Abstract
Background and Aims
It is essential to illuminate the evolutionary history of crop domestication in order to understand further the origin and development of modern cultivation and agronomy; however, despite being one of the most important crops, the domestication origin and bottleneck of soybean (Glycine max) are poorly understood. In the present study, microsatellites and nucleotide sequences were employed to elucidate the domestication genetics of soybean.
Methods
The genomes of 79 landrace soybeans (endemic cultivated soybeans) and 231 wild soybeans (G. soja) that represented the species-wide distribution of wild soybean in East Asia were scanned with 56 microsatellites to identify the genetic structure and domestication origin of soybean. To understand better the domestication bottleneck, four nucleotide sequences were selected to simulate the domestication bottleneck.
Key Results
Model-based analysis revealed that most of the landrace genotypes were assigned to the inferred wild soybean cluster of south China, South Korea and Japan. Phylogeny for wild and landrace soybeans showed that all landrace soybeans formed a single cluster supporting a monophyletic origin of all the cultivars. The populations of the nearest branches which were basal to the cultivar lineage were wild soybeans from south China. The coalescent simulation detected a bottleneck severity of K′ = 2 during soybean domestication, which could be explained by a foundation population of 6000 individuals if domestication duration lasted 3000 years.
Conclusions
As a result of integrating geographic distribution with microsatellite genotype assignment and phylogeny between landrace and wild soybeans, a single origin of soybean in south China is proposed. The coalescent simulation revealed a moderate genetic bottleneck with an effective wild soybean population used for domestication estimated to be ≈2 % of the total number of ancestral wild soybeans. Wild soybeans in Asia, especially in south China contain tremendous genetic resources for cultivar improvement.
Keywords: Wild soybean, Glycine soja, cultivated soybean, G. max, microsatellite, nucleotide sequence, domestication origin, domestication bottleneck
INTRODUCTION
The origin of agriculture known as the Neolithic revolution in human history has resulted in the transition from a hunter-gather mode to an agricultural-based society (Diamond, 2002; Salamini et al., 2002). Domestication of animal and plant has played an essential role in the rising of agriculture (Diamond, 2002; Salamini et al., 2002). Making the genetics of domestication clearer could lead to a more comprehensive understanding of the evolution and history of crops and provide valuable information not only on the increasing demand of improvement of yield and quality, but also on the origin of agriculture (Jones and Brown, 2000; Diamond, 2002).
Domestication was an evolutionary process in which several characters such as loss of seed dispersal, increase in grain size and synchronous ripening and so on, were adaptively evolved and selected by human beings (Brown et al., 2009; Glémin and Bataillon, 2009). Modern crops, especially landraces that descended from the wild-relative populations should maintain many of the features from wild populations. Therefore, genotypic comparison between cultivars and wild relatives from the natural distribution areas would indicate the direct ancestors of the crops (Brown et al., 2009). Recently, molecular genetics has been used to trace the evolutionary origin and domestication history of crops, such as common bean, sunflower, einkorn wheat, rice, maize and potato (Heun et al., 1997; Matsuoka et al., 2002; Harter et al., 2004; Chacón S et al., 2005; Spooner et al., 2005; Doebley et al., 2006; Londo et al., 2006; Smith, 2006).
Domestication was accompanied by a reduction in genetic diversity, as well as loss of useful traits reserved in wild relatives. During domestication, lines that contained agronomically important characters were selected, which resulted in a genome-wide reduction of genetic diversity or selective sweep in domesticated crops (Tanksley and McCouch, 1997; Buckler et al., 2001; Diamond, 2002). For example, it was suggested that several grasses had about two-thirds of the genetic diversity of their wild relatives (Buckler et al., 2001). Domestication bottleneck greatly and rapidly reduced the number of rare alleles, and then the genetic diversity of crops, which resulted in a narrowing of genetic basis for crop improvement. The domestication bottleneck was usually elucidated by the bottleneck severity coefficient (K′) with the hypothesis of single domestication (Wright et al., 2005). For instance, domestication bottleneck simulation revealed a more severe bottleneck for rice than maize (Tenaillon et al., 2004; Zhu et al., 2007).
As one of the economically important crops in the world, soybean (Glycine max) can provide most of the vegetable oil and protein for humans and animals (Singh and Hymowitz, 1999; Boerma and Specht, 2004). Evidence from morphological, cytogenetic and molecular analyses has indicated that soybean was domesticated from wild soybean (G. soja) in China (Broich and Palmer, 1980; Kollipara et al., 1997; Doebley et al., 2006). The geographic distribution of wild soybeans limited to East Asia covers wide areas of China as well as adjacent regions, including Russian Far East, the Korean Peninsula and Japan (Singh and Hymowitz, 1999; Boerma and Specht, 2004). During expansion and adaptation, beneficial traits have accumulated, such as pest and disease resistance, increased yield, improved quality, male sterility and fertility restoration (Hajjar and Hodgkin, 2007). Great genetic variation in these areas has been serving as an important gene pool for cultivar improvement (Singh and Hymowitz, 1999). Understanding the domestication genetics of soybean will greatly facilitate the discovery and utilization of rare but potentially important alleles in these resources. However, where and how soybean originated from the wild progenitors is still under intense debate.
