SUMMARY
X-linked inhibitor of apoptosis (XIAP) is a potent antagonist of caspase apoptotic activity. XIAP also functions as an E3 ubiquitin ligase, targeting caspases for degradation. However, molecular pathways controlling XIAP activities remain unclear. Here we report that nitric oxide (NO) reacts with XIAP by S-nitrosylating its RING domain (forming SNO-XIAP), thereby inhibiting E3 ligase and antiapoptotic activity. NO-mediated neurotoxicity and caspase activation have been linked to several neurodegenerative disorders, including Alzheimer’s, Parkinson’s, and Huntington’s diseases. We find significant SNO-XIAP formation in brains of patients with these diseases, implicating this reaction in the etiology of neuronal damage. Conversely, S-nitrosylation of caspases is known to inhibit apoptotic activity. Unexpectedly, we find that SNO-caspase transnitrosylates (transfers its NO group) to XIAP, forming SNO-XIAP, and thus promotes cell injury and death. These findings provide unique insights into the regulation of caspase activation in neurodegenerative disorders mediated, at least in part, by nitrosative stress.
INTRODUCTION
Neuronal cell injury and death are prominent features of neurodegenerative disorders such as Alzheimer’s, Huntington’s, and Parkinson’s diseases (Mattson, 2000; Friedlander, 2003). While acute fulminant insults result in osmotic swelling and necrosis, chronic degenerative disorders can produce apoptotic cell death (Ankarcrona et al., 1995; Bonfoco et al., 1995), often mediated by the caspase family of cysteine proteases (Chan and Mattson, 1999; Lu et al., 2000). During degenerative processes, for example in Alzheimer’s disease, activated caspases may induce proteolysis of β-amyloid precursor protein (APP) or synaptic proteins, which may contribute to synaptic dysfunction and neuronal cell death. Inhibitor of apoptosis proteins (IAPs) represent important regulators of apoptosis through their ability to associate with active caspases and repress their catalytic activity (Eckelman et al., 2006; Salvesen and Duckett, 2002). In particular, XIAP interacts with active caspases-3/7/9 in the cytosol and is thought to be the most potent endogenous caspase inhibitor among the IAPs. XIAP harbors three copies of the baculovirus IAP repeat (BIR) domain and one RING domain. Characteristic BIR and RING folds contain zinc ions coordinated by histidine and cysteine residues. Biochemical and structural analyses indicate that BIR domains and their flanking sequences bind and inhibit the catalytic activity of apoptotic caspases (Fuentes-Prior and Salvesen, 2004). Additionally, the RING domain of XIAP can act as an E3 ligase, functioning in ubiquitination and subsequent degradation of heterologous substrates in vivo (caspases, and other IAP proteins) as well as XIAP itself (MacFarlane et al., 2002; Schile et al., 2008; Suzuki et al., 2001; Vaux and Silke, 2005; Yang et al., 2000).
Nitric oxide (NO) is also known to contribute to neuronal cell damage and death when present at excessive levels, but can promote neuronal survival under physiological conditions (Beckman, 1990; Dawson et al., 1991; Lipton et al., 1993). NO exerts its effects in large part through stimulation of guanylate cyclase or via protein S-nitrosylation, representing the covalent attachment of NO to cysteine thiol or more properly thiolate anion (Hess et al., 2005; Stamler et al., 1997). S-Nitrosylation has recently emerged as an important regulator of redox signaling, comparable in controlling protein function to other posttranslational modifications such as phosphorylation or acetylation. Physiological levels of NO can be neuroprotective, in part, via S-nitrosylation-mediated inhibition of N-methyl-d-aspartate (NMDA)-type glutamate receptor activity and caspase activity (Choi et al., 2000; Dimmeler et al., 1997; Lipton et al., 1993; Mannick et al., 1999; Melino et al., 1997; Tenneti et al., 1997). However, excess NO production in neurons results in activation of cell death signaling cascades that underlie many neurodegenerative disorders (Dawson et al., 1991; Lipton et al., 1993; Huang et al., 1994; Leist et al., 1997; Gu et al., 2002; Hara et al., 2005). Recent evidence directly links S-nitrosylation to protein misfolding and neuronal cell death (Chung et al., 2004; Yao et al., 2004; Uehara et al., 2006).
Here we report that S-nitrosylation of the RING domain of XIAP decreases its E3 ubiquitin ligase activity both in vitro and in intact cells, thereby blocking its ability to inhibit apoptosis by degrading caspases. We found that S-nitrosylated XIAP (SNO-XIAP) accumulates in neurons stimulated with pathophysiologically relevant levels of NMDA and in the brains of patients exhibiting neurodegeneration. Moreover, transnitrosylation of XIAP by SNO-caspase provides an additional mechanism for proapoptotic signaling. These results indicate that SNO-XIAP regulates caspase activity and contributes to neuronal injury or death in a number of neurodegenerative diseases.
RESULTS
S-Nitrosylation of XIAP In Vitro and in Intact Cells
Since we and others have found that the E3 ubiquitin ligase, parkin, is S-nitrosylated via cysteine thiol in its RING domain (Chung et al., 2004; Yao et al., 2004), we asked whether another RING domain-containing E3 ligase, XIAP, was also a target of S-nitrosylation. To answer this question, we employed a specific fluorescence assay for S-nitrosothiols (Gu et al., 2002; Wink et al., 1999) to detect SNO-XIAP in vitro. Incubation of a GST-XIAP fusion protein with a physiological NO donor, S-nitrosocysteine (SNOC), induced S-nitrosothiol formation, indicating the presence of SNO-XIAP (Figure 1A). To ascertain whether the RING domain of XIAP was S-nitrosylated, we performed mapping studies using truncated forms of XIAP. The SNO-fluorescence assay indicated that XIAP fragments containing BIR2 or BIR2/3 domains were not S-nitrosylated, whereas a fragment expressing the BIR2/3 and RING domains (BIR2-3-RING) was S-nitrosylated, suggesting that the ubiquitin-associated (UBA) or RING domain of XIAP contains the principal S-nitrosylated residue(s) (Figure 1B).
Figure 1. S-Nitrosylation of XIAP In Vitro and in Intact Cells.
(A) Recombinant protein GST-XIAP (0.5 µM) was incubated with the physiological NO donor SNOC (200 µM) at RT. After 30 min, S-nitrosylated XIAP was assessed by release of NO, causing conversion of 2,3-diaminonaphthalene (DAN) to the fluorescent compound 2,3-naphthotriazole (NAT). The degree of NAT fluorescence from GST-XIAP was set at 100% (n = 3–4; *p < 0.001 by ANOVA).
(B) S-Nitrosylation of XIAP-RING domain in vitro. Recombinant truncated XIAP proteins, His-BIR2 (BIR2), His-BIR2-3 (BIR2-3), and His-BIR2-3-RING (BIR2-3-RING) (0.2 µM each) were incubated with SNOC. Thirty minutes later, S-nitrosylated protein was assessed by fluorescence assay. The degree of NAT fluorescence from the BIR-2-3-RING domains was set at 100% (n = 3–5; *p < 0.01).
