance in our prior experience. Supersignal West Pico Chemiluminescent (Thermo Scientific, #34080) was used for developing western blots.
Cell Culture, Lysis, and Sample Preparation for Mass Spectrometry
HCT116, HT29, KM12, and SW620 colon adenocarcinoma cell lines were purchased from DCTD Tumor Cell Line Repository (NCI at Frederick, MD). The SW480 colon adenocarcinoma cell line was purchased from ATCC (American Type Culture Collection, Manassas, VA). The KM12C, KM12L4A and KM12SM colon adenocarcinoma cell lines were obtained from MD Anderson Cancer Center (from the laboratory of Dr. I. Fidler).74 Cell lines are cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, and incubated in a 5% CO2 atmosphere at 37 °C.
For LC-MRM screens, colon cancer cell lines, HCT116 and HT29, were used to predict and monitor the proteins in Wnt signaling pathway. Ten million cells were harvested from each cell line, and washed twice with cold PBS. The cell pellets were lysed in aqueous 8 M urea/100 mM ammonium bicarbonate buffer on ice, sonicated, and centrifuged at 14,000 × g for 10 minutes. The resulting supernatant (corresponding to 100,000 cells) was denatured by boiling in SDS-PAGE loading buffer and separated in 10% Bis-Tris gels (Criterion XT, Biorad, Hercules, CA) for 30 minutes, and then visualized with colloidal Coomassie Brilliant Blue G-250 (Bio-Rad, Hercules, CA). Based on the predicted migration of the target proteins, the gel lane was excised into 8 bands. Target proteins inside each excised gel band were reduced with tris(2-carboxyethyl)phosphine (2 mM), alkylated with iodoacetamide (20 mM), and digested with sequencing grade trypsin (Promega, Madison, WI) at 37°C overnight. The digested peptide solutions were concentrated by vacuum centrifugation, and then resuspended in 20 μl 2% acetonitrile with 0.1% formic acid (LC solvent A) for mass analysis. Both adjacent gel sections were subjected to screening for peptides of the target proteins expected in each region of the gel.
For protein quantification in cell lines, protein extracts from 200,000 cells of KM12, KM12C, KM12SM, KM12L4A, SW480, and SW620 were fractionated by SDS-PAGE, digested in-gel, and analyzed with LC-MRM. Protein extracted from 50,000 cells was injected for each analysis. The appropriate concentration of each internal standard has been decided by calibration curves before spiking into the biological samples (data published online at http://proteome.moffitt.org/QUAD).
For serial dilution quantification experiments in the cell line SW480, the equivalent of 400, 2,000, 4,000, 20,000, 40,000, 100,000 cells were fractionated by SDS-PAGE and 25% of each sample was analyzed in each LC-MRM measurement.
Western Blot Analysis
Cell lysates were prepared in a urea buffer containing 8 M urea and 100 mM ammonium bicarbonate. Protein extracts prepared from an equal number of cells for each cell line were separated by SDS-PAGE and transferred to nitrocellulose membrane. Membranes were blocked by PBS containing 0.05% tween-20, 5% powdered milk for 1 hour and probed with primary antibodies overnight in blocking solution. The primary antibodies targeted the following proteins: β-catenin (diluted 1:5000), c-Src (diluted 1:500), c-Myc (diluted 1:500), PP2A catalytic subunit (diluted 1:10,000), CD44 (diluted 1:5000), α-catenin (diluted 1:500), and cortactin (diluted 1:2000). Blots were washed by PBS containing 0.05% Tween-20 and probed for 2 hours with the appropriate secondary antibody conjugated with horseradish peroxidase diluted in blocking buffer (Donkey anti-Mouse, diluted 1:15,000 and donkey anti-rabbit, diluted 1:3000). Blots were washed by PBS containing 0.05% Tween-20, developed and exposed to film.
Frozen Tissue Sectioning, LCM, and Sample Preparation for Mass Spectrometry
After review by a board certified pathologist (AN/MB), both adenocarcinoma of the colon and normal tissue were examined from the same patients (n = 3). Whole colon tissue sections (25 μm thick) imbedded in optimal cutting temperature medium (OCT) were washed with 200 μl 95% ethanol, shaken for 10 min at ambient temperature, and centrifuged at 14,000 × g for 2 minutes at 20°C. After removal of the supernatant, the tissue sections are homogenized in 30 μl 8 M urea in 100 mM aqueous ammonium bicarbonate. The same steps described above in sample preparation for cell lines are used here. Tissue homogenate (containing 28 μg protein as measured by Bradford assay) is loaded on 10% SDS-PAGE gels prior to band excised and protein digestion.
From a different group of colon cancer patients (n = 3), frozen sections (5 μm thick) are mounted on uncharged/uncoated glass slides for laser capture microdissection (LCM). Subsequent dehydration is performed using graded alcohols and xylene treatments as follows: 100% ethanol for 30 seconds, 70% ethanol for 30 seconds, deionized water for 5–10 dips in a coplin jar, hematoxylin for 1 minute, then rinsed lightly until the water runs clear, 70% ethanol for 30 seconds, 95% ethanol for 5–10 dips, 100% ethanol for 5–10 dips then 30 seconds, and xylene for 2 minutes (2 times). Slides are then dried in a hood prior to LCM (AutoPix LCM system, Arcturus, Sunnyvale, CA). Based on the size of the tissue section, 25,000 shots using the 23.2 μm diameter near infrared laser beam are utilized to obtain cells on Macro LCM Caps (Arcturus CapSure, #09F10A) for each sample. After microdissection, LCM caps are inserted into an Eppendorf tube containing 40 μL of 2% SDS, 10% glycerol, 2% 2-mercaptoethanol, 0.02% bromophenol blue, 50 mM Tris HCl buffer (pH 6.8). The tube is placed upside down and incubated at 4 °C for 1 hour. After removing the cap, the thermoplastic resin is obtained from the LCM cap using forceps and placed into the sample solution, which is then sonicated and centrifuged at 14,000 × g for 10 minutes. The supernatant is denatured by boiling and separated by 10% Criterion XT Bis-Tris Gel for 30 minutes. The gel bands corresponding to the proteins of interest were systematically excised (as shown in Supplementary Figure 1), reduced, alkylated, and digested. To compare LCM results to results from manually macrodissected tissue, the same region of the dehydrated tissue section was collected using a #21 scalpel (Personna Medical, #73-0121) and processed using the same steps as tissue samples, as described above.
Peptide Selection
Prediction of target peptides and transitions is performed with Pinpoint (Thermo, San Jose, CA) or Skyline, which has been developed by MacCoss and coworkers.75 Doubly charged peptides within the lengths from 7 to 25 amino acids are selected; peptides containing the redox reactive residues, cysteine and methionine, are typically excluded. Initial LC-MRM screens are set to detect all y fragment ions starting from y3 or yx > peptide m/z and ending with y(n−1). Methods are developed for Xcalibur 2.0 SR2 and TSQ Quantum 1.4 software versions.
Liquid Chromatography coupled to Multiple Reaction Monitoring
A nanoLC system (either U3000, Dionex, Sunnyvale, CA or EasynLC, Proxeon, Denmark) is connected to a triple quadrupole mass spectrometer (TSQ Quantum Ultra, Thermo, San Jose, CA) equipped with a nanoelectrospray ion source operated in the positive ion mode. Reverse phase C18 trap columns with 300 μm inner diameter and 0.5 cm length and C18 analytical columns with 75 μm inner diameter, 15 cm length, 3 μm particle size, and 100 Å pore size (PepMap100, Dionex, Sunnyvale, CA) with are used for chromatography. The solvent system includes aqueous 2% acetonitrile with 0.1% formic acid (solvent A) and aqueous 90% acetonitrile with 0.1% formic acid (solvent B). Samples (5 μl) are loaded on the trapping column at 6 μL/min and desalted by washing with solvent A for 5 minutes. The LC gradient from 5 to 50% solvent B is delivered at 300nl/min over 35 minutes. MRM settings include 2,500 V nanoelectrospray from 10 μm tips (New Objective, Woburn, MA) with 200°C transfer tube temperature, and 12 V skimmer offset. The Q1 is set to filter m/z 0.2 in width, and Q3 is set to filter m/z 0.7 in width. Fragmentation is obtained with 1.5 mTorr argon. Each transition is monitored for 20 milliseconds.