The genetic diversity and structure of wild and cultivated soybeans have been reported in many studies (Xu et al., 2002; Lee et al., 2008; Li et al., 2008; Li et al., 2009). A severe genetic bottleneck during soybean domestication was also found in several independent analyses (Xu et al., 2002; Hyten et al., 2006; Kuroda et al., 2006). There is supporting evidence for both single and multiple domestication events (Hymowitz and Kaizuma, 1981; Fukuda, 1993; Zhuang et al., 1994; Gai et al., 2000; Xu et al., 2002; Xu and Gai, 2003; Dong et al., 2004; Zhao and Gai, 2004). Furthermore, the inconsistency in former studies was mainly due to the limitation of sampling or small number of molecular markers. Here, landrace (endemic cultivated soybean) and wild soybeans that cover the entire distribution range in East Asia were collected to investigate the domestication genetics of soybean based on 56 microsatellites and four nucleotide sequences. By integrating genotype assignment, phylogenetic analysis and bottleneck simulation it is possible to (a) elucidate the genetic diversity and structure of wild and landrace soybeans, (b) identify the domestication origin of soybean and (c) investigate the bottleneck severity during domestication.
MATERIALS AND METHODS
Plant materials
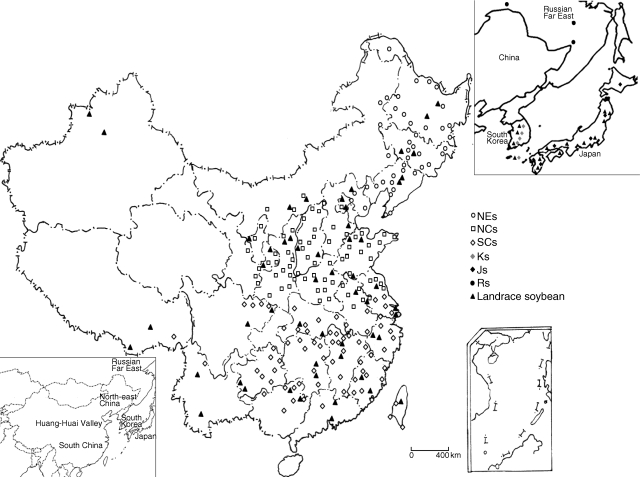
The genus Glycine includes two sub-genera: glycine and soja. Subgenus soja contains the wild soybean (Glycine soja) and soybean (G. max). The sample consisted of 310 individuals that included 231 wild soybeans and 79 landrace soybeans (endemic cultivated soybeans; see Table S1 in Supplementary data, available online) that were selected based on the even distribution pattern across the natural distribution area of wild and cultivated soybean (Fig. 1). The 231 wild soybeans covered the natural distribution areas in East Asia including China (216 accessions), Russian Far East (five accessions), South Korea (five accessions) and Japan (five accessions). A total of 79 landrace soybeans covering the whole distribution area of China (60 accessions), South Korea (five accessions) and Japan (14 accessions) were collected. In these 310 individuals, 62 landraces and 62 wild soybeans with one or two individuals from each province or autonomous region in China and three to five individuals from South Korea and Japan were selected for coalescent simulation. Glycine tomentella (eight accessions) collected from Taiwan, was employed to root the phylogenetic tree. Pure-line seeds of all Chinese accessions were obtained from the soybean germplasm bank of the Chinese Academy of Agricultural Sciences, and seeds of all accessions from other countries were provided by the US Department of Agriculture Soybean Germplasm Collection.
Fig. 1.
The geographic distribution of wild soybeans from north-east China (NEs), the Huang-Huai Valley (NCs), south China (SCs), South Korea (Ks), Japan (Js) and Russian Far East (Rs), and landrace soybeans.
DNA extraction and genotyping
The seeds of each accession were collected for germination, and then used for DNA extraction following the CTAB method (Doyle and Doyle, 1987). Based upon soybean genetic linkage maps, 134 microsatellites covering 20 linkage groups with 20 cM between each pair of loci were chosen (Cregan et al., 1999; Song et al., 2004). After a pre-amplification test, 56 primer pairs with good amplification results were selected for genotype analysis (Table S2 in Supplementary data). The PCR protocol followed was described in Cregan et al. (1999) with a final volume of 10 µL containing 10 mm Tris–HCl (pH 8·4), 1·5 mm MgCl2, 50 mm (NH4)2SO4, 1 unit Taq polymerase (Fermentas, Vilnius, Lithuania), 0·2 mm dNTPs, 0·2 µm each primer, and 50 ng of genomic DNA. PCR products were separated on the 6 % PAGE, visualized with silver staining, and scored according to a 25-bp DNA marker ladder (Promega, Madison, WI, USA).