(C) S-Nitrosylation of XIAP in intact cells. SH-SY5Y cells were exposed to 200 µM SNOC, and S-nitrosylated XIAP (SNO-XIAP) was detected by the NO-biotin switch assay. Equal amounts of total XIAP in the cell lysate (Input) were verified. SNO-XIAP was detected by the NO-biotin switch assay (Eluate). The “Control” sample was subjected to decayed (old) SNOC under the same conditions. MMTS, methyl methane thiosulfonate; w/o Ascorbate, w/o MMTS, and w/o Biotin represent controls without reducing agent, thiol blocking agent, or biotin linker, respectively; arrowhead, XIAP; *, 60 kDa.
(D) S-Nitrosylation of XIAP-RING domain in intact cells. HEK293T cells were transfected with GFP-tagged WT-XIAP or XIAPΔRING (ΔRING) and exposed to SNOC. Control WT was exposed to decayed SNOC. S-Nitrosylation of transfected XIAP (SNO-GFP-XIAP) was detected with the NO-biotin switch assay; equal loading was confirmed (Input GFP-XIAP). Data are presented as mean + SEM.
We then asked whether XIAP is S-nitrosylated in intact cells using the NO-biotin switch method, a modified immunoblot to detect nitrosothiols (Jaffrey et al., 2001). After exposing neuroblastoma SH-SY5Y cells to SNOC, we detected S-nitrosylation of endogenous XIAP using a specific anti-XIAP antibody (Figure 1C and Figure S1). Next, using the NO-biotin switch assay after expressing XIAP fragments in intact cells, we found that the RING domain of XIAP is the predominant location of S-nitrosylated residues (Figure 1D and Figure S1B), confirming our earlier finding on recombinant preparations in vitro.
Identification of Cys450 as the S-Nitrosylated Cysteine Residue of XIAP
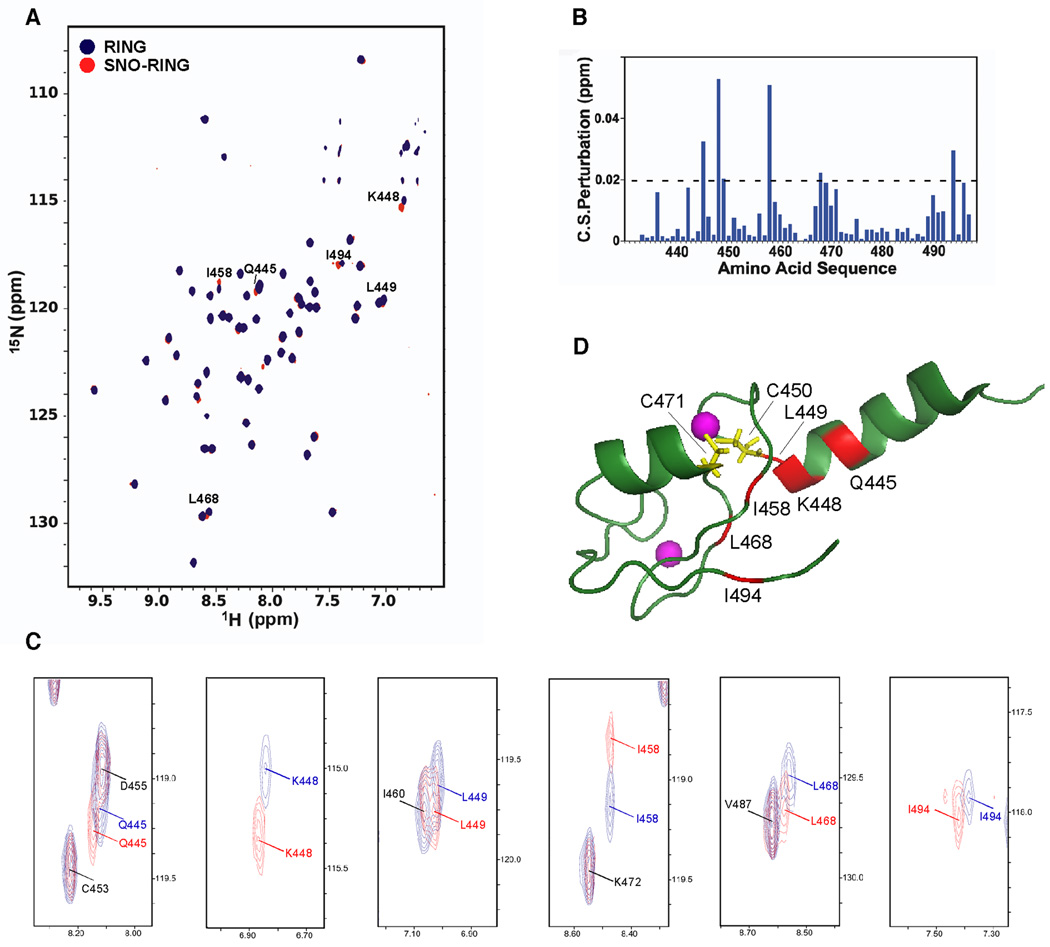
In order to identify the principal site of S-nitrosylation in the RING domain of XIAP, we employed three complementary approaches, using nuclear magnetic resonance (NMR) techniques to direct additional NO-biotin switch assays and mass spectrometry analyses. We labeled recombinant RING domain (residues 432–497 of XIAP) with 15N, and then acquired 2D 1H-15N transverse relaxation-optimized spectroscopy (TROSY)-NMR spectra to reveal the target cysteine of S-nitrosylation by chemical shift mapping (Pervushin et al., 1997). Comparison of the NMR spectra between native- and SNOC-treated RING domains revealed small but reproducible chemical shifts of several peaks corresponding to the backbone 1H-15N groups of amino-acid residues that were located in close proximity to Cys450 and Cys471 (Figure 2 and Figures S2A and S2B). These TROSY-NMR experiments were consistent with the notion that among the seven cysteines in the RING domain either Cys450 or Cys471 (or possibly both) were the target cysteine residues for S-nitrosylation. Moreover, these experiments suggested that formation of S-nitrosothiol on the RING domain induced minor conformational perturbations to proximate amino-acid residues (e.g., Lys448, Leu449, Ile458, and Leu468), and that nitrosylation did not result in substantial unfolding of the RING (Figure 2D and Figure S2B). Since we also found that alanine substitution of two of these residues (Lys448 or Leu449) resulted in a decrease in E3 ligase activity (Figure S2C), it is tempting to speculate that S-nitrosylation-induced conformational changes can affect the function of the RING domain. Consistent with this notion, we found that exposure to SNOC could effect release of zinc from the RING domain (Figure S2D).
Figure 2. NMR Analysis of the XIAP-RING Domain.
(A) [1H-15N] TROSY NMR spectra of native and S-nitrosylated XIAP-RING domain. Chemical shift (CS) differences are shown between native RING (RING; blue) and S-nitrosylated RING (SNO-RING; red). Cross peaks with significant CS changes caused by S-nitrosylation (>0.02 ppm) are labeled with one letter amino-acid codes.
(B) CS perturbations induced by S-nitrosylation versus amino acid sequence.
(C) Higher-powered views of cross peaks with significant CS changes caused by S-nitrosylation (>0.02 ppm). Cross peaks are marked in either red (S-nitrosylated form) or blue (native form). Cross peaks without significant CS changes (<0.02 ppm) are labeled with black one letter amino-acid codes.