Synthesis of Peptide Standards
After LC-MRM screening, the best peptide candidates are selected by intensity and number of transitions for further evaluation. Assays are developed for peptides with the fewest predicted interferences for both the biological and stable isotope labeled peptide as determined using an in-house software, In silico Peptide Interference Predictor, 76 described in the supplementary materials. Ease of synthesis for each sequence is also a consideration. Peptide standards are created at the 25 micromole scale using standard FMOC chemistry (Symphony, Protein Technologies, Tucson, AZ). Matrix assisted laser desorption MS establishes peptide purity and label incorporation prior to sequence verification with tandem time-of-flight MS/MS (4700, Applied Biosystems, Framingham, MA).
Results and Discussion
LC-MRM Screening for the Components in Wnt/β-catenin Signaling Pathway
LC-MRM assays were developed for twenty-two proteins involved in the Wnt/β-catenin signaling pathway, including components in structural interactions (E-cadherin, β-catenin, α-catenin, and c-Src), the β-catenin regulatory/destruction complex (Axin1, APC protein, GSK3β, PP2A catalytic subunit, PP2A 65kDa regulatory subunit A α, PP2A 65kDa regulatory subunit A β, PP2A 55kDa regulatory subunit B α, Casein kinase I, Casein kinase II α, Casein kinase II α′, Casein kinase II β), regulation of transcription (TCF7), and downstream protein expression (CD44, Cyclin D1, MMP-7, c-Myc, Cortactin, Axin2). These proteins and peptides are then worked up as targets for LC-MRM assays monitoring components of the Wnt/β-catenin signaling pathway as shown in Figure 1 and Supplementary Figures 2–4. Analysis of eight gel bands (for the excision template see Supplementary Figure 1) enabled the detection of proteotypic peptides in the cell lysate (from 25,000 of either HCT116 or HT29 colon cancer cells). Four or more transitions were detected and subsequently monitored for each target peptide. Quantification is achieved using the sum of the peak areas for all transitions.
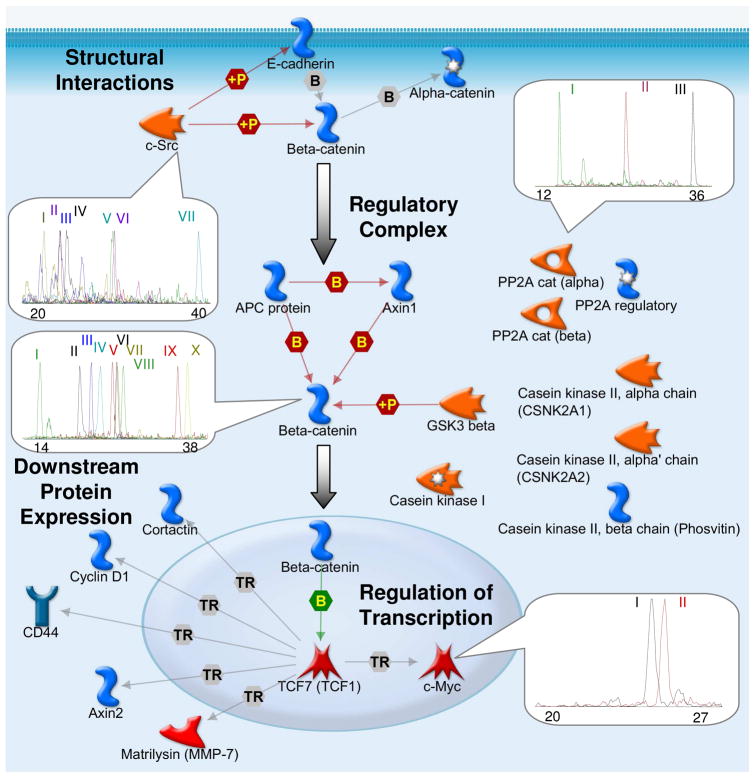
Figure 1. LC-MRM Screening to Select Peptides for Assay Development to Monitor Wnt/β-catenin Signaling Components and Downstream Protein Expression.
A GeneGO pathway map is shown to illustrate the interactions between the selected proteins. The results of LC-MRM screens used to select peptide candidates for quantitative assay development are shown for four selected proteins, β-catenin, c-Src, c-Myc, and the catalytic subunit of PP2A. Each extracted ion chromatogram plot shows peptide signal over time. Using these data, one peptide standard is developed for quantification, but all peptides detected in these screens are monitored in biological experiments. The Roman numerals link to the peptide sequences listed in Table 1.
As shown in Table 1, a minimum of one peptide standard was synthesized for each protein with either stable isotope labels or single amino acid replacement. The other peptides detected in these screens are also monitored in all biological experiments. For example, ten peptides passed the initial LC-MRM screening for β-catenin: HQEAEMAQNAVR (I), AIPELTK (II), LLNDEDQVVVNK (III), SGGIPALVK (IV), VAAGVLCELAQDK (V), LHYGLPVVVK (VI), LLWTTSR (VII), LVQLLVR (VIII), EGLLAIFK (IX), NEGVATYAAAVLFR (X). Among these peptides, AIPELTK and EGLLAIFK were selected for synthesis due to their signal intensity. Leucine was stable isotope labeled (13C6, 15N) in the peptide standard, AIPELTK; glutamic acid was replaced with aspartic acid in the peptide standard, DGLLAIFK, to avoid pyroglutamic acid formation and obviate the need for a stable isotope label. β-Catenin can be quantified by spiking known amounts of these two peptides in each biological sample. The appropriate concentration of each internal standard is determined using calibration curves prior to spiking into the biological samples.
Table 1. The Target Peptide(s), Corresponding Internal Standards and Selected Transitions For Monitoring Protein Components of the Wnt/beta-Catenin Signaling Pathway.
Residues underlined in bold font indicate the sites of incorporation of stable isotope labels or single amino acid replacements (e.g. alanine to glycine). Roman numerals link to the corresponding peptide signals in Figure 1.
| Assay Development | ||||
|---|---|---|---|---|
| Protein | Target Peptide | Label/Modification | Transitions | Additional Monitored Peptides |
| E-cadherin | VTEPLDR (II) | L5: 13C6, 15N | y3 - y6 | NTGVISVVTTGLDR (I) GLDARPEVTR (III) |
| α-catenin | SDALNSAIDK (III) | A7→G | y4 - y8 | NTSDVISAAK (I) DVDGLDR (II) VVEDGILK IV) IAEQVASFQEEK (V) NAGNEQDLGIQYK (VI) LVYDGIR (VII) |
| β-catenin |
EGLLAIFK (IX) AIPELTK (II) |
E1→D L5: 13C6, 15N |
y2 - y5 y3 - y6 |
HQEAEMAQNAVR (I) LLNDEDQVVVNK (III) SGGIPALVK (IV) VAAGVLCELAQDK (V) LHYGLPVVVK (VI) LLWTTSR (VII) LVQLLVR (VIII) NEGVATYAAAVLFR (X) |
| c-Src | LLLNAENPR (IV) | P8: 13C5, 15N | y4 - y7 | EVLDQVER (I) GPSAAFAPAAAEPK (II) TETDLSFK (III) LFGGFNSSDTVTSPQR (V) WTAPEAALYGR (VI) GSLLDFLK (VII) |
| Axin1 | LLLETAAPR (II) | P8: 13C5, 15N | y3 - y8 | EQVEAEATR (I) SDIYLEYTR (III) SASTEVPGASEDAEK (IV) |
| APC protein | YSDEQLNSGR (I) | G9→A | y3 - y9 | TPPPPPQTAQTK (II) ESTGYLEELEK (III) HVDQPIDYSLK (IV) YATDIPSSQK (V) QNQLHPSSAQSR (VI) |
| GSK3β | QTLPVIYVK (II) | V8: 13C5, 15N | y3 - y8 | LLEYTPTAR (I) |
| PP2A catalytic subunit α/β | YSFLQFDPAPR (III) | P10: 13C5, 15N | y4 - y7 | QLSESQVK (I)* GGWGISPR (II) |
| PP2A 65 kDa regulatory subunit A α | AVGPEITK (I) | A1→G | y3 - y6 | VLELDNVK (II) EWAHATIIPK (III) |
| PP2A 65 kDa regulatory subunit A β | VLELDSVK (I) | V7: 13C5, 15N | y3 - y7 | FGTEWAQNTIVPK (II) LASGDWFTSR (III) |
| PP2A 55 kDa regulatory subunit B α | ILHTAWHPK (I) | P8: 13C5, 15N | y3 - y7 | DITLEASR (II) NAAQFLLSTNDK (III) INLWHLEITDR (IV) |
| Casein kinase I | LFLIDFGLAK (II) | A9→G | y3 - y8 | HPQLLYESK (I) AEFIVGGK (III) ILQGGVGIPHIR (IV) |
| Casein kinase II α | QLYQTLTDYDIR (III) | D10→E | y3 - y10 | YNIELDPR (I) VYTDVNTHRPR (II) |
| Casein kinase II α′ | VLGTEELYGYLK (II) | L11: 13C6, 15N | y3 - y10 | VYAEVNSLR (I) |
| Casein kinase II β | FNLTGLNEQVPHYR (II) | G5→A | y3 - y10 | RPANQFVPR (I) |
| CD44 | FAGVFHVEK (II) | V7: 13C5, 15N | y3 - y8 | LVINSGNGAVEDR (I) YGFIEGHVVIPR (III) |
| Cyclin D1 | FLSLEPVK (I) | V7: 13C5, 15N | y3 - y7 | |
| MMP-7 | DLPHITVDR (I) | V7: 13C5, 15N | y4 - y7 | FYLYDSETK (II) |
| Cortactin | ANFENLAK (VI) | L6: 13C6, 15N | y3 - y7 | YGVQADR (I) FGVQTDR and FGVQSER (II) FGVEQDR (III) TVPVEAVTSK (IV) ENVFQEHQTLK (V) VDQSAVGFEYQGK (VII) LPSSPVYEDAASFK (VIII) YGLFPANYVELR (IX) |
| Axin2 | STETVDSGYR (II) | G8→A | y4 - y8 | ANGQVSLPHFPR (I) NEDGLGEPEGR (III) |
| c-Myc | DQIPELENNEK (I) | E5→D | y6 - y9 | SSDTEENVK (II) |
| TCF7(TCF1) | LPEPLEDGLK (I) | L9: 13C6, 15N | y3 - y9 | |
Sequence QLSESQVK (I) is unique to PP2A catalytic subunit α.