Several nuclear genes were selected from previous studies for sequencing. Among them four nuclear genes which showed homozygosity in all accessions were used for bottleneck simulation: BG406170 was selected from soybean cDNA sequences homologous to Solanaceae conserved orthologous sequences (COSII) (E value < e−10) (Wu et al., 2006; Cheng and Strömvik, 2008); AF105221, J02746 and AJ003246 with high polymorphism were selected from previous reports (Zhu et al., 2003; Van et al., 2005; Hyten et al., 2006; Table S3 in Supplementary data). PCR was performed based on the original description with a final volume of 50 µL containing 10 mm Tris–HCl (pH 8·4), 50 mm (NH4)2SO4, 2 mm MgCl2, 2·5 unit Taq polymerase (Fermentas), 2 µm dNTPs, 0·8 µm each primer, and 100 ng of genomic DNA. Sequences for each locus were successfully obtained after direct sequencing of the PCR products. PCR amplifications and sequencing were repeated twice for each product to confirm the sequence variation. Sequences were aligned and edited by software Clustal_X 1·81 (Thompson et al., 1997) and Bioedit version 7.0.1 (Hall, 1999). Alignment sequences were deposited in GenBank with accession numbers GU112110–GU112178.
Data analysis
GENALEX6 (Peakall and Smouse, 2006) and FSTAT2.9.3 (Goudet, 2001) were used to calculate the genetic diversity index. The STRUCTURE2·2 program was used to analyse the genetic structure of samples using the model-based method (Pritchard et al., 2000; Falush et al., 2003). The admixture model was employed to infer population structure of wild and landrace soybeans for a number of clusters with K of 1–10 (Pritchard et al., 2000; Falush et al., 2003). The ad hoc statistic ΔK which was calculated based on the rate of change of the log-likelihood for the present K value was employed to identify the optimal number of populations present in the data set following the recommendation of Evanno et al. (2005). The genetic structure was then plotted with the DISTRUCT1·1 program based on the optimal number of K (Rosenberg, 2004). The prior population model incorporated with the inferred population of wild soybeans was used to assign the landrace soybeans to wild populations to infer the genotype origin of landrace soybeans (Pritchard et al., 2000; Falush et al., 2003; Harter et al., 2004). The number of pre-defined populations of wild soybeans was set to three according to the admixture model analysis results. For each procedure, at least five independent runs were processed based on runs of 100 000 iterations, following a burn-in period of 50 000 iterations.
Genetic distances of microsatellite genotypes were calculated using 1 minus the proportion of shared alleles (1 – ‘Dps’) which was widely used in multilocus microsatellite data by MICROSAT with 1000 replications (Minch et al., 1996; Rosenberg et al., 2001; Matsuoka et al., 2002). Phylogenies among soybeans were reconstructed based on the neighbor-joining (NJ) method in the PHYLIP3·67 program (Felsenstein, 2004).
DNA polymorphism parameters (nucleotide diversity, θ; and number of segregating sites, S) were calculated using DnaSP (Rozas et al., 2003). Coalescent simulation as previously described for maize and rice was employed to model the domestication bottleneck (Eyre-Walker et al., 1998; Tenaillon et al., 2004; Wright et al., 2005; Zhu et al., 2007). Tenaillon et al. (2004), Wright et al. (2005) and Zhu et al. (2007) proved a positive correlation between the bottleneck population size (Nb) and the duration (d), thus the bottleneck stringency value K′ = Nb/d (Wright et al., 2005) was employed to describe the severity of the bottleneck during domestication. Sequences of four nuclear genes were employed for the simulation using MS software (Hudson, 2002). Values of d (100, 500, 1000, 1500, 2000 and 3000) were chosen referring to the analysis of rice and maize (Wright et al., 2005; Zhu et al., 2007), and K′ values of range 0·1–20 with a total of 300 scenarios for each locus were examined with 10 000 simulations (Table S4 in Supplementary data). The number of segregating sites (S) was employed to evaluate simulations and used to calculate the maximal likelihood of bottleneck severity by integrating multilocus analysis (Tenaillon et al., 2004; Wright et al., 2005).
RESULTS
Genetic diversity of wild and landrace soybeans
There were 1498 alleles detected across 56 microsatellites in 231 wild and 79 landrace soybeans with an average of 26·8 alleles per locus, ranging from 5 (BE021153) to 56 (Satt243). The overall genetic diversity of all loci ranged from 0·384 (BE021153) to 0·969 (Sat_093) (Table S2 in Supplementary data). The unbiased expected heterozygosity (UHE) and allelic richness (NR), regardless of sample size, were employed to calculate genetic diversity of wild and landrace soybean populations. Genetic diversity in wild soybean (UHE = 0·843 and NR = 20·3) was significantly higher than that in landrace soybean (UHE = 0·719 and NR = 11·8) (Table 1). The analysis based on four nucleotide sequences also revealed that wild soybeans (θ ranging from 0·003 in BG406170 and 0·023 in AJ003246) had a significantly higher polymorphism than that in landrace soybean (θ ranging from 0·001 in BG406170 and 0·011 in AJ003246). Compared with wild soybeans, landraces lost most of the nucleotide variation, which suggested bottleneck during domestication (Table 1).
Table 1.
Genetic diversity of 56 microsatellites and four nuclear genes in wild and landrace soybean
| Species | N* | Microsatellite |
Nuclear gene |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| UHE† | NR‡ |
BG406170 |
AF105221 |
J02746 |
AJ003246 |
||||||
| θ§ | S¶ | θ | S | θ | S | θ | S | ||||
| Wild soybean | 231 (62) | 0·843 | 20·3 | 0·003 | 6 | 0·004 | 7 | 0·004 | 8 | 0·023 | 49 |
| Landrace soybean | 79 (62) | 0·719 | 11·8 | 0·001 | 2 | 0·002 | 5 | 0·002 | 4 | 0·011 | 26 |
* The number of individuals used for microsatellite genotype with the number of nucleotide loci in parenthesis.