(D) NMR structure of XIAP-RING domain (PDB: 2ECG). Residues manifesting CS changes after S-nitrosylation are displayed in red (>0.02 ppm). Two cysteines (C450 and C471) located proximate to the chemically shifted/perturbed residues are colored yellow.
To further investigate the specific site of S-nitrosylation on XIAP under physiological conditions, we next expressed wild-type (WT)-XIAP, XIAP(C450H), or XIAP(C471H) in HEK293 cells stably expressing neuronal NO synthase (HEK-nNOS) (Bredt et al., 1991). This approach allowed us to detect S-nitrosylation of XIAP engendered by endogenous NO. We chose histidine to replace cysteine in these constructs because, similar to cysteine, histidine can coordinate the zinc ion in the RING structure. The NO-biotin switch assay revealed that the C450H mutation significantly diminished endogenous NO-mediated S-nitrosylation, pointing to Cys450 in the RING domain as the primary site of S-nitrosylation of XIAP (Figure 3A). Interestingly, a characteristic acid-base motif for S-nitrosylation (Asp455, Lys448, and Lys451) surrounds the Cys450 residue (Stamler et al., 1997; Hess et al., 2005).
Figure 3. Identification of the Predominant S-Nitrosylation Site on XIAP and Formation of SNO-XIAP in Vivo.
(A) S-Nitrosylation of XIAP at Cys450 by NO-biotin switch assay. HEK-nNOS cells were transfected with WT, C450H, or C471H XIAP constructs and exposed to the Ca2+ ionophore A23187 (5 µM) to activate endogenous nNOS.
(B) ETD-MS/MS spectra of XIAP-RING domain. To obtain MS/MS spectra, charge +6 precursors ([M+6H]+6) of unmodified RING (RING, top; 1053.68 m/z) and, after exposure to 10 µM SNOC, S-nitrosylated RING (SNO-RING, bottom; 1058.40 m/z) were isolated for the ETD experiment by a linear ion trap (LTQ) analyzer. Amino-acid sequence of the XIAP-RING domain is listed at the top of the RING spectrum. c and z ions present in both modified and unmodified proteins are indicated in blue, while ions detected only in the SNO-RING are in red. m/z values for C11+3,C13+2, and C16+3 ions in the SNO-RING spectrum are 425.17, 755.36, and 632.17, respectively. Additional MS data (LTQ Orbitrap XL-MS and ETD-MS/MS) are available in Figures S3A and S3B.
(C) SNO-XIAP in brains of patients with neurodegenerative diseases. Postmortem brain tissues from patients with neurodegenerative and non-CNS conditions (controls) were subjected to the NO-biotin switch assay. Images separated by a black line are from the same gel.
Next, we performed a high-resolution Orbitrap mass spectrometry analysis of the wild-type XIAP-RING domain after exposure to SNOC in order to determine if S-nitrosylation of XIAP occurs at Cys450. To keep this post-translational modification intact during the acquisition of MS/MS spectra, we employed an innovative top-down approach together with electron transfer dissociation (ETD) technology, which, compared to conventional collision-induced dissociation methods, allows us to preserve labile modifications on larger peptides (Han et al., 2008; Mikesh et al., 2006). Consistent with the results of our NO-biotin switch assay, the LTQ Orbitrap XL-ETD MS/MS data detected an NO adduct predominantly at Cys450 of the XIAP-RING domain (Figure 3B and Figure S3). Taken together, these results strongly suggest that S-nitrosylation occurs on the RING domain of XIAP, specifically at Cys450, and that formation of SNO-XIAP at this site modulates the surrounding local conformation or chemical environment.
S-Nitrosylation of XIAP In Vivo
Caspase activation and excessive NO generation have been associated with several human neurodegenerative conditions, including Parkinson’s with diffuse Lewy body disease (DLBD), Alzheimer’s disease (AD), and Huntington’s disease (HD). Our findings from in vitro and cell-based experiments raised the possibility that XIAP activity could be affected by S-nitrosylation in these disorders. Therefore, to determine whether XIAP is S-nitrosylated in humans in vivo, we performed an NO-biotin switch assay with brain extracts prepared from postmortem brains of patients with sporadic DLBD, AD, HD, and control brains (patients who died of non-CNS disorders) (Uehara et al., 2006). We observed that these brains contained significantly higher levels of SNO-XIAP than controls (Figure 3C, Figure S3C, and Table S1), supporting our hypothesis that SNO-XIAP levels are positively correlated with disease pathogenesis. Interestingly, we did not find elevated levels of S-nitrosylated caspase-3 in these diseased brains (Figure S3D and Table S1). In addition to XIAP, other inhibitors of apoptosis include cIAP1 and cIAP2. However, SNO-cIAP1 and SNO-cIAP2 were undetectable under our conditions (Figure S3E and S3F, and Table S1). Taken together, these results are in accord with previous findings that downregulation of XIAP but not cIAP1 or cIAP2 is essential in regulating neuronal apoptosis (Potts et al., 2003).
S-Nitrosylation of XIAP Downregulates its E3 Ligase Activity
Recent studies have shown that XIAP and caspases are substrates for XIAP-mediated ubiquitination in vitro and in intact cells (Morizane et al., 2005; Schile et al., 2008; Shin et al., 2003; Suzuki et al., 2001; Yang et al., 2000). Our NMR data suggested that NO could perturb the structural or chemical environment of the RING domain in a region that would be expected to affect ubiquitin enzymatic activity. Thus, we asked whether S-nitrosylation of the XIAP-RING domain affects its E3 ligase activity. Initially, we examined auto-ubiquitination of XIAP in vitro to confirm a direct effect of NO on XIAP E3 ubiquitin ligase activity. When recombinant GST-XIAP was incubated in vitro with ubiquitin-activating enzyme E1, ubiquitin conjugating enzyme E2 (UbcH5b), and ubiquitin (Ub), a smear of proteins representing polyubiquitinated XIAP on immunoblots was identified by reaction with anti-ubiquitin antibody (Figures 4A and S4A). Pretreatment of recombinant XIAP with SNOC starting 30 min prior to addition of-E1,-E2, and Ub proteins markedly reduced XIAP auto-ubiquitination, suggesting that NO directly interferes with XIAP E3 ligase activity. Although NO dissipates from SNOC quickly (within ~5 min under our experimental conditions), we found that S-nitrosylation of cysteine thiol on XIAP and certain other proteins lasts for more than 30 min (Gu et al., 2002; Lei et al., 1992). This time frame allowed us to exclude a direct effect of NO on E1, E2, or ubiquitin since all of the NO from SNOC was dissipated by the time that these proteins were added to the reaction milieu, supporting the notion that NO directly inactivated the E3 ligase activity of XIAP (Figure 4A and Figure S4A). Additionally, control treatment with inactive SNOC (old SNOC from which NO had been dissipated) had no effect on auto-ubiquitination (Figure 4A and Figure S4A).
Figure 4. Regulation of XIAP E3 Ligase Activity by S-Nitrosylation in Vitro and in Intact Cells.
(A) In vitro S-nitrosylation of XIAP downregulates auto-ubiquitination. Purified recombinant GST-XIAP was incubated with SNOC (200 µM) or old SNOC, and 30 min later subjected to in vitro ubiquitination at RT. All samples were incubated with ATP. SNOC decreased XIAP auto-ubiquitination, as detected by anti-XIAP antibody.