LC-MRM Assay for Protein Expression in Colon Cancer Cell Lines and Comparison with Western Blots
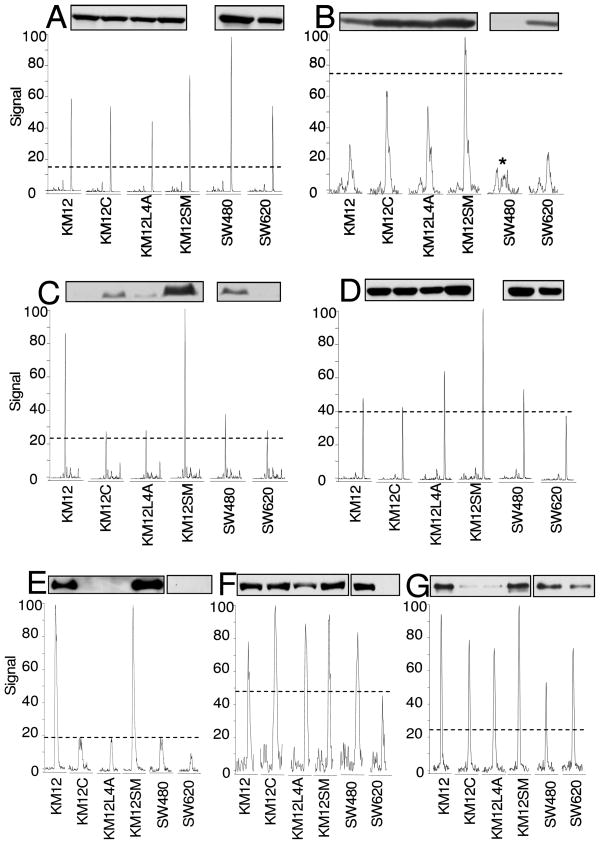
Protein expression in the Wnt/β-catenin signaling pathway was monitored in six colon cancer cell lines including KM12, KM12C, KM12L4A, KM12SM, SW480, and SW620 by LC-MRM. Out of twenty-two targets, seventeen assays were successful when analyzing protein extracted from 50,000 cells. Western blotting was performed for seven proteins: β-catenin, c-Src, c-Myc, PP2A catalytic subunit, CD44, α-catenin, and cortactin to enable initial comparisons with the qualitative measurements typically made in cancer biology. As shown in Figure 2, LC-MRM raw data of each peptide from seven proteins in six cell lines are consistent with Western blot analyses with the exception of c-Myc in the KM12 cell lines and cortactin in SW480 and SW620; these differences could be due to the selectivity of the antibody, because no post-translational modifications are expected and no mutations are reported for the peptides detected with LC-MRM.
Figure 2. Comparison of Protein Expression Measured by Western Blot and LC-MRM.
Western blot and extracted ion chromatograms from LC-MRM quantification of protein expression in colon cancer cell lines KM12, KM12C, KM12L4A, KM12SM, SW480, SW620 are shown for (A) EGLLAIFK from β-catenin (15–45 min), (B) LLLNAENPR from c-Src (22–27 min), (C) DQIPELENNEK from c-Myc (15–35 min), (D) YSFLQFDPAPR from PP2A Catalytic subunit (20–40 min), (E) FAGVFHVEK from CD44 (23–28 min), (F) SDALNSAIDK from α-catenin (20–23.2 min), and (G) ANFENLAK from Cortactin (21–26 min). Dashed lines show the signal intensity from internal standards (see Table 1) which are used for normalization. The asterisk indicates the correct peak from SW480, which was verified by fragment ion signal ratios in the composite tandem mass spectrum.
Comparison of antigens selected for development of the antibodies used for Western blots and selected peptide targets for LC-MRM has been shown in Supplementary Table 1 to assess the comparison of LC-MRM and Westerns in more detail. For β-catenin, the antibody has a long antigen (a.a. 571–781) which includes two MRM targets VAAGVLCELAQDK (a.a. 613–625) and NEGVATYAAAVLFR (a.a. 648–661) as well as additional LC-MRM targets for both nearby and distant (Figure 2A). For PP2A catalytic subunit, consistent results between Western blots and LC-MRM are also shown in Figure 2D for an antibody developed against long antigen (a.a. 153–309) that includes peptides, such as GGWGISPR (a.a. 207–214) and YSFLQFDPAPR (a.a. 284–294). For c-Src, the antigen (a.a. 82–169) also includes one of the monitored peptides, TETDLSFK (a.a. 99–106); neighboring sequences, LFGGFNSSDTVTSPQR (a.a. 63–78) and LLLNAENPR (a.a. 164–172), also show consistency with Western blots (Figure 2B). However, for c-Myc Western blots using an antibody developed using a relatively short antigen in C-terminal (a.a. 408–439) show slightly different results when compared to LC-MRM of two monitored peptides (a.a. 347–355 and a.a. 379–389), see Figure 2C; note that the two LC-MRM measurements were consistent with each other. These data suggest that variation between Westerns and LC-MRM may occur when the antibodies are developed against short epitopes in the protein. However, these results only suggest potential pitfalls; rules for antibody or LC-MRM peptide selection could be developed only after comparison of a much larger body of assays for protein expression. For the other proteins tested here, comparable results are all shown between the two methods regardless of the length of the antigen and the proximity of the MRM targets to that epitope.
The improved sensitivity of LC-MRM is noted in cases of negative Western blot analysis, such as c-Src in SW480 and SW620 as well as full length CD44 in KM12C, KM12L4A, SW480, and SW620 (see Figure 2). For positive results in Western blotting that are similar, LC-MRM can better discern differences in protein expression between the cell lines, e.g. β-catenin and PP2A catalytic subunit. Expression measurements in the same group of cell lines for other proteins involved in Wnt/β-catenin signaling using LC-MRM are shown in Supplementary Figure 5; these proteins have not been analyzed with Westerns.