† Unbiased expected heterozygosity.
‡ Allelic richness.
§ Nucleotide diversity.
¶ Number of segregating sites.
Genetic structure of wild and landrace soybeans based on microsatellites
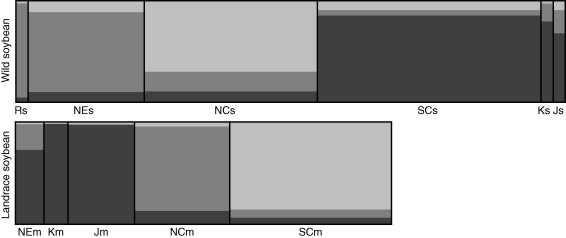
Admixture model from the STRUCTURE2·2 program was used to infer the population structure of wild soybean in which the cultivars could be assigned. A single solution was found for K = 2 based on the ΔK statistic (Evanno et al., 2005) with most individuals separated into two clusters, individuals from the Huang-Huai Valley (NCs) were clustered independently from other regions including south China (SCs), north-east China (NEs), Russian Far East (Rs), South Korea (Ks) and Japan (Js). At K = 3, individuals from SCs, NEs, Rs, Ks and Js further split into two clusters with Rs clustered together with NEs, while Ks and Js clustered with SCs (Fig. 2). However, the genetic structure of landrace soybeans differed from wild soybeans, in which the highest likelihood of genetic structure of landrace soybean was obtained when K = 3 based on ΔK statistics. It was found that individuals from South Japan (Jm) and Korea (Km) were clustered with individuals from north-east China (NEm), which was different from the pattern in wild soybean. Individuals from south China (SCm) and the Huang-Huai Valley (NCm) formed independent clusters (Fig. 2). The genotype cluster for wild and landrace soybeans from China was mostly consistent with previous studies, in which three eco-regions were proposed, based on morphological and physiological analysis (Bu and Pan, 1982; Gai and Wang, 2001).
Fig. 2.
Genetic structure of wild soybeans and landrace soybeans inferred from the admixture model in the STRUCTURE2·2 program based on 56 microsatellites with K = 3. Wild soybean: Rs, Russian Far East; NEs, north-east China; NCs, Huang-Huai Valley; SCs, south China; Ks, South Korea; Js, Japan. Landrace soybean: NEm, north-east China; Km, Korea; Jm, South Japan; NCm, Huang-Huai Valley; SCm, south China.
Domestication origin of soybean based on microsatellites
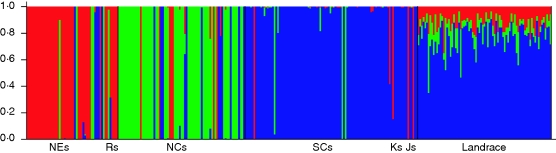
Using the population structure of wild soybeans, individuals were defined from SCs plus Ks and Js, NEs plus Rs, and NCs as three independent ancestral source clusters which could be used to model the assignment of landraces using the prior information model in the STRUCTURE2·2 program. Most of the landrace genotypes (77·3 %) were assigned to the cluster of SCs plus Ks and Js; 12·3 % of the genotypes were assigned to the cluster of NEs plus Rs, and 10·4 % to NCs (Fig. 3). This indicated that landrace soybeans were genetically similar to wild soybeans from SCs plus Ks and Js. However, >20 % of the genome of landrace soybeans were similar to those of NEs plus Rs and NCs.
Fig. 3.
Genotype assignment of landrace soybeans to the three wild soybean source clusters. For abbreviations see Fig. 2.