(B) S-Nitrosylation of XIAP reduces auto-ubiquitination in HEK293T cells. HEK 293T cells co-transfected with HA-tagged ubiquitin plus Myc-tagged XIAP or Myc-tagged XIAPΔRING were incubated with SNOC to S-nitrosylate XIAP. After 2 to 12 hr, cell lysates were subjected to immunoprecipitation with anti-myc followed by immunoblot analysis with anti-HA to detect ubiquitinated proteins.
(C, D) S-Nitrosylation of XIAP in vitro down-regulates caspase-3 and -9 ubiquitination. Recombinant GST-XIAP was incubated with SNOC or old SNOC and then subjected to in vitro ubiquitination for 90 min at RT. All samples were incubated with ubiquitin and ATP. High molecular weight forms of caspases-3 and -9, apparently representing polyubiquitinated proteins, were detected with specific antibodies against these caspases.
(E) S-Nitrosylation of XIAP reduces caspase-3 ubiquitination in HEK293T cells. HEK293T cells co-transfected with HA-tagged ubiquitin, Fas, Myc-tagged mutant caspase-3(C285A), and His-tagged XIAP were exposed to SNOC. After 6 hr, cell lysates were immunoprecipitated with anti-myc antibody followed by immunoblot analysis with the indicated antibodies. Arrowhead, procaspase-3; *, immunoglobulin light chain.
(F) S-Nitrosylation of XIAP reduces caspase-9 ubiquitination in HEK293T cells. HEK 293T cells co-transfected with HA-tagged ubiquitin, Bax, FLAG-tagged mutant caspase-9(C285A), and Myc-tagged XIAP were exposed to SNOC. After 6 hr, cell lysates were immunoprecipitated with anti-FLAG antibody followed by immunoblot analysis with the indicated antibodies. Arrowhead, XIAP; *, immunoglobulin heavy chain.
To further verify the effect of NO on XIAP, we compared the E3 ligase activity of wild-type (WT)-XIAP before and after nitrosylation in HEK293T cells. HEK293T cells transfected with myc-tagged XIAP and hemagglutinin (HA)-tagged ubiquitin exhibited a significant, time-dependent decrease in XIAP auto-ubiquitination after exposure to SNOC (Figure 4B and Figures S4B and S4C.). Lysates of cells transfected with XIAPΔRING did not exhibit auto-ubiquitination.
Next, we asked if S-nitrosylation of XIAP affected its function in a pathophysiologically relevant manner. Since a contribution of XIAP auto-ubiquitination to antiapoptotic activity has not been clearly demonstrated (Shin et al., 2003; Yang et al., 2000), we studied whether XIAP-mediated ubiquitination of caspases-3 and -9, which has recently been reported to be important in modulating apoptosis in vivo (Schile et al., 2008), was affected by NO. We found that pretreatment of XIAP with SNOC resulted in a decrease in polyubiquitination of caspases-3 and -9 in vitro (Figures 4C and 4D). Next, to verify that SNO-XIAP manifests decreased E3 ligase activity in intact cells, we cotransfected HEK293T cells with various combinations of XIAP, ubiquitin, and catalytic mutants of caspases-3 or -9. We and others had previously shown that NO can inhibit activity of caspases via S-nitrosylation of the catalytic cysteine (Choi et al., 2000; Dimmeler et al., 1997; Hoffmann et al., 2001; Kim and Tannenbaum, 2004; Kim et al., 1997; Mannick et al., 1999; Melino et al., 1997; Mitchell and Marletta, 2005; Tenneti et al., 1997). We therefore used cysteine mutants of caspases [caspase-3(C285A) and caspase-9(C285A)] in this assay to exclude potential effects of SNO-caspases on their ubiquitination. To induce caspase cleavage and activation, the pro-apoptotic constructs Fas and Bax were co-transfected into HEK293T cells in order to stimulate the extrinsic and intrinsic apoptotic pathways, respectively (Deveraux et al., 1999; Ghatan et al., 2000). Co-transfection of XIAP caused a substantial increase in the amount of ubiquitinated caspase-3 compared to untransfected cells, consistent with previous reports (Suzuki et al., 2001). Exposure to SNOC 6 hr prior to harvesting the transfected cells abrogated the increase in caspase-3 ubiquitination (Figure 4E and Figure S4B), while the NO donor failed to downregulate the E3 ligase activity of the S-nitrosylation-deficient mutant XIAP(C450H) (Figure S4D). We performed similar experiments analyzing the analogous caspase-9 mutation (C285A) and found that SNOC dramatically decreased this ubiquitination (Figure 4F and Figure S4B). Finally, we observed that either exposure to SNOC or knockdown of XIAP, attained using short hairpin RNAs (shRNAs) targeting XIAP, inhibited ubiquitination of endogenous caspase-3 (Figure S4E-G). Endogenously produced NO also suppressed caspase-3 ubiquitination (Figure S4H-J). In a control assay, we found that physiological levels of NO did not affect XIAP interaction with caspase-3; only extremely high, non-physiological levels of an NO donor (≥0.5 mM influenced XIAP binding to caspase) (Figure S4K and S4L). Together, these results are consistent with the notion that at physiological levels NO inhibit ubiquitination of caspases-3 and -9 through S-nitrosylation of XIAP, rather than affecting caspase/XIAP interactions via the BIR domains.
NO Results in Increased Levels of Cleaved/Activated Caspase-3 in Response to Apoptotic Stimuli
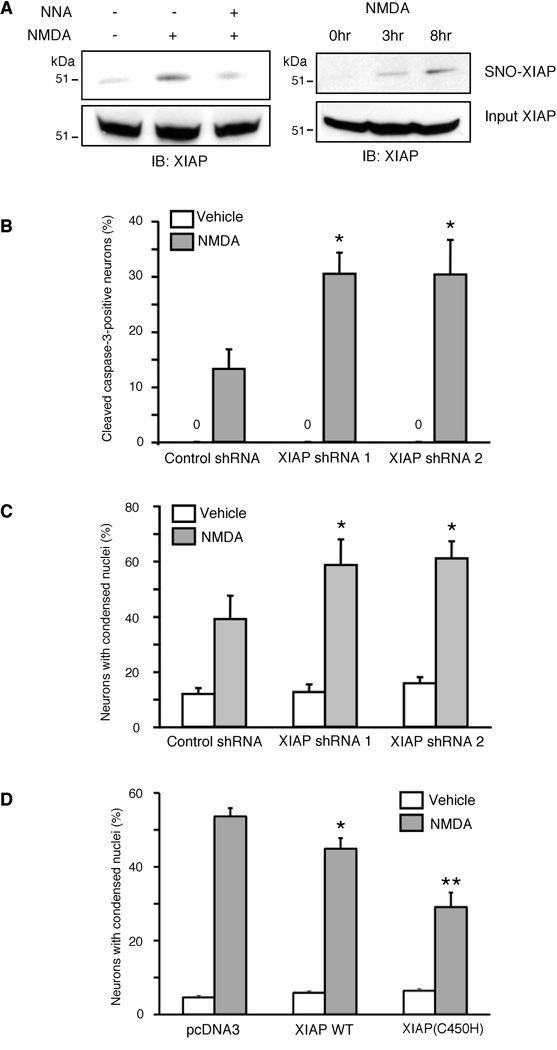
Because S-nitrosylation inhibits XIAP E3 ligase activity, we expected that NO would also suppress proteasomal degradation of active caspases, leading to their accumulation in apoptotic cells. To investigate the effect of endogenous NO on the active/cleaved form of caspase-3, we used HEK-nNOS cells. Treatment of HEK-nNOS cells with the calcium ionophore A23187 stimulates nNOS activity and leads to rapid NO generation (Tenneti et al., 1997), enabling us to monitor the effect of physiologically-produced NO on caspase-3. We found that the endogenous NO thus generated promoted S-nitrosylation of XIAP within 2 hr of A23187 exposure, and this increase was abrogated by the NOS inhibitor N-nitro-l-arginine (NNA, Figure 5A).