Absolute Quantification in Colon Cancer Cell Lines
Another advantage of LC-MRM over Western blotting is the ability to use known amounts of internal standards to determine the copy number for each protein within the cell. In addition to the relative quantification shown in Figure 2 and Supplementary Figure 5, The absolute amount of proteins from each of the six cell lines has also been calculated and presented in Supplementary Table 2. These values should be viewed as minimum estimates due to the potential for incomplete recovery during digestion and peptide extraction from the gel. Protein expression was detected across three orders of magnitude, from thousands of copies per cell to millions of copies per cell. While these proteins would typically be described to be present in “low abundance,” the dysregulation in colon cancer is expected to result in such high expression levels. For comparison, the expression level of a kinase driving glioblastoma multiforme, EGFRvIII, has been measured to be as high as 3 × 106 copies per cell.77
Reproducibility of LC-MRM Analysis
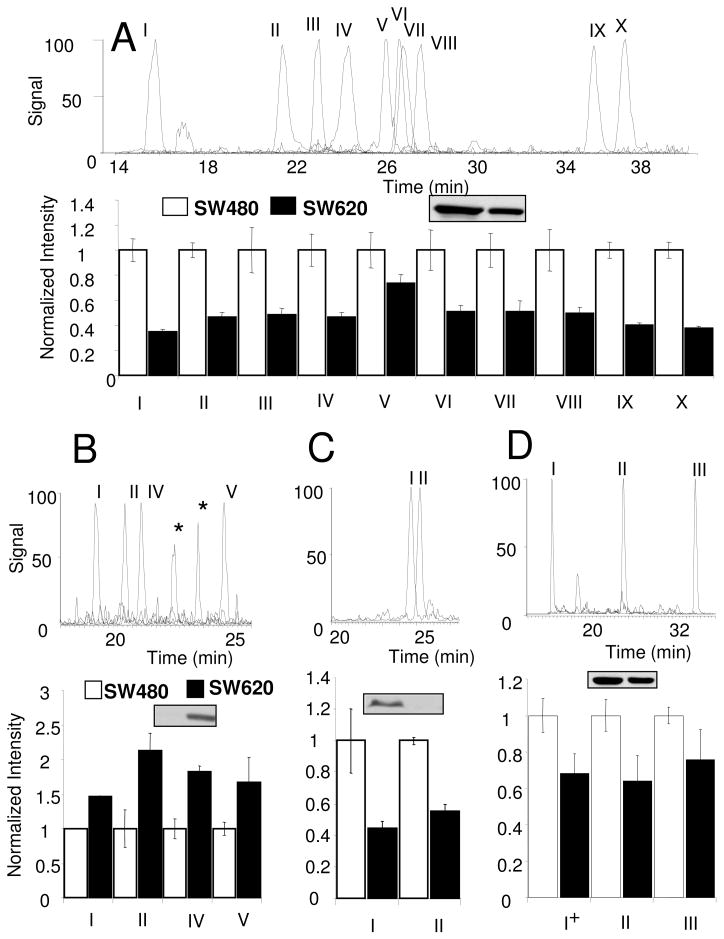
The consistency of all detected peptides from β-catenin, c-Src, c-Myc, and PP2A catalytic subunit in SW480 and SW620 colon cancer cell lines of four biological replicates is shown in Figure 3. These proteins were selected to enable comparison of data from proteins with multiple peptides that give high intensity signal (β-catenin and PP2A) and proteins that have few peptides with lower amounts of signal (c-Myc and c-Src). For β-catenin (Figure 3A), ten peptides are monitored in each LC-MRM analysis. Two synthetic peptides (described in Table 1 for peptides II and IX) are used as internal standards. To examine the relative expression of β-catenin in SW480 and SW620 colon cancer cells, the ten peptides are monitored in four biological replicates; the data are normalized relative to SW480 expression. LC-MRM measurements of nine peptides give similar SW620/SW480 expression ratios of ~0.45; the sole exception is the peptide VAAGVLCELAQDK (peptide V), which is contained in β-catenin and γ-catenin. Relative standard deviation values (RSD) for each peptide are between 4.7% and 17.9%; the variability does not correlate with the signal intensity. As an example, using comparison of peak areas normalized by internal standards, HQEAEMAQNAVR (1.20E6 ± 1.09E5 for SW480 and 6.77E5 ± 3.19E4 for SW620) is detected with less than 10% of signal intensity observed for AIPELTK (2.00E7 ± 1.17E6 for SW480 and 1.35E7 ± 1.25E6 for SW620), but the measurements of HQEAEMAQNAVR yielded RSD values of 9.1%(SW480) and 4.7%(SW620) similar to AIPELTK (RSD value 5.8% for SW480 and 9.3% for SW620). Therefore, variability is based on the peptide itself as well as its signal intensity. For PP2A (Figure 3D), three peptides are monitored which indicate SW620/SW480 expression ratios from 0.64 to 0.76 with RSD from 4.5% to 21.7%. RSD values increase with decreasing signal. For c-Src (Figure 3B), four peptides are monitored in LC-MRM analysis. SW620/SW480 expression ratios for each peptide vary from 1.47 to 2.12 with RSD values between 0.25% and 27.6%. For c-Myc (Figure 3C), two peptides are monitored with SW620/SW480 expression ratios from 0.45 to 0.55 and RSD values ranging from 2.3% to 20.3%.
Figure 3. Quantitative Assessment of β-Catenin, c-Src, c-Myc and PP2A from SW480 and SW620 Colon Cancer Cell Lines.
Extracted ion chromatograms of detected peptides and normalized bar graph of the average and standard deviation from four biological replicates in SW480 and SW620 for each peptide (See Table 1) as well as Western blot results from (A) β-catenin, (B) c-Src, (C) c-Myc, and (D) PP2A Catalytic subunit are shown. The asterisks indicate interference in transitions from the peptide, LFGGFNSSDTVTSPQR, from c-Src; however, the correct peak can be selected by comparison of fragment ion ratios. The + symbol indicates the correct signal for the peptide, QLSESQVK (I), belonging to PP2A Catalytic subunit α.
From the measurements of different biological replicates used for these reproducibility measurements, absolute quantification of these four proteins is consistent with the values listed in Supplementary Table 2, which reports the expression in six colon cancer cell lines. After spiking internal standard peptide, replicate LC-MRM analyses injecting protein extracted from 50,000 cells were used to determine the amount of the protein in 200,000 SW480 and SW620 cells. In SW480, quantification of β-catenin indicated 152.58 ± 8.92 fmol total, corresponding 4.59 × 105 molecules/cell. In SW620, 70.58 ± 6.54 fmol of β-catenin was detected, indicating 2.12 × 105 molecules/cell. Total PP2A catalytic subunit expression was measured to be 496.30 ± 22.28 fmol in SW480 (1.49E6 molecules/cell) and 375.83 ± 81.24 fmol in SW620 cells (1.13E6 molecules/cell). c-Src was detected at lower levels: only 2.79 ± 0.38 fmol total in SW480 (8.41 × 103 molecules/cell) and 5.12 ± 0.22 fmol from the same amount of SW620 cells (1.54 × 104 molecules/cell), consistent with prior Western analysis.78 The expression of c-Myc in 200,000 cells was measured to be 128.06 ± 26.04 fmol in SW480 (3.85E5 molecules/cell) and 57.23 ± 5.58 fmol in SW620 (1.72E5 molecules/cell).
Limits of Detection and Quantification to Evaluate Assay Translation to Clinical Specimens
The sensitivity of each assay has been tested using serial dilutions of SW480 cell lysate (using the protein extracted from 100, 1,000, 5,000, 10,000, and 25,000 cells). As shown in Table 2, limits of detection for β-catenin, c-Src, c-Myc, and the PP2A catalytic subunit are given in terms of signal-to-noise ratio for the lowest number of cells where each peptide could still be detected by LC-MRM. Limits of quantification were obtained by plotting the peak area obtained for each amount of cells and applying a linear fit to the data acquired from the triplicate LC-MRM experiments. The minimum amount required for quantification is determined from the x-intercept. The peptides selected for internal standard synthesis in each protein have the lowest limits of detection and quantification. For β-catenin, both sequences, AIPELTK (II) and EGLLAIFK (IX), can be detected from 100 cells; based on the S/N values: 411 ± 147, and 136 ± 19, the limit of detection is actually lower. For PP2A, limits of detection and quantification are as low as 100 cells based on the peptide, YSFLQFDPAPR. For c-Src and c-Myc, the limits of detection and quantification start from 1,000 to 10,000 cells and 5,000 to 10,000 cells, depending on which peptide is monitored. These differences could be due to lower abundance of these proteins than β-catenin and the PP2A catalytic subunit in colon cancer cells or to poorer peptide response in LC-MRM analysis.
Table 2. Limits of Detection and Quantification for Selected Peptides from β-Catenin, c-Src, c-Myc and PP2A in SW480 Colon Cancer Cells.