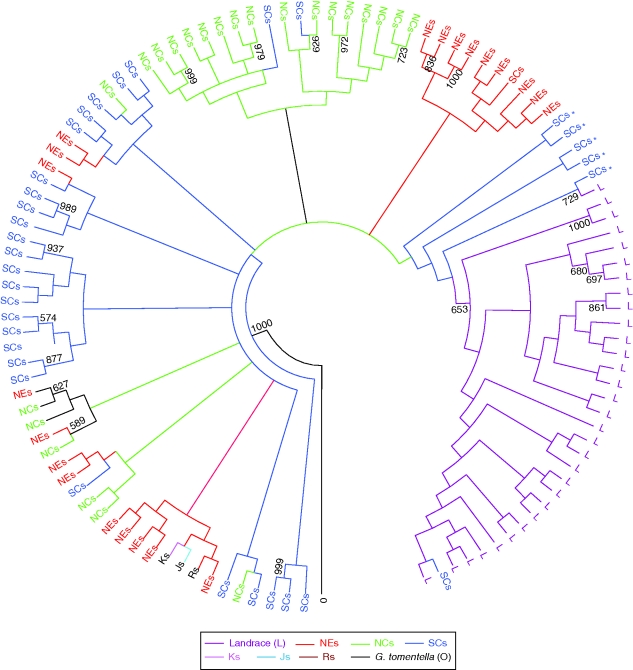
To clarify further the phylogenetic relationship between wild and landrace soybeans, the NJ method from PHYLIP3·67 program was employed to construct the phylogenetic tree. All 310 individuals were classified into 112 geographically defined groups for bootstrap resampling in which individuals from China within approx. 2 ° of latitude and longitude were classified into one group and individuals from Russian Far East, South Korea and Japan were classified into distinct groups (Matsuoka et al., 2002). The phylogenetic analyses from microsatellite genotypes showed all landrace soybeans formed a monophyletic lineage which was coincident with maize based on analysis from microsatellites (Matsuoka et al., 2002). This indicated that landrace soybeans had a single domestication origin such as maize (Fig. 4). The regions containing those wild populations that are phylogenetically close with cultivars could be proposed as the domestication region of crops (Matsuoka et al., 2002; Spooner et al., 2005). In this study, the populations of the nearest branches which were basal to the landrace soybean lineage were wild soybeans from south China, especially those from Zhejiang, Hubei and Taiwan provinces. Combining the present results with genotype assignment of landrace soybeans, it was proposed that soybeans might have been domesticated only once in south China, probably in the middle and lower regions of the Yangtze River basin and on the south-east coast where great genetic diversity for both wild and landrace soybeans is preserved.
Fig. 4.
Neighbor-joining phylogenetic relationships of wild and landrace soybeans based on 56 microsatellites. Pairwise genetic distance of wild and landrace soybeans populations were calculated using the distance 1 – ‘Dps’ based on 56 nuclear microsatellites. The asterisks indicate the wild soybean populations from south China that are basal to all the landrace soybeans.
Bottleneck coalescent simulation based on nucleotide sequences
It was reported that allelic diversity was reduced faster than heterozygosity for a population with recent reduction (Cornuet and Luikart, 1996). Comparison based on microsatellites revealed that the expected heterozygosity in landrace soybeans was 14·7 % lower than wild soybeans, but contained just 41·9 % of allelic richness in wild soybeans (Table 1). This indicated that landraces lost most of the rare alleles during domestication in which landraces contained 81·8 % of different alleles with a frequency of >5 %, but contained just 13·6 % of the private alleles in wild soybeans (data not shown). The nucleotide polymorphism of the four sequences also showed that the nucleotide diversity in landrace soybean was about half of that in wild soybean, which strongly suggested genetic bottleneck associated with domestication.
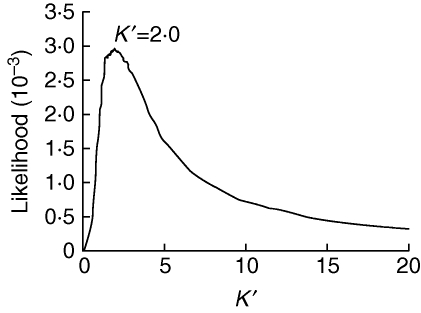
Given that a single domesticated origin was detected for landrace soybean, four nuclear genes were obtained to explore further the bottleneck severity during soybean domestication based on the two-subpopulation model previously described in maize and rice (Eyre-Walker et al., 1998; Tenaillon et al., 2004; Wright et al., 2005; Zhu et al., 2007; Gao and Innan, 2008). Tajima's D test revealed that all the four genes evolved neutrally in both wild and landrace soybeans except locus J02764 in landrace soybean. Multilocus integration analysis was employed to narrow the confidence regions (Tenaillon et al., 2004). The statistics produced a likelihood peak value for bottleneck severity of K′ = 2·0 (Fig. 5), indicating that if domestication lasted for 1000 years, the domesticated population was composed of 2000 effective ancestral individuals. The domestication process of soybean was proposed to start 5000 years ago and cultivars were introduced to other countries from the first century ad (about 2000 years ago) based on linguistic, geographical and historical evidence (Hymowitz and Newell, 1981; Hymowitz, 1990; Singh and Hymowitz, 1999). This suggested that the maximum duration of domestication was 3000 years. Thus, a maximum effective population of 6000 wild soybean individuals during soybean domestication was speculated.
Fig. 5.
Estimation of maximum likelihood of severity parameter K′ during soybean domestication based on the four nuclear genes.
DISCUSSION
Genetic structure of landrace soybean in East Asia
The distribution of wild and landrace soybeans in East Asia serves as a very important gene pool for cultivar improvement. Although wild soybeans from limited resources had been analysed, the genetic structure of wild soybeans covering the whole distribution area in East Asia was rarely discussed (Gai and Wang, 2001; Kuroda et al., 2006; Lee et al., 2008; Li et al., 2008). In this study, species-wide individuals were sampled to carry out an analysis, based on genome-wide distributed microsatellites, of the genetic structure of wild and landrace soybeans in East Asia. Three genetic structure clusters of landrace soybean were detected in East Asia in which NEm plus Km and Jm formed a cluster, and individuals from NCm and SCm formed independent clusters, implying that landrace soybeans from South Korea and Japan were genotypically similar to those from north-east China (Fig. 2). This was different from the genetic structure of wild soybean in which SCs plus Ks and Js formed a cluster, NEs plus Rs formed a cluster, and NCs formed a cluster (Fig. 2). Wild soybeans are naturally distributed in East Asia and might have existed there before the Ice Age. Thus, the distribution of wild soybean might have been influenced by the climate changes and construction of the land-bridge during the Ice Age (J. Guo et al., WBGCAS, Wuhan, China, unpubl. res.). Compared with wild soybean, the expansion of landrace soybean is more likely to be a recent event. Unlike the evolution of wild soybean, domestication of cultivated crops is a more recent process which began approx. 12 000 years ago in the Fertile Crescent or more recently in other ancestral agriculture centres (Brown et al., 2009; Glémin and Bataillon, 2009). During this time, human activity with exchange of seeds resulted in gene flow of cultivated soybean within north-east China, the Korean Peninsula and Japan.