Figure 5. S-Nitrosylation of XIAP Augments Levels of Active Caspase-3 and Impairs XIAP Protective Function.
(A) S-Nitrosylation of XIAP by endogenous NO. HEK-nNOS cells were exposed to 5 µM A23187 to activate nNOS in the presence or absence of the NOS inhibitor, Nω-nitro-l-arginine (NNA), and SNO-XIAP was assessed by NO-biotin switch assay.
(B) Endogenous NO stabilizes cleaved caspase-3. HEK-nNOS cells co-transfected with myc-tagged caspase-3, myc-tagged XIAP or XIAPΔRING, and Bax were incubated with A23187 for 6 hr. Active caspase-3 (mycCaspase-3) was assessed by immunoblotting with anti-myc antibody. Endogenous c-myc detected by anti-myc antibody served as a loading control. SF, small fragment of cleaved caspase-3.
(C) NO reduces the inhibitory effect of XIAP on caspase activity. SH-SY5Y cells were transfected with a Bax expression plasmid. After 17 hr, cells were exposed to SNOC, and DEVDase activity was measured 7 hr later. Co-expression of XIAP attenuated Bax-induced caspase activation, an effect reversed by SNOC. XIAPΔR was less efficient than XIAP in inhibiting caspase activity triggered by Bax expression, and SNOC had no effect on XIAPΔR. As a control, the caspase inhibitor zVAD-fmk (25 µM) completely blocked Bax-induced caspase activation (n = 3–7).
(D) NO reduces the antiapoptotic effect of XIAP. A Bax expression plasmid was transfected into SH-SY5Y cells along with the indicated XIAP constructs. Apoptotic cell death was monitored by counting the number of nuclei with condensed or fragmented chromatin with Hoechst staining. Co-expression of XIAP attenuated Bax-induced apoptosis, an effect reversed by SNOC. XIAPΔR was less efficient than XIAP in protecting cells from cell death triggered by Bax expression, and SNOC had no effect on XIAPΔR. zVAD.fmk (25 µM) completely blocked Bax-induced cell death (n ≥ 3; *p < 0.01 by ANOVA). Data are presented as mean + SEM.
We then sought to determine whether endogenous NO could further increase the level of cleaved caspase-3 produced in response to the apoptotic stimuli Fas or Bax. This finding would be consistent with the notion that S-nitrosylation of XIAP was inhibiting its E3 ubiquitin ligase activity and thus the degradation of the caspase. In these experiments, we found that in the absence of exogenous XIAP, transfection of Bax augmented the level of cleaved caspase-3 (Figure 5B and Figure S5A). Transfection with WT-XIAP abrogated this increase in caspase-3 processing (Figure 5B). In contrast, cells transfected with XIAP shRNAs exhibited increased levels of cleaved caspase-3 (Figure S5C). This effect was mimicked by XIAP RING domain mutant (XIAPΔR; Figure 5B, lane 5). These results are consistent with the presence of XIAP-mediated degradation of endogenous caspase under our conditions. Importantly, exposure of HEK-nNOS cells to A23187 or UV (to generate endogenous NO) resulted in an increase in cleaved caspase-3 levels, presumably due to the decreased E3 ligase activity of SNO-XIAP (Figure 5B, lane 7 and Figure S5D, lane 2). Co-treatment with NNA (to inhibit the generation of endogenous NO) prevented this increase in cleaved caspase-3. Additionally, exposure to the proteasome inhibitor MG132 resulted in increased levels of cleaved caspase-3 (Figure S5B, lane 5), indicating that proteasome-dependent degradation of the caspase would otherwise have occurred after its ubiquitination by XIAP in the absence of endogenous NO. Taken together, these results strongly suggest that down-regulation of the E3 ligase activity of XIAP by S-nitrosylation promotes accumulation of cleaved caspase-3.
NO Abrogates XIAP-Mediated Inhibition of Caspase Activity and Apoptosis
To further define the mechanism and pathophysiological relevance of S-nitrosylation on XIAP function, we analyzed the effect of SNO-XIAP on caspase activity and cell death. Translocation of Bax from the cytosol to mitochondria is thought to contribute to the pathogenesis of neurodegeneration by triggering the disruption of mitochondrial membrane potential, cytochrome c release, and caspase activation (Ghatan et al., 2000). We monitored caspase activity and cell viability induced by Bax expression under conditions promoting SNO-XIAP formation in neuroblastoma SH-SY5Y cells. We chose SH-SY5Y cells for these experiments because, unlike primary neurons, exposure to NO by itself does not affect death in this cell line under basal conditions (Figure S5E) (Chung et al., 2004; Uehara et al., 1999; Uehara et al., 2006). Thus, we were able to not only obtain NO-induced SNO-XIAP formation without triggering cell death but also monitor the effect of SNO-XIAP on Bax-induced cell death. We found that cells co-expressing WT-XIAP and Bax showed a significant decrease in caspase activity compared to cells expressing Bax alone, as measured by cleavage of the fluorogenic peptide substrate Ac-DEVD-Afc by executioner caspases (Figure 5C and Figure S5F). Exposure to SNOC partially abolished this inhibitory effect of XIAP under conditions in which NO produces S-nitrosylation and thus inhibition of XIAP E3 ligase activity (Figures 1C, 4E, and 4F). In contrast, transfection with XIAPΔRING manifested a little if any inhibitory effect on caspase activity and was insensitive to NO (Figure 5C), consistent with the notion that NO regulated XIAP function via reaction with cysteine in the RING domain. These results also imply that, under these conditions, RING domain-dependent effects of XIAP, presumably reflecting XIAP E3 ligase activity, exert control over caspase function separate from the effect of XIAP binding to either caspase-3 via BIR2 or caspase-9 via BIR3 domains, as observed elsewhere (Deveraux et al., 1999).
As a control for these experiments, immunoblots indicated that the increase in caspase activity, observed after either exposure to SNOC or expression of XIAPΔR, was not due to different levels of XIAP expression (Figure S5G). Moreover, consistent with these effects on caspase activity, we found that Bax-induced apoptosis was ameliorated by WT-XIAP; however, this protective effect was abrogated by exposure to SNOC (Figure 5D). Overall, these results suggest that S-nitrosylation of the XIAP-RING domain attenuates the protective function of XIAP against neuronal cell death.