Peptides in bold font indicate the best sequences for monitoring the corresponding proteins based on the signal intensity and consistent data acquired from the triplicate LC-MRM experiments.
| Protein | Peptide | Limit of Detection (Cells) | Limit of Quantification (Cells) |
|---|---|---|---|
| β-catenin | AIPELTK (II) | 100 (S/N 411 ± 147) | 1000 |
| LLNDEDQVVVNK (III) | 500 (S/N 61 ± 11) | 1000 | |
| SGGIPALVK (IV) | 500 (S/N 128 ± 30) | 1000 | |
| LHYGLPVVVK (VI) | 5000 (S/N 136 ± 30) | 5000 | |
| LLWTTSR (VII) | 100 (S/N 33 ± 8) | 100 | |
| LVQLLVR (VIII) | 500 (S/N 72 ± 17) | 1000 | |
| EGLLAIFK (IX) | 100 (S/N 136 ± 19) | 1000 | |
| NEGVATYAAAVLFR (X) | 1000 (S/N 332 ± 95) | 1000 | |
| c-Src | EVLDQVER (I) | 5000 (S/N 58 ± 5) | 5000 |
| GPSAAFAPAAAEPK (II) | 10000 (S/N 36 ± 11) | 10000 | |
| TETDLSFK (III) | 5000 (S/N 107 ± 19) | 5000 | |
| LLLNAENPR (IV) | 5000 (S/N 20 ± 2) | 5000 | |
| LFGGFNSSDTVTSPQR (V) | 10000 (S/N 35 ± 6) | 10000 | |
| WTAPEAALYGR (VI) | 5000 (S/N 100 ± 24) | 5000 | |
| GSLLDFLK (VII) | 5000 (S/N 506 ± 75) | 5000 | |
| c-Myc | DQIPELENNEK (I) | 1000 (S/N 27 ± 7) | 5000 |
| SSDTEENVK (II) | 5000 (S/N 43 ± 17) | 5000 | |
| PP2A catalytic subunit | GGWGISPR (II) | 500 (S/N 32 ± 10) | 500 |
| YSFLQFDPAPR (III) | 100 (S/N 41 ± 1) | 100 |
As determined above, the low amounts of tumor material required for LC-MRM are compatible with the amounts that could be obtained from even the most precious samples, including core biopsies or fine needle aspirates. In the case of colon cancer, surgery is usually the primary option, particularly for early stage cases where it is curative for most patients. With full resection of the tumor, the quantity may not be the limiting factor; rather, the heterogeneity of the tumor specimen may complicate sampling. Colon adenocarcinoma frequently has low tumor purity, and laser capture microdissection is frequently required to enrich tumor cells. Therefore, in the following paragraphs, the translation of these assays to individual frozen sections and laser captured cells is described.
LC-MRM Assay Application to the Frozen Tissue Sections
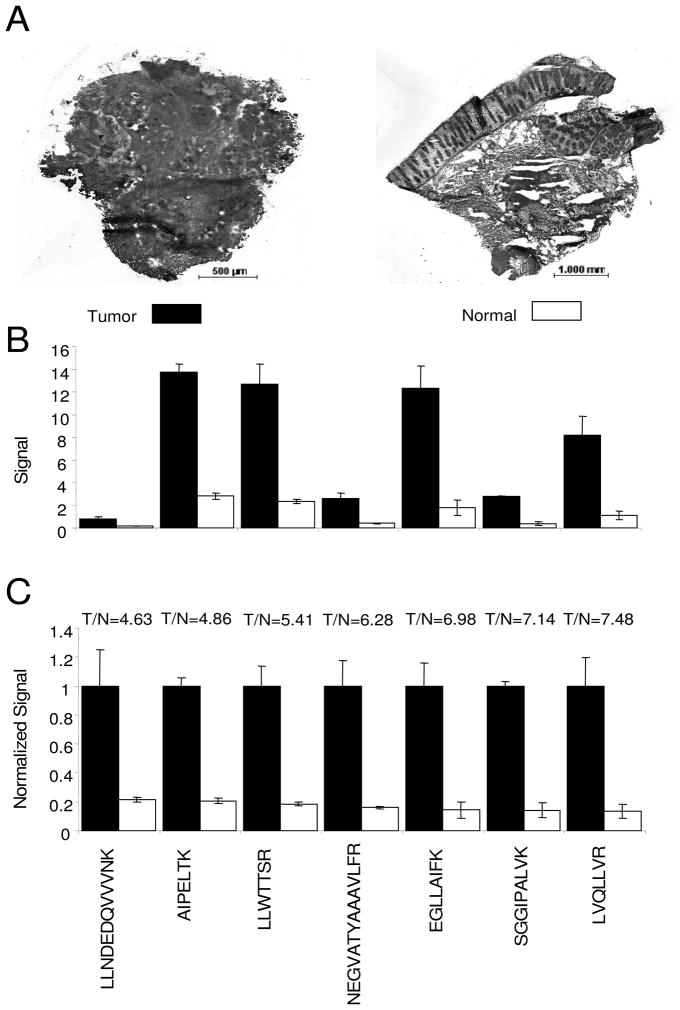
LC-MRM quantification of β-catenin, c-Src, c-Myc and PP2A catalytic subunit has been successful in individual frozen sections of colon tumors and normal tissue from the same patient (n = 3). Images of hematoxylin and eosin (H&E) stained colon cancer tumor and normal tissue sections from a single patient are shown in Figure 4A. The tumor section has been reviewed and confirmed by board certified pathologist (AN), to be 99% adenocarcinoma, while the normal section mainly contains colonic mucosa and submucosa. As shown in Figure 4B, β-catenin can be detected by LC-MRM in single frozen tissue sections using seven peptides; of these, the ion signals corresponding to AIPELTK, LLWTTSR, and EGLLAIFK are the highest. In Figure 4C, all the peak ratios of each peptide from tumor samples are normalized for viewing on the same scale. Tumor to normal ratios range from 4.63 to 7.48, with RSD ranging from 2.9% to 38.4%, values that are similar to previous results obtained in cell lines. Detection of c-Src, c-Myc, and PP2A catalytic subunit, in tumor and normal tissue sections is shown in Supplementary Figure 6. Tumor to normal ratio values were calculated to be 1.65 for c-Src (RSD 12.1% and 13.6%), 1.00 for c-Myc (RSD 32.8% and 8.5%), 1.85 for PP2A (RSD 1.1% and 3.8%). Comparison of equal amounts of total protein (28 μg), the protein expression in frozen sections of tumor tissue and normal colon can be compared. Amounts of the protein in tumor tissue homogenate are measured as 68.53 ± 3.73 fmol of β-catenin, 12.62 ± 1.53 fmol of c-Src, 16.09 ± 5.28 fmol of c- Myc, and 32.39 ± 0.37 fmol of PP2A. Using the same amount of protein from homogenized normal tissue sections, 14.10 ± 1.40 fmol of β-catenin, 7.65 ± 1.04 fmol of c-Src, 16.00 ± 1.36 fmol of c-Myc, and 17.46 ± 0.65 fmol of PP2A are detected. If protein expression is solely intracellular, comparison to the amounts quantified in cell lines would indicate that approximately 90,000 cells are contained in the amount of tumor tissue that produces 28 μg of total protein.
Figure 4. Successful LC-MRM Detection of β-catenin in Individual Frozen Tissue Sections.
Images of H&E stained colon cancer tumor (40× magnification) and normal tissue (20× magnification) from same patient (A). Signal intensity comparison for detected peptides from β-catenin from tumor and normal tissues (B). Normalized results for tumor and normal tissues with ratios for different peptides from β-catenin are also shown (C).
LC-MRM Assay Application to Laser Capture Microdissected Tumor Cells
LC-MRM quantification of β-catenin, c-Src, c-Myc and PP2A catalytic subunit has also been successful using laser capture microdissected colon cancer tumor cells. Around 25,000 shots (23.2 μm laser diameter) with 75% (or greater) efficiency per cap are achieved for the each tumor section. The volume of each capture (V) can be calculated by Equation 1 if each capture is assumed to be a cylinder with 5 μm height (L) and 23.2 μm diameter (d).
| (Equation 1) |
Considering each cell as a sphere of 25 μm diameter (D), the estimated number of cells (C) can be calculated using equation 2 using the number of laser shots (n) with the average efficiency of the capture (f) and the ratio of volume for cylinder over the sphere.