Origin of soybean domestication
Both phylogenetic and assignment analysis indicated that landrace soybeans might have originated from their wild progenitors in south China (Figs 3 and 4). The genetic diversity of wild and landrace soybean populations also showed that south China had the highest level of genetic diversity in previous and present studies (Table S5 in Supplementary data; Shimamoto et al., 1998, 2000; Xu et al., 1999; Ding et al., 2008; Li et al., 2008). Vavilov (1992) considered that the genetic diversity centre was the origin centre for major crops. Moreover, gene introgression is very difficult for self-pollinated crops, especially soybean with an outcross rate of 1·8 % (Ray et al., 2003). In addition, a recent archaeological discovery revealed that rice domestication occurred in Zhejiang about 6600–6900 years ago in the lower Yangtze River basin (Fuller et al., 2009). Prolific agricultural activities and abundant wild resources in south China further proved domestication and cultivation of major crops, including soybean, might have occurred in this area.
The domestication of soybean was similar to maize (Matsuoka et al., 2002), potato (Spooner et al., 2005) and sunflower (Harter et al., 2004) that have single domestication events, but different from rice (Londo et al., 2006) and common bean (Chacón S et al., 2005) that are proposed to have multiple domestication origins. The origin of soybean domestication has been the subject of intense debate with both single and multiple domestication hypotheses (Hymowitz and Kaizuma, 1981; Fukuda, 1993; Zhuang et al., 1994; Gai et al., 2000; Xu et al., 2002; Xu and Gai, 2003; Zhao and Gai, 2004). As semi-domesticated soybean grows prolifically in north-east China, Fukuda (1993) suggested that soybean originated in this region, but most of the evidence indicated that the origin of soybean domestication was in the Huang-Huai Valley or south China (Hymowitz and Newell, 1981; Zhuang et al., 1994; Gai et al., 2000; Dong et al., 2004; Zhao and Gai, 2004). The Huang-Huai Valley and the Yangtze River basin have both been suggested as important areas where agriculture originated in China. Dong et al. (2004) proposed that cultivated soybean might be domesticated from wild soybean downstream of the Huang-Huai Valley, given that the greatest morphological variation is preserved in this region. Considering that morphological characters are sensitive to climate and environmental change, archaeological and morphological evidence for the Huang-Huai Valley origin might be insufficient for speculation on the origin of soybean domestication in this region (Hymowitz and Newell, 1981; Dong et al., 2004). In addition to a single domestication origin, molecular evidence indicated that soybean might have been domesticated independently in different regions (Xu et al., 2002; Xu and Gai, 2003). The difference between multiple origins and a single origin in these previous studies might be due to a limited number of either molecular markers or samples. Here, genotype comparison of 56 genome-wide distributed microsatellites revealed that landrace soybeans were genotypically similar to wild soybeans from south China, especially from the middle and downstream areas of the Yangtze River basin. As a result, it was speculated that soybeans were domesticated in south China and then disseminated to the northern areas. During south–north dissemination, flowering time loci and other genes were selected as adaptations to local environments, thus forming different eco-topic genotypes. Though great genetic diversity was found in south China, prolific morphological diversity preserved in the Huang-Huai Valley (Dong et al., 2004) and richness resources in other regions in East Asia (Kuroda et al., 2006; Lee et al., 2008; Li et al., 2008) could provide great adaptive characters for cultivar improvement.
Bottleneck severity of soybean during domestication
Intensive selection during domestication was proposed to result in extensive narrowing of genetic base of the cultivated crops (Tanksley and McCouch, 1997; Buckler et al., 2001; Diamond, 2002). Hyten et al. (2006) reported that landraces retained 48·9 % of the nucleotide diversity of 102 wild soybean-based genes. In this study, comparison of genetic diversity between wild and landrace soybeans indicated a great bottleneck during domestication in which landrace soybean retained approximately half of the allelic richness and nucleotide diversity based on both microsatellites and nucleotide sequences (Table 1) (Cornuet and Luikart, 1996).