Endogenous XIAP in Rat Cerebrocortical Neurons Inhibits NMDA Receptor-Mediated Excitotoxic Cell Death
It is well known that overstimulation of neuronal NMDA receptors (NMDARs) can trigger intracellular signaling pathways, including excessive NO generation by nNOS, that lead to neuronal cell death. Indeed, NMDAR-mediated excitotoxicity may contribute to the pathogenesis of a number of neurodegenerative diseases (Dawson et al., 1991; Lipton and Rosenberg, 1994). To study the potential relevance of SNO-XIAP formation to these pathophysiological conditions, we employed rat primary cerebrocortical cell cultures. Using the NO-biotin switch assay, we found that NMDA elicited S-nitrosylation of XIAP in a time- and NOS-dependent manner prior to the manifestation of neurotoxicity (Figure 6A). Since we had already shown that S-nitrosylation of XIAP inhibits its antiapoptotic activity, we next sought to determine in primary cerebrocortical neurons if attenuation of XIAP activity affected cell death in a pathophysiologically-relevant context.
Figure 6. XIAP Provides Neuroprotection from NMDAR-Mediated Excitotoxicity.
(A) SNO-XIAP was detected in cultured rat cerebrocortical neurons after exposure to NMDA by NO-biotin switch analysis. The NOS inhibitor NNA inhibited the formation of SNO-XIAP.
(B, C) Reduction of XIAP levels by shRNA sensitizes cortical neurons to NMDA. Cortical neurons were transfected with shRNAs, exposed to NMDA, and after 6 hr immunostained with anti-NeuN (to identify neurons) and anti-cleaved caspase-3 (to detect active caspase-3). The percentage of active caspase-3-positive neurons increased in XIAP shRNA-transfected neurons compared to control neurons (B). In parallel, the number of apoptotic neurons increased 8–14 hr later (C).
(D) Enhanced antiapoptotic activity of XIAP(C450H). Cortical neurons were transfected with XIAP or XIAP(C450H), exposed to NMDA, and after 12 hr immunostained with anti-NeuN. Apoptotic cell death was monitored by counting the number of Hoechst-stained nuclei containing condensed/fragmented chromatin (n ≥ 500 neurons scored in 3 independent experiments; *p < 0.05, **p < 0.01 by ANOVA). Data are presented as mean + SEM.
In cortical neurons, we previously found (Budd et al., 2000; Tenneti and Lipton, 2000) that the caspase-dependent component of NMDA-induced cell death required activation of caspase-3. To assess the antiapoptotic activity of endogenous XIAP in cortical neurons, we employed two shRNAs targeting rat XIAP (shRNA-1 and- 2; Figures S4D and S4E). We then determined the effect of XIAP knockdown induced by RNAi on accumulation of cleaved caspase-3 in the neurons. Fluorescent immunocytochemistry showed enhanced levels of cleaved caspase-3 in XIAP-depleted neurons 6 hr after exposure to NMDA (Figure 6B and Figure S6). In parallel, XIAP-shRNA increased the degree of NMDA-evoked neuronal cell death that subsequently developed in the cultures (Figure 6C and Figure S6). Importantly, XIAP(C450H), which lacks the S-nitrosylation site in the RING domain, enhanced antiapoptotic activity during NMDA-triggered neuronal cell death (Figure 6D). These results suggest that under these conditions XIAP protected primary cortical neurons from NMDAR-mediated neurotoxicity. Taken together, since NO inhibits XIAP activity via S-nitrosylation, these findings are consistent with the notion that the formation of SNO-XIAP may render primary cortical neurons more susceptible to excitotoxic cell death. Additionally, using a previously developed technique involving quantitative immunoblots30, we found evidence that levels of SNO-XIAP in human brains with neurodegenerative diseases are of pathophysiological significance (Figure S6C and Table S1).
Transnitrosylation from Caspase-3 to XIAP
NO has been reported to constitutively S-nitrosylate the pro-form of caspase-3. Activation of cell death pathways induces both cleavage and selective denitrosylation of the catalytic cysteine residue of caspase-3, resulting in activation (Mannick et al., 1999). S-Nitrosylation of caspase-3 has been demonstrated in several cell types, including primary cortical neurons (Dimmeler et al., 1997; Melino et al., 1997; Tenneti et al., 1997). Interestingly, SNO-thioredoxin (Trx) can transnitrosylate caspase-3, possibly representing an anti-apoptotic activity of Trx (Mitchell and Marletta, 2005). Since XIAP can also be nitrosylated and interacts with caspase-3, we asked if a mechanism for caspase denitrosylation and activation could involve transnitrosylation, i.e., transfer of the NO group from caspase-3 to XIAP? To determine this, we first exposed purified caspase-3 or XIAP to SNOC to allow the formation of SNO-caspase-3 or SNO-XIAP, respectively. Then, each of these S-nitrosylated proteins was mixed with their unlabeled counterpart, i.e., SNO-caspase-3 with XIAP or SNO-XIAP with caspase-3 to induce potential transnitrosylation. Subsequently, we detected the transfer of NO from caspase-3 to XIAP by the biotin-switch assay, i.e., we observed transnitrosylation from caspase-3 to XIAP but not vice-versa (Figure 7A). These results suggest that NO is preferentially transferred from caspase-3 to XIAP. As a control, we showed that residual SNOC was not the nitrosylating agent because NO had been totally dissipated by SNOC at the time that the transnitrosylation reaction was initiated by adding the potential substrate (XIAP in this case). Additionally, we found that SNO-caspase-3 could not transnitrosylate mutant XIAP(D148A), which was impaired in its interaction with caspase-3 (Figure 7B). This latter result indicates that transnitrosylation requires physical interaction between caspase-3 and XIAP. Furthermore, we confirmed that S-transnitrosylation only occurs between XIAP and the catalytically processed (cleaved) form of caspase-3 since the addition of SNO-procaspase-3 to XIAP did not generate SNO-XIAP formation (Figure 7C). Importantly, we found evidence that the transnitrosylation reaction from caspase-3 to XIAP would proceed under in vivo conditions by calculating the relative redox potential (ΔE o′) using a modification of the Nernst equation and the associated change in Gibbs free energy (ΔGo′) (see Figure S7 and Supplemental Experimental Procedures).
Figure 7. Transnitrosylation of XIAP by SNO-Caspase-3.
(A) SNO-caspase-3 transnitrosylates XIAP but not vice-versa. Transnitrosylation reactions were performed as described in the Supplemental Data. Amounts (Input) of XIAP and caspase-3 were verified in each reaction. S-Nitrosylated proteins were detected by the NO-biotin switch assay. All panels depicted are from the same gel.
(B) SNO-caspase-3 does not transnitrosylate an XIAP mutant lacking the caspase-3 binding motif [XIAP(D148A)].
(C) Cleaved SNO-caspase-3 transnitrosylates XIAP. Caspase cleavage in HEK 293T lysates was activated with dATP and cytochrome c (dATP/CytC). Caspase-3 was then immunoprecipitated and exposed to SNOC. The resulting SNO-capase-3 was then incubated with recombinant XIAP to test for possible transnitrosylation. S-Nitrosylated proteins were detected by the NO-biotin switch assay and revealed transnitrosylation from cleaved (rather than full-length) SNO-caspase-3 to XIAP.