| (Equation 2) |
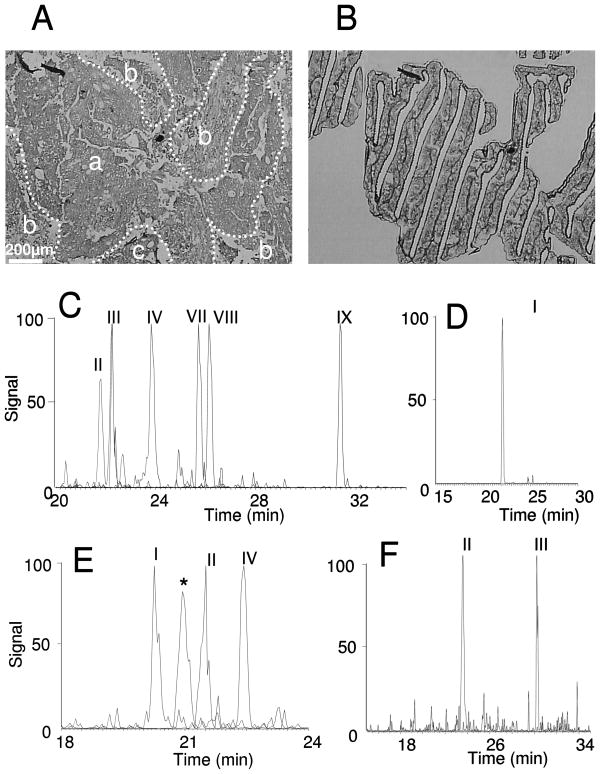
Therefore, the number of tumor cells acquired with laser capture microdissection is approximately 5,000, exceeding the amount of cells indicated in the cell line experiments to determine the limit of quantification for the target proteins, β-catenin, c-Src, c-Myc and PP2A catalytic subunit. An image of a colon adenocarcinoma tissue section prior to LCM is shown in Figure 5A with the target location for colonic adenocarcinoma (a) and the areas excluded from the capture, such as tumor necrosis (b), and stroma (c). The captured cells on the cap are shown in Figure 5B. The extracted ion chromatograms of the detected peptides from β-catenin, c-Src, c-Myc and PP2A catalytic subunit are also shown in Figure 5. In LCM tumor samples, ion signals corresponding six peptides of β-catenin (Figure 5C) can be detected, including AIPELTK (II), LLNDEDQVVVNK (III), SGGIPALVK (IV), LLWTTSR (VII), LVQLLVR (VIII), EGLLAIFK (IX). The extracted ion chromatogram for the peptide, DQIPELENNEK (I), from c- Myc is shown (Figure 5D). Three peptides, EVLDQVER (I), GPSAAFAPAAAEPK (II), and LLLNAENPR (IV) from c-Src (Figure 5E) can be monitored; GGWGISPR (II) and YSFLQFDPAPR (III) of PP2A catalytic subunit (Figure 5F) are also detected. Therefore, successful LC-MRM detection can be used to quantify β-catenin, c-Src, c-Myc and PP2A catalytic subunit in ~5,000 laser capture microdissected colon cancer tumor cells. The expression levels of the four target proteins in these LCM tumor cells are 1.50 × 105 molecules/cell for β-catenin, 5.32 × 104 molecules/cell for c-Src, 1.65 × 105 molecules/cell for c-Myc, and 1.14 × 106 molecules/cell for PP2A, which are consistent with the measurements in cell lines (see Supplementary Table 2). This result is important, as it shows that cell line models are appropriate sources of material and reasonable test cases for LC-MRM assay development.
Figure 5. LC-MRM Detection of β-catenin, c-Src, c-Myc and PP2A in Laser Capture Microdissected Tumor Cells.
Optical microscope photographs (40× magnification) of colon adenocarcinoma tissue sections (A) stained with hematoxylin and dehydrated prior to LCM and extracted cancer cells on the LCM cap (B). In panel A, areas with colonic adenocarcinoma (a), tumor necrosis (b), and stroma (c) are indicated. Extracted ion chromatograms of detected peptides (see Table 1) from β-catenin (C), c-Myc (D), c-Src (E), and PP2A Catalytic subunit (F) are shown. The asterisks indicate interference in transitions from peptide, LLLNAENPR, from c-Src; however, the correct peak can be determined by comparison of fragment ion ratios in the composite tandem mass spectra.
Conclusions
The initial steps in the development of a reaction monitoring mass spectrometry platform for intratumoral elucidation of colon cancer biology have been illustrated using β-catenin signaling as the model. As noted in the introduction, the upregulation and downregulation of several targets of this set of assays play important roles in colon cancer biology and are associated with the clinical outcome for each patient. The ultimate goal is to develop quantitative LC-MRM assays for protein mutation, expression, and modification that will be applied to patient assessment to characterize the biological background of each tumor. The resulting molecular information may prove useful to guide therapy and follow up regimens. Proof of principle has been demonstrated for expression measurement of proteins that are involved with β-catenin signaling. Relative quantification can be used to examine preclinical models in colon cancer cell lines, and absolute quantification is possible for cell lines (in copies per cell),_frozen tumor sections (in femtomoles per microgram of tissue) and laser capture microdissected cells (in copies per cell).
The assessment of protein expression of multiple components in a signaling pathway with LC-MRM analysis is reported with comparisons to Western blots for selected proteins, and illustration of the translation to patient samples. Using SDS-PAGE separation coupled to LC-MRM, twenty-two components from Wnt/β-catenin signaling can be monitored and quantified from various colon cancer cell lines; these proteins include structural interacting proteins, members of the β-catenin regulatory/destruction complex, interacting transcription factors, and downstream proteins whose expression is regulated by those transcription factors. Results are comparable to Western blots with better quantification and improved sensitivity in LC-MRM. Biological replicates are performed for two colon cancer cell lines, SW480 and SW620, show the consistent ratio of protein expression between the two cell lines and low variability in most measurements (RSD values: 4.7%–17.9% for β-catenin, 4.5%–21.7% for PP2A catalytic subunit, 0.3%–27.6% for c-Src, and 2.3%–20.3% for c-Myc) as well as consistency for all the peptides detected from the same protein. The sensitivity of each assay has been tested using serial dilutions of cell lysate before translation to individual frozen tissue sections and laser capture microdissected cells. All selected target proteins can be quantified from as few as 5,000 cells; some can be detected or even quantified from 100 cells, such as β-catenin and PP2A catalytic subunit. As shown for β-catenin, c-Src, c-Myc and PP2A catalytic subunit, LC-MRM assays have been successfully translated to individual frozen tissue sections and laser capture microdissected cells.
The versatility of LC-MRM approaches is a critical advantage; novel discoveries can be easily added to this platform. For example, the involvement of Bruton’s tyrosine kinase with β-catenin has recently been elucidated,79 so the addition of this protein to the current assays could lead to improvement of this LC-MRM method for tissue assessment. Other future additions to this LC-MRM platform include selection of proteins for tissue quality analysis/quality control, which will quantify the components of each tissue section, including tumor cells, epithelial cells, infiltrating immune cells, extracellular matrix/stroma, blood cells & blood plasma, and muscle. The implementation of these assays may provide a method to interrogate whole tissue sections, rather than relying on laser capture microdissection. In addition, to make use of archived specimens with extensive medical histories, translation of these assays to formalin-fixed paraffin-embedded (FFPE) tissue samples will also provide an important step for clinical implementation. Prior experiments using shotgun proteomic analysis 80,81 and LC-MRM targeted quantification 82 will guide the development of analytical workflows for archived FFPE tissue, which must be implemented at the peptide level. Using either frozen or FFPE tissue, this module of assays focused around β-catenin will be useful to establish the biological background of each colon tumor. These assays can be paired with other critical pathways to elucidate the biology of the tumor, evaluate metastatic risk, and stratify patients for adjuvant therapies using specific molecular information.
Supplementary Material
Acknowledgments
Moffitt Proteomics is supported by the US Army Medical Research and Materiel Command under Award No. DAMD17-02-2-0051 for a National Functional Genomics Center, the National Cancer Institute under Award No. P30-CA076292 as a Cancer Center Support Grant, and the Moffitt Foundation. The authors would like to thank the Tissue Core and Analytic Microscopy for their support of the project. The triple quadrupole mass spectrometer was purchased with a shared instrument grant from the Bankhead-Coley Cancer Research program of the Florida Department of Health (06BS-02-9614). This work has been funded by the University of Florida-Moffitt Collaborative Partnership and a subcontract from the institutional National Functional Genomics Center grant awarded by the Department of Defense.