Genetic diversity analysis and coalescent simulation both revealed a moderate reduction in genetic diversity during soybean domestication compared with maize and rice, in which the bottleneck severity of soybean domestication (2·0) was less severe than that of rice (0·2 and 0·5 for japonica and indica, respectively), but more than that of maize (4·0–5·0) (Buckler et al., 2001; Tenaillon et al., 2004; Hyten et al., 2006; Zhu et al., 2007). Compared with outbreeding maize, the predominant self-pollination of soybean might have strengthened the bottleneck during domestication (Ray et al., 2003). However, preservation of genetic diversity in soybean could be comparable to sunflower in which 40–50 % of wild sunflower nucleotide diversity appeared to be preserved in the cultivar due to a limited sample of individuals used in the analysis (Liu and Burke, 2006). Thus, the sampling strategy and the mating system of a species might explain most of the differences in the rate of genetic diversity reduction between the wild relative and the cultivar. Extensive sampling and various molecular markers used in this study elucidated a moderate domestication bottleneck of soybean.
It was suggested that establishment of the cultivated traits was probably a slow process which took over 1000–2000 years or more (Tanno and Willcox, 2006; Fuller, 2007). Evidence based on linguistic, geographical and historical data supported the suggestion that the duration of soybean domestication is no more than 3000 years (Hymowitz and Newell, 1981; Hymowitz, 1990; Singh and Hymowitz, 1999). Based on the coalescent simulation with bottleneck severity of 2·0, the sequence diversity found in the landrace soybean could be explained by a founding population of no more than 6000 wild soybean individuals, which was much larger than the self-pollinated rice either based on nucleotide sequences (600 individuals for japonica rice and 1500 for indica rice) (Zhu et al., 2007) or microsatellites (2700 individuals for japonica rice and 4500 for indica rice) (Gao and Innan, 2008). Because nuclear variation in wild soybean was approximately one-third of that in the wild progenitor of maize, teosinte, an ancestral wild soybean population size of ≈300 000 was speculated given a population size of ≈900 000 for teosinte (Eyre-Walker et al., 1998; Gao and Innan, 2008). Therefore, the effective wild soybean population used for domestication accounted for ≈2 % of the ancestors. Although it experienced a moderate level of genetic diversity reduction compared with other crops, microsatellite diversity analysis and bottleneck simulation based on nucleotide sequences implied that soybean had lost most of the rare alleles in wild soybean during domestication.
Conclusions
In this study, microsatellites and a nuclear gene were employed to analyse the genetic structure and the domestication genetics of landrace soybean. Phylogenetic and genotype assignment analysis showed that landrace soybeans formed a single cluster and genotype similar to wild soybeans from south China, suggesting a single origin of landrace soybeans in south China. A moderate genetic bottleneck was proposed during domestication with the maximum likelihood of the ratio of domesticated individuals and duration of bottleneck of 2·0. The origin of soybeans in south China, combined with the bottleneck simulation, implies that the wild soybean resources in south China could be a valuable gene pool for soybean breeding. Considering that many wild soybean populations have diminished or disappeared due to human activities and habitat deterioration, a great conservation effort for wild soybean populations, especially those in south China, is urgently needed to ensure the sustainable utilization of soybean resources.
SUPPLEMENTARY DATA
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by The National Natural Science Foundation of China (Project 30670158), the CAS/CAFEA International Partnership Program for Creative Research Teams (Project 0921101001) and 100 Talents Program of Chinese Academy of Sciences. Special thanks to Prof. Xinan Zhou, the Chinese Academy of Agricultural Sciences and United States Department of Agriculture for assisting in sample collection. We are also indebted to Profs Steve D. Tanksley, Scott Jackson and Ting Wang, Drs Alexandra Casa, Sharon Mitchell, Qihui Zhu, Yifei Liu, Xiaohong Yao, Chen Lu and Guopei Chen for useful suggestions on the data analysis and manuscript preparation.
LITERATURE CITED
- Boerma HR, Specht JE. Soybeans: improvement, production and uses. Madison, WI: American Society of Agronomy; 2004. [Google Scholar]
- Broich SL, Palmer RG. A cluster analysis of wild and domesticated soybean phenotypes. Euphytica. 1980;29:23–32. [Google Scholar]
- Brown TA, Jones MK, Powell W, Allaby RG. The complex origins of domesticated crops in the Fertile Crescent. Trends in Ecology & Evolution. 2009;24:103–109. doi: 10.1016/j.tree.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Bu MH, Pan TF. Discussion of Chinese soybean cultivation region. Soybean Science. 1982;1:105–121. [Google Scholar]
- Buckler ES, Thornsberry JM, Kresovich S. Molecular diversity, structure and domestication of grasses. Genetics Research. 2001;77:213–218. doi: 10.1017/s0016672301005158. [DOI] [PubMed] [Google Scholar]
- Chacón SMI, Pickersgill B, Debouck DG. Domestication patterns in common bean (Phaseolus vulgaris L.) and the origin of the Mesoamerican and Andean cultivated races. Theoretical and Applied Genetics. 2005;110:432–444. doi: 10.1007/s00122-004-1842-2. [DOI] [PubMed] [Google Scholar]