(D) Schematic Illustration of the Mechanism of SNO-XIAP—Mediated Neuronal Cell Death. Pathway (1): Under normal conditions, XIAP efficiently blocks caspases. Additionally, XIAP serves as an E3 ligase that ubiquitinates caspases and thus targets caspases for proteasomal degradation. Pathway (2): Under nitrosative conditions, NO inactivates the E3 ligase activity of XIAP via S-nitrosylation, thus stabilizing caspases, and sensitizing neurons to apoptotic stimuli. Pathway (3): Constitutively S-nitrosylated caspases serve as an additional mechanism to produce SNO-XIAP via transnitrosylation in neurons undergoing apoptotic cell death.
DISCUSSION
In the present study, we demonstrate that endogenous levels of NO are capable of S-nitrosylating the RING domain of XIAP, thus suppressing its E3 ligase activity in neuronal cells. Point mutation of the cysteine residue at position 450 (C450H) in the RING domain virtually eliminated S-nitrosylation of full-length XIAP. Since caspase-3 is normally a substrate for XIAP E3 ligase, less caspase-3 is degraded when XIAP is S-nitrosylated and thus caspase-dependent cell death pathways are further activated and contribute to neuronal cell death (Figure 7D). Our finding that SNO-XIAP accumulates in the brains of human patients with neurodegenerative diseases at pathophysiologically relevant levels supports a role for SNO-XIAP in neurodegeneration.
We initially found in SH-SY5Y cells that S-nitrosylation of XIAP not only decreased its E3 ligase activity but also rendered cells susceptible to cell death triggered by Bax. We also demonstrated in primary cerebrocortical neurons that direct reduction of XIAP levels by RNAi led to increased caspase-3 activity and subsequent cell death elicited by NMDA. These findings are consistent with the notion that decreased XIAP activity via nitrosylation leads to SNO-XIAP-mediated caspase activation, which may at least in part underlie the neuronal loss seen in several neurodegenerative diseases. Additionally, we observed evidence for transnitrosylation in this system, resulting in the transfer of NO from SNO-caspase-3 to XIAP, providing another mechanism for SNO-XIAP formation to increase neuronal vulnerability. Mechanistic details of this reaction and its effect on XIAP activity are discussed further in the Supplemental Data.
While this work was under review, another group found that XIAP is S-nitrosylated in animal models of PD and human PD brains (Tsang et al., 2009). These authors reported that very high (500 µM) concentrations of exogenous NO produced S-nitrosylation of cysteine residues in the BIR domains of XIAP to influence caspase binding and activity. We feel that the discrepancy with our observation that XIAP is S-nitrosylated in the RING domain rather than the BIR domains can be explained by our use of lower (physiological) levels of endogenous NO to characterize the critical protein thiol undergoing reaction; very high (non-physiological) amounts of exogenous NO can react indiscriminately with other protein thiols to produce nitrosylation of virtually any cysteine residue, and therefore may affect BIR domain cysteines in this manner (Tsang et al., 2009).
In conclusion, in this study, we demonstrate that NO negatively regulates the antiapoptotic function of XIAP by decreasing E3 ligase activity. We find evidence for specific S-nitrosylation of XIAP (to form SNO-XIAP) and posit that this reaction may be relevant to a number of pathophysiological conditions mediated at least in part by nitrosative or oxidative stress. The elucidation of this pathway to S-nitrosylation and inhibition of XIAP activity also provides a potential target for therapies directed at neurodegenerative disorders and possibly other diseases associated with nitrosative stress.
EXPERIMENTAL PROCEDURES
Plasmids
Most XIAP and caspase plasmids used in this study were previously published (Deveraux et al., 1999; Shin et al., 2003; Yao et al., 2004). For details of other plasmids, see Supplemental Experimental Procedures.
Generation of Recombinant Proteins
Recombinant proteins were purified from BL21 (DE3) E. coli as described previously (Deveraux et al., 1999) with minor modifications (Supplemental Experimental Procedures).
NO-Biotin Switch Assays
The NO-biotin switch assay was performed as described previously (Jaffrey et al., 2001; Yao et al., 2004) with some modifications (Supplemental Experimental Procedures).
Human Brain Tissue
Brains were obtained postmortem from subjects whose age, postmortem interval, and gender have been described in part previously (Uehara et al., 2006); additional characteristics are presented in Table S1. Human brain samples were analyzed with Institutional permission under state of California and NIH guidelines. Informed consent was obtained according to procedures approved by Institutional Review Boards at the University of California, San Diego and the Sanford-Burnham Medical Research Institute, La Jolla.
Supplementary Material
ACKNOWLEDGMENTS
We thank Xiaoqun Fang and Traci Fang Newmeyer for assistance with primary cultures, Elizer Masliah for providing human brain tissues, and Dean Jones for helpful discussions. This work was supported in part by a JSPS Postdoctoral Fellowship for Research Abroad (T.N.), NIH Fellowship grant T32 CA77109 (B.P.E), NIH grants P01 HD29587, R01 EY09024, R01 EY05477, R01 NS41207, P30 NS057096 (S.A.L) and R01 NS37878 (G.S.S.), and the American Parkinson’s Disease Association, San Diego Chapter. During the course of this work, S.A.L. was a Senior Scholar in Aging Research of the Ellison Medical Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information include Supplemental Results, Discussion, Experimental Procedures, seven figures, and one table can be found with this article online at http://cell.com/molecular-cell/supplemental/XXXX.