References
- 1.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.Bienz M, Clevers H. Linking colorectal cancer to Wnt signaling. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 3.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 4.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Su LK, Vogelstein B, Kinzler KW. Association of the APC tumor suppressor protein with catenins. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 6.Rubinfeld B, Souza B, Albert I, Müller O, Chamberlain SH, Masiarz FR, Munemitsu S, Polakis P. Association of the APC gene product with beta-catenin. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 7.Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 8.Gehrke I, Gandhirajan RK, Kreuzer KA. Targeting the WNT/beta-catenin/TCF/LEF1 axis in solid and haematological cancers: Multiplicity of therapeutic options. Eur J Cancer. 2009;45:2759–2767. doi: 10.1016/j.ejca.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Yanagawa S, Matsuda Y, Lee JS, Matsubayashi H, Sese S, Kadowaki T, Ishimoto A. Casein kinase I phosphorylates the Armadillo protein and induces its degradation in Drosophila. EMBO J. 2002;21:1733–1742. doi: 10.1093/emboj/21.7.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci U S A. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrens J, Jerchow BA, Würtele M, Grimm J, Asbrand C, Wirtz R, Kühl M, Wedlich D, Birchmeier W. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280(5363):596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 12.Choi HJ, Huber AH, Weis WI. Thermodynamics of beta-catenin-ligand interactions: the roles of the N- and C-terminal tails in modulating binding affinity. J Biol Chem. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- 13.Ha NC, Tonozuka T, Stamos JL, Choi HJ, Weis WI. Mechanism of phosphorylation-dependent binding of APC to beta-catenin and its role in beta-catenin degradation. Mol Cell. 2004;15:511–521. doi: 10.1016/j.molcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Xing Y, Hinds TR, Zheng J, Xu W. The third 20 amino acid repeat is the tightest binding site of APC for beta-catenin. J Mol Biol. 2006;360:133–144. doi: 10.1016/j.jmb.2006.04.064. [DOI] [PubMed] [Google Scholar]
- 15.Su Y, Fu C, Ishikawa S, Stella A, Kojima M, Shitoh K, Schreiber EM, Day BW, Liu B. APC is essential for targeting phosphorylated beta-catenin to the SCFbeta-TrCP ubiquitin ligase. Mol Cell. 2008;32:652–661. doi: 10.1016/j.molcel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 16.Marikawa Y, Elinson RP. beta-TrCP is a negative regulator of Wnt/beta-catenin signaling pathway and dorsal axis formation in Xenopus embryos. Mech Dev. 1998;77:75–80. doi: 10.1016/s0925-4773(98)00134-8. [DOI] [PubMed] [Google Scholar]
- 17.Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K, Nakamichi I, Kikuchi A, Nakayama K, Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, Polakis P. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 19.Polakis P. Mutations in the APC gene and their implications for protein structure and function. Curr Opin Genet Dev. 1995;5:66–71. doi: 10.1016/s0959-437x(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 20.Daniels DL, Weis WI. ICAT inhibits beta-catenin binding to Tcf/Lef-family transcription factors and the general coactivator p300 using independent structural modules. Mol Cell. 2002;10:573–584. doi: 10.1016/s1097-2765(02)00631-7. [DOI] [PubMed] [Google Scholar]
- 21.Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 22.Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 23.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 24.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destrée O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Yost HJ, Virshup DM, Seeling JM. Protein phosphatase 2A and its B56 regulatory subunit inhibit Wnt signaling in Xenopus. EMBO J. 2001;20:4122–4131. doi: 10.1093/emboj/20.15.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seeling JM, Miller JR, Gil R, Moon RT, White R, Virshup DM. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science. 1999;283:2089–2091. doi: 10.1126/science.283.5410.2089. [DOI] [PubMed] [Google Scholar]
- 27.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–6. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 28.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 29.Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol. 2010;30:1411–20. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brabletz T, Jung A, Dag S, Hlubek F, Kirchner T. beta-catenin regulates the expression of the matrix metalloproteinase-7 in human colorectal cancer. Am J Pathol. 1999;155:1033–8. doi: 10.1016/s0002-9440(10)65204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YY, Yu CP, Lin CK, Nieh S, Hsu KF, Chiang H, Jin JS. Expression of survivin and cortactin in colorectal adenocarcinoma: association with clinicopathological parameters. Dis Markers. 2009;26:9–18. doi: 10.3233/DMA-2009-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li AF, Hsu PK, Tzao C, Wang YC, Hung IC, Huang MH, Hsu HS. Reduced axin protein expression is associated with a poor prognosis in patients with squamous cell carcinoma of esophagus. Ann Surg Oncol. 2009;16:2486–2493. doi: 10.1245/s10434-009-0593-3. [DOI] [PubMed] [Google Scholar]
- 33.Xu HT, Wei Q, Liu Y, Yang LH, Dai SD, Han Y, Yu JH, Liu N, Wang EH. Overexpression of axin downregulates TCF-4 and inhibits the development of lung cancer. Ann Surg Oncol. 2007;14:3251–3259. doi: 10.1245/s10434-007-9555-9. [DOI] [PubMed] [Google Scholar]
- 34.Dehm SM, Bonham K. SRC gene expression in human cancer: the role of transcriptional activation. Biochem Cell Biol. 2004;82:263–74. doi: 10.1139/o03-077. [DOI] [PubMed] [Google Scholar]
- 35.Iravani S, Mao W, Fu L, Karl R, Yeatman T, Jove R, Coppola D. Elevated c-Src protein expression is an early event in colonic neoplasia. Lab Invest. 1998;78:365–71. [PubMed] [Google Scholar]
- 36.Irby RB, Yeatman TJ. Increased Src activity disrupts cadherin/catenin-mediated homotypic adhesion in human colon cancer and transformed rodent cells. Cancer Res. 2002;62:2669–74. [PubMed] [Google Scholar]
- 37.Aligayer H, Boyd DD, Heiss MM, Abdalla EK, Curley SA, Gallick GE. Activation of Src kinase in primary colorectal carcinoma: an indicator of poor clinical prognosis. Cancer. 2002;94:344–51. doi: 10.1002/cncr.10221. [DOI] [PubMed] [Google Scholar]
- 38.Bondi J, Bukholm G, Nesland JM, Bukholm IR. Expression of non-membranous beta-catenin and gamma-catenin, c-Myc and cyclin D1 in relation to patient outcome in human colon adenocarcinomas. APMIS. 2004;112:49–56. doi: 10.1111/j.1600-0463.2004.apm1120109.x. [DOI] [PubMed] [Google Scholar]
- 39.Park CH, Hahm ER, Lee JH, Jung KC, Rhee HS, Yang CH. Ionomycin downregulates beta-catenin/Tcf signaling in colon cancer cell line. Carcinogenesis. 2005;26:1929–1933. doi: 10.1093/carcin/bgi145. [DOI] [PubMed] [Google Scholar]
- 40.Park CH, Chang JY, Hahm ER, Park S, Kim HK, Yang CH. Quercetin, a potent inhibitor against beta-catenin/Tcf signaling in SW480 colon cancer cells. Biochem Biophys Res Commun. 2005;328:227–234. doi: 10.1016/j.bbrc.2004.12.151. [DOI] [PubMed] [Google Scholar]
- 41.Gwak J, Song T, Song JY, Yun YS, Choi IW, Jeong Y, Shin JG, Oh S. Isoreserpine promotes beta-catenin degradation via Siah-1 up-regulation in HCT116 colon cancer cells. Biochem Biophys Res Commun. 2009;387:444–9. doi: 10.1016/j.bbrc.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 42.Bos CL, Kodach LL, van den Brink GR, Diks SH, van Santen MM, Richel DJ, Peppelenbosch MP, Hardwick JC. Effect of aspirin on the Wnt/beta-catenin pathway is mediated via protein phosphatase 2A. Oncogene. 2006;25:6447–56. doi: 10.1038/sj.onc.1209658. [DOI] [PubMed] [Google Scholar]
- 43.Bos CL, Diks SH, Hardwick JC, Walburg KV, Peppelenbosch MP, Richel DJ. Protein phosphatase 2A is required for mesalazine-dependent inhibition of Wnt/beta-catenin pathway activity. Carcinogenesis. 2006;27:2371–82. doi: 10.1093/carcin/bgl071. [DOI] [PubMed] [Google Scholar]
- 44.Lepourcelet M, Chen Y-NP, France DS, Wang H, Crews P, Petersen F, Bruseo C, Wood AW, Shivdasani RA. Small-molecule antagonists of the oncogenic Tcf/[beta]-catenin protein complex. Cancer Cell. 2004;5:91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 45.Gandhirajan RK, Staib PA, Minke K, Gehrke I, Plickert G, Schlösser A, Schmitt EK, Hallek M, Kreuzer KA. Small molecule inhibitors of Wnt/beta-catenin/lef-1 signaling induces apoptosis in chronic lymphocytic leukemia cells in vitro and in vivo. Neoplasia. 2010;12:326–335. doi: 10.1593/neo.91972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo Q, Wu M, Lian P, Liao M, Xiao Z, Wang X, Shen S. Synergistic effect of indomethacin and NGX6 on proliferation and invasion by human colorectal cancer cells through modulation of the Wnt/beta-catenin signaling pathway. Mol Cell Biochem. 2009;330:71–81. doi: 10.1007/s11010-009-0102-9. [DOI] [PubMed] [Google Scholar]