REFERENCES
- Ankarcrona M, Dypbukt JM, Bonfoco E, Zhivotovsky B, Orrenius S, Lipton SA, Nicotera P. Glutamate-induced neuronal death: a succession of necrosis or apoptosis depending on mitochondrial function. Neuron. 1995;15:961–973. doi: 10.1016/0896-6273(95)90186-8. [DOI] [PubMed] [Google Scholar]
- Beckman JS. Ischaemic injury mediator. Nature. 1990;345:27–28. doi: 10.1038/345027b0. [DOI] [PubMed] [Google Scholar]
- Bonfoco E, Krainc D, Ankarcrona M, Nicotera P, Lipton SA. Apoptosis and necrosis: two distinct events induced, respectively, by mild and intense insults with N-methyl-D-aspartate or nitric oxide/superoxide in cortical cell cultures. Proc. Natl. Acad. Sci. USA. 1995;92:7162–7166. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome P-450 reductase. Nature. 1991;351:714–718. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- Budd SL, Tenneti L, Lishnak T, Lipton SA. Mitochondrial and extramitochondrial apoptotic signaling pathways in cerebrocortical neurons. Proc. Natl. Acad. Sci. USA. 2000;97:6161–6166. doi: 10.1073/pnas.100121097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Mattson MP. Caspase and calpain substrates: roles in synaptic plasticity and cell death. J. Neurosci. Res. 1999;58:167–190. [PubMed] [Google Scholar]
- Choi YB, Tenneti L, Le DA, Ortiz J, Bai G, Chen HS, Lipton SA. Molecular basis of NMDA receptor-coupled ion channel modulation by S-nitrosylation. Nat. Neurosci. 2000;3:15–21. doi: 10.1038/71090. [DOI] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Dawson VL, Dawson TM, London ED, Bredt DS, Snyder SH. Nitric oxide mediates glutamate neurotoxicity in primary cortical cultures. Proc. Natl. Acad. Sci. USA. 1991;88:6368–6371. doi: 10.1073/pnas.88.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveraux QL, Leo E, Stennicke HR, Welsh K, Salvesen GS, Reed JC. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmeler S, Haendeler J, Nehls M, Zeiher AM. Suppression of apoptosis by nitric oxide via inhibition of interleukin-1beta-converting enzyme (ICE)-like and cysteine protease protein (CPP)-32-like proteases. J. Exp. Med. 1997;185:601–607. doi: 10.1084/jem.185.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander RM. Apoptosis and caspases in neurodegenerative diseases. N. Engl. J. Med. 2003;348:1365–1375. doi: 10.1056/NEJMra022366. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatan S, Larner S, Kinoshita Y, Hetman M, Patel L, Xia Z, Youle RJ, Morrison RS. p38 MAP kinase mediates bax translocation in nitric oxide-induced apoptosis in neurons. J. Cell Biol. 2000;150:335–347. doi: 10.1083/jcb.150.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- Han X, Aslanian A, Yates JR., 3rd Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Hoffmann J, Haendeler J, Zeiher AM, Dimmeler S. TNFalpha and oxLDL reduce protein S-nitrosylation in endothelial cells. J. Biol. Chem. 2001;276:41383–41387. doi: 10.1074/jbc.M107566200. [DOI] [PubMed] [Google Scholar]
- Huang Z, Huang PL, Panahian N, Dalkara T, Fishman MC, Moskowitz MA. Effects of cerebral ischemia in mice deficient in neuronal nitric oxide synthase. Science. 1994;265:1883–1885. doi: 10.1126/science.7522345. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Kim JE, Tannenbaum SR. S-Nitrosation regulates the activation of endogenous procaspase-9 in HT-29 human colon carcinoma cells. J. Biol. Chem. 2004;279:9758–9764. doi: 10.1074/jbc.M312722200. [DOI] [PubMed] [Google Scholar]
- Kim YM, Talanian RV, Billiar TR. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J. Biol. Chem. 1997;272:31138–31148. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- Lei SZ, Pan ZH, Aggarwal SK, Chen HS, Hartman J, Sucher NJ, Lipton SA. Effect of nitric oxide production on the redox modulatory site of the NMDA receptor-channel complex. Neuron. 1992;8:1087–1099. doi: 10.1016/0896-6273(92)90130-6. [DOI] [PubMed] [Google Scholar]
- Leist M, Volbracht C, Kuhnle S, Fava E, Ferrando-May E, Nicotera P. Caspase-mediated apoptosis in neuronal excitotoxicity triggered by nitric oxide. Mol. Med. 1997;3:750–764. [PMC free article] [PubMed] [Google Scholar]
- Lipton SA, Choi YB, Pan ZH, Lei SZ, Chen HS, Sucher NJ, Loscalzo J, Singel DJ, Stamler JS. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- Lipton SA, Rosenberg PA. Excitatory amino acids as a final common pathway for neurologic disorders. N. Engl. J. Med. 1994;330:613–622. doi: 10.1056/NEJM199403033300907. [DOI] [PubMed] [Google Scholar]
- Lu DC, Rabizadeh S, Chandra S, Shayya RF, Ellerby LM, Ye X, Salvesen GS, Koo EH, Bredesen DE. A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor. Nat. Med. 2000;6:397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- MacFarlane M, Merrison W, Bratton SB, Cohen GM. Proteasome-mediated degradation of Smac during apoptosis: XIAP promotes Smac ubiquitination in vitro. J. Biol. Chem. 2002;277:36611–36616. doi: 10.1074/jbc.M200317200. [DOI] [PubMed] [Google Scholar]
- Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Apoptosis in neurodegenerative disorders. Nat. Rev. Mol. Cell Biol. 2000;1:120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- Melino G, Bernassola F, Knight RA, Corasaniti MT, Nistico G, Finazzi-Agro A. S-nitrosylation regulates apoptosis. Nature. 1997;388:432–433. doi: 10.1038/41237. [DOI] [PubMed] [Google Scholar]
- Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marletta MA. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat. Chem. Biol. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- Morizane Y, Honda R, Fukami K, Yasuda H. X-linked inhibitor of apoptosis functions as ubiquitin ligase toward mature caspase-9 and cytosolic Smac/DIABLO. J. Biochem. 2005;137:125–132. doi: 10.1093/jb/mvi029. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts PR, Singh S, Knezek M, Thompson CB, Deshmukh M. Critical function of endogenous XIAP in regulating caspase activation during sympathetic neuronal apoptosis. J. Cell Biol. 2003;163:789–799. doi: 10.1083/jcb.200307130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen GS, Duckett CS. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 2002;3:401–410. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- Schile AJ, Garcia-Fernandez M, Steller H. Regulation of apoptosis by XIAP ubiquitin-ligase activity. Genes Dev. 2008;22:2256–2266. doi: 10.1101/gad.1663108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Okada K, Wilkinson JC, Solomon KM, Duckett CS, Reed JC, Salvesen GS. Identification of ubiquitination sites on the X-linked inhibitor of apoptosis protein. Biochem. J. 2003;373:965–971. doi: 10.1042/BJ20030583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Nakabayashi Y, Takahashi R. Ubiquitin-protein ligase activity of X-linked inhibitor of apoptosis protein promotes proteasomal degradation of caspase-3 and enhances its anti-apoptotic effect in Fas-induced cell death. Proc. Natl. Acad. Sci. USA. 2001;98:8662–8667. doi: 10.1073/pnas.161506698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenneti L, D'Emilia DM, Lipton SA. Suppression of neuronal apoptosis by S-nitrosylation of caspases. Neurosci. Lett. 1997;236:139–142. doi: 10.1016/s0304-3940(97)00780-5. [DOI] [PubMed] [Google Scholar]
- Tenneti L, Lipton SA. Involvement of activated caspase-3-like proteases in N-methyl-D-aspartate-induced apoptosis in cerebrocortical neurons. J. Neurochem. 2000;74:134–142. doi: 10.1046/j.1471-4159.2000.0740134.x. [DOI] [PubMed] [Google Scholar]
- Tsang AH, Lee YI, Ko HS, Savitt JM, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM, Chung KK. S-nitrosylation of XIAP compromises neuronal survival in Parkinson's disease. Proc. Natl. Acad. Sci. USA. 2009;106:4900–4905. doi: 10.1073/pnas.0810595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Kikuchi Y, Nomura Y. Caspase activation accompanying cytochrome c release from mitochondria is possibly involved in nitric oxide-induced neuronal apoptosis in SH-SY5Y cells. J. Neurochem. 1999;72:196–205. doi: 10.1046/j.1471-4159.1999.0720196.x. [DOI] [PubMed] [Google Scholar]
- Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Wink DA, Kim S, Coffin D, Cook JC, Vodovotz Y, Chistodoulou D, Jourd’heuil D, Grisham MB. Detection of S-nitrosothiols by fluorometric and colorimetric methods. Methods Enzymol. 1999;301:201–211. doi: 10.1016/s0076-6879(99)01083-6. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, et al. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.