- 47.Howells LM, Neal CP, Brown MC, Berry DP, Manson MM. Indole-3-carbinol enhances anti-proliferative, but not anti-invasive effects of oxaliplatin in colorectal cancer cell lines. Biochem Pharmacol. 2008;75:1774–1782. doi: 10.1016/j.bcp.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 48.Hakim SG, Kosmehl H, Sieg P, Trenkle T, Jacobsen HC, Attila Benedek G, Ribbat J, Driemel O. Altered expression of cell-cell adhesion molecules beta-catenin/E-cadherin and related Wnt-signaling pathway in sporadic and syndromal keratocystic odontogenic tumors. Clin Oral Investig. 2010 Feb 27; doi: 10.1007/s00784-010-0388-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 49.Bond D, Primrose DA, Foley E. Quantitative evaluation of signaling events in Drosophila s2 cells. Biol Proced Online. 2008;10:20–28. doi: 10.1251/bpo139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markovic D, Punn A, Lehnert H, Grammatopoulos DK. Intracellular mechanisms regulating corticotropin-releasing hormone receptor-2beta endocytosis and interaction with extracellularly regulated kinase 1/2 and p38 mitogen-activated protein kinase signaling cascades. Mol Endocrinol. 2008;22:689–706. doi: 10.1210/me.2007-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun H, Chua MS, Yang D, Tsalenko A, Peter BJ, So S. Antibody Arrays Identify Potential Diagnostic Markers of Hepatocellular Carcinoma. Biomark Insights. 2008;3:1–18. doi: 10.4137/bmi.s595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathur P, Kaga S, Zhan L, Das DK, Maulik N. Potential candidates for ischemic preconditioning-associated vascular growth pathways revealed by antibody array. Am J Physiol Heart Circ Physiol. 2005;288:H3006–3010. doi: 10.1152/ajpheart.01203.2004. [DOI] [PubMed] [Google Scholar]
- 53.Ramirez AB, Loch CM, Zhang Y, Liu Y, Wang X, Wayner EA, Sargent JE, Sibani S, Hainsworth E, Mendoza EA, Eugene R, Labaer J, Urban ND, McIntosh MW, Lampe PD. Use of a single chain antibody library for ovarian cancer biomarker discovery. Mol Cell Proteomics. 2010 May 13; doi: 10.1074/mcp.M900496-MCP200. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loebke C, Sueltmann H, Schmidt C, Henjes F, Wiemann S, Poustka A, Korf U. Infrared-based protein detection arrays for quantitative proteomics. Proteomics. 2007;7:558–564. doi: 10.1002/pmic.200600757. [DOI] [PubMed] [Google Scholar]
- 55.Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff M, Mills GB, Kornblau SM. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 56.Sheehan KM, Calvert VS, Kay EW, Lu Y, Fishman D, Espina V, Aquino J, Speer R, Araujo R, Mills GB, Liotta LA, Petricoin EF, 3rd, Wulfkuhle JD. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol Cell Proteomics. 2005;4:346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 58.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci U S A. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin LL, Tong J, Prakash A, Peterman SM, St-Germain JR, Taylor P, Trudel S, Moran MF. Measurement of Protein Phosphorylation Stoichiometry by Selected Reaction Monitoring Mass Spectrometry. J Proteome Res. 2010 Mar 24; doi: 10.1021/pr100024a. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 61.Held JM, Danielson SR, Behring JB, Atsriku C, Britton DJ, Puckett RL, Schilling B, Campisi J, Benz CC, Gibson BW. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol Cell Proteomics. 2010 Mar 16; doi: 10.1074/mcp.M900643-MCP200. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt C, Lenz C, Grote M, Lührmann R, Urlaub H. Determination of protein stoichiometry within protein complexes using absolute quantification and multiple reaction monitoring. Anal Chem. 2010;82:2784–2796. doi: 10.1021/ac902710k. [DOI] [PubMed] [Google Scholar]
- 63.Jiang H, Ramos AA, Yao X. Targeted quantitation of overexpressed and endogenous cystic fibrosis transmembrane conductance regulator using multiple reaction monitoring tandem mass spectrometry and oxygen stable isotope dilution. Anal Chem. 2010;82:336–342. doi: 10.1021/ac902028f. [DOI] [PubMed] [Google Scholar]
- 64.Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, Sheng M, Peng J. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Whiteaker JR, Zhang H, Zhao L, Wang P, Kelly-Spratt KS, Ivey RG, Piening BD, Feng LC, Kasarda E, Gurley KE, Eng JK, Chodosh LA, Kemp CJ, McIntosh MW, Paulovich AG. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- 66.Armenta JM, Perez M, Yang X, Shapiro D, Reed D, Tuli L, Finkielstein CV, Lazar IM. Fast proteomic protocol for biomarker fingerprinting in cancerous cells. J Chromatogr A. 2010;1217:2862–2870. doi: 10.1016/j.chroma.2010.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang H, Chang-Wong T, Tang HY, Speicher DW. Comparison of extensive protein fractionation and repetitive LC-MS/MS analyses on depth of analysis for complex proteomes. J Proteome Res. 2010;9:1032–1040. doi: 10.1021/pr900927y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu BJ, Li J, Beauchamp RD, Shyr Y, Li M, Washington MK, Yeatman TJ, Whitehead RH, Coffey RJ, Caprioli RM. Identification of early intestinal neoplasia protein biomarkers using laser capture microdissection and MALDI MS. Mol Cell Proteomics. 2009;8:936–45. doi: 10.1074/mcp.M800345-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Engmann O, Campbell J, Ward M, Giese KP, Thompson AJ. Comparison of a protein-level and peptide-level labeling strategy for quantitative proteomics of synaptosomes using isobaric tags. J Proteome Res. 2010;9:2725–2733. doi: 10.1021/pr900627e. [DOI] [PubMed] [Google Scholar]
- 70.Seibert C, Davidson BR, Fuller BJ, Patterson LH, Griffiths WJ, Wang Y. Multiple-approaches to the identification and quantification of cytochromes P450 in human liver tissue by mass spectrometry. J Proteome Res. 2009;8:1672–1681. doi: 10.1021/pr800795r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 72.Lin Y, Buckhaults PJ, Lee JR, Xiong H, Farrell C, Podolsky RH, Schade RR, Dynan WS. Association of the actin-binding protein transgelin with lymph node metastasis in human colorectal cancer. Neoplasia. 2009;11:864–873. doi: 10.1593/neo.09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawrie LC, Curran S, McLeod HL, Fothergill JE, Murray GI. Application of laser capture microdissection and proteomics in colon cancer. Mol Pathol. 2001;54:253–8. doi: 10.1136/mp.54.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morikawa K, Walker SM, Jessup JM, Fidler IJ. In vivo selection of highly metastatic cells from surgical specimens of different primary human colon carcinomas implanted into nude mice. Cancer Res. 1988;48:1943–1948. [PubMed] [Google Scholar]
- 75.https://brendanx-uw1.gs.washington.edu/labkey/project/home/software/Skyline/begin.view)
- 76.Available at http://proteome.moffitt.org/
- 77.Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci U S A. 2007;104:12867–72. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maa MC, Lee JC, Chen YJ, Chen YJ, Lee YC, Wang ST, Huang CC, Chow NH, Leu TH. Eps8 facilitates cellular growth and motility of colon cancer cells by increasing the expression and activity of focal adhesion kinase. J Biol Chem. 2007;282:19399–409. doi: 10.1074/jbc.M610280200. [DOI] [PubMed] [Google Scholar]
- 79.James RG, Biechele TL, Conrad WH, Camp ND, Fass DM, Major MB, Sommer K, Yi X, Roberts BS, Cleary MA, Arthur WT, MacCoss M, Rawlings DJ, Haggarty SJ, Moon RT. Bruton’s tyrosine kinase revealed as a negative regulator of Wnt-beta-catenin signaling. Sci Signal. 2009;2:ra25. doi: 10.1126/scisignal.2000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sprung RW, Brock JC, Tanksley JP, Li M, Washington MK, Slebos RC, Liebler DC. Equivalence of Protein Inventories Obtained from Formalin-fixed Paraffin-embedded and Frozen Tissue in Multidimensional Liquid Chromatography-Tandem Mass Spectrometry Shotgun Proteomic Analysis. Molecular & Cellular Proteomics. 2009;8:1988–1998. doi: 10.1074/mcp.M800518-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawamura T, Nomura M, Tojo H, Fujii K, Hamasaki H, Mikami S, Bando Y, Kato H, Nishimura T. Proteomic analysis of laser-microdissected paraffin-embedded tissues: (1) Stage-related protein candidates upon non-metastatic lung adenocarcinoma. J Proteomics. 2010;73:1089–1099. doi: 10.1016/j.jprot.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 82.Nishimura T, Nomura M, Tojo H, Hamasaki H, Fukuda T, Fujii K, Mikami S, Bando Y, Kato H. Proteomic analysis of laser-microdissected paraffin-embedded tissues: (2) MRM assay for stage-related proteins upon non-metastatic lung adenocarcinoma. J Proteomics. 2010;73:1100–1110. doi: 10.1016/j.jprot.2009.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.