- Cheng KCC, Strömvik MV. SoyXpress: a database for exploring the soybean transcriptome. BMC Genomics. 2008;9:368. doi: 10.1186/1471-2164-9-368. doi:10.1186/1471-2164-9-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, et al. An integrated genetic linkage map of the soybean genome. Crop Science. 1999;39:1464–1490. [Google Scholar]
- Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- Ding YL, Zhao TJ, Gai JY. Genetic diversity and ecological differentiation of Chinese annual wild soybean (Glycine soja) Biodiversity Science. 2008;16:133–142. [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Dong YS, Zhao LM, Liu B, Wang ZW, Jin ZQ, Sun H. The genetic diversity of cultivated soybean grown in China. Theoretical and Applied Genetics. 2004;108:931–936. doi: 10.1007/s00122-003-1503-x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS. Investigation of the bottleneck leading to the domestication of maize. Proceedings of the National Academy of Sciences of the USA. 1998;95:4441–4446. doi: 10.1073/pnas.95.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3·6. 2004 University of Washington. http://evolution.genetics.washington.edu/phylip.html . [Google Scholar]
- Fukuda Y. Cytogenetical studies on the wild and cultivated Manchurian soybeans (Glycine L.) Japanese Journal of Botany. 1993;6:489–506. [Google Scholar]
- Fuller DQ. Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the Old World. Annals of Botany. 2007;100:903–924. doi: 10.1093/aob/mcm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DQ, Qin L, Zheng YF, et al. The domestication process and domestication rate in rice: spikelet bases from the lower Yangtze. Science. 2009;323:1607–1610. doi: 10.1126/science.1166605. [DOI] [PubMed] [Google Scholar]
- Gai JY, Wang YS. A study on the varietal eco-regions of soybeans in China. Scienta Agricultura Sinica. 2001;34:139–145. [Google Scholar]
- Gai JY, Xu DH, Gao Z, et al. Studies on the evolutionary relationship among eco-types of G. max and G. soja in China. Acta Agronomica Sinica. 2000;26:513–520. [Google Scholar]
- Gao LZ, Innan H. Nonindependent domestication of the two rice subspecies, Oryza sativa ssp. indica and ssp. japonica, demonstrated by multilocus microsatellites. Genetics. 2008;179:965–976. doi: 10.1534/genetics.106.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S, Bataillon T. A comparative view of the evolution of grasses under domestication. New Phytologist. 2009;183:273–290. doi: 10.1111/j.1469-8137.2009.02884.x. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3) Switzerland: Lausanne University; 2001. http://www2.unil.ch/popgen/softwares/fstat.htm . [Google Scholar]
- Hajjar R, Hodgkin T. The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica. 2007;156:1–13. [Google Scholar]
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Harter AV, Gardner KA, Falush D, Lentz DL, Bye RA, Rieseberg LH. Origin of extant domesticated sunflowers in eastern North America. Nature. 2004;430:201–205. doi: 10.1038/nature02710. [DOI] [PubMed] [Google Scholar]
- Heun M, Schafer-Pregl R, Klawan D, et al. Site of einkorn wheat domestication identified by DNA fingerprinting. Science. 1997;278:1312–1314. [Google Scholar]
- Hudson RR. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hymowitz T. Soybeans: the success story, advances in new crops. Portland, OR: Timber Press; 1990. [Google Scholar]
- Hymowitz T, Kaizuma N. Soybean seed protein electrophoresis profiles from 15 Asian countries or regions: hypotheses on paths of dissemination of soybeans from China. Economic Botany. 1981;35:10–23. [Google Scholar]
- Hymowitz T, Newell CA. Taxonomy of the genus Glycine, domestication and uses of soybeans. Economic Botany. 1981;35:272–288. [Google Scholar]
- Hyten DL, Song Q, Zhu Y, et al. Impacts of genetic bottlenecks on soybean genome diversity. Proceedings of the National Academy of Sciences of the USA. 2006;103:16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Brown T. Agricultural origins: the evidence of modern and ancient DNA. The Holocene. 2000;10:769–776. [Google Scholar]
- Kollipara KP, Singh RJ, Hymowitz T. Phylogenetic and genomic relationships in the genus Glycine Willd. based on sequences from the ITS region of nuclear rDNA. Genome. 1997;40:57–68. doi: 10.1139/g97-008. [DOI] [PubMed] [Google Scholar]
- Kuroda Y, Kaga A, Tomooka N, Vaughan DA. Population genetic structure of Japanese wild soybean (Glycine soja) based on microsatellite variation. Molecular Ecology. 2006;15:959–974. doi: 10.1111/j.1365-294X.2006.02854.x. [DOI] [PubMed] [Google Scholar]
- Lee JD, Yu JK, Hwang YH, et al. Genetic diversity of wild soybean (Glycine soja Sieb. and Zucc.) accessions from South Korea and other countries. Crop Science. 2008;48:606–616. [Google Scholar]
- Li XH, Wang KJ, Jia JZ. Genetic diversity and differentiation of Chinese wild soybean germplasm (G. soja Sieb. & Zucc.) in geographical scale revealed by SSR markers. Plant Breeding. 2009;128:658–664. [Google Scholar]
- Li YH, Guan RX, Liu ZX, et al. Genetic structure and diversity of cultivated soybean (Glycine max (L.) Merr.) landraces in China. Theoretical and Applied Genetics. 2008;117:857–871. doi: 10.1007/s00122-008-0825-0. [DOI] [PubMed] [Google Scholar]
- Liu A, Burke JM. Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics. 2006;173:321–330. doi: 10.1534/genetics.105.051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proceedings of the National Academy of Sciences of the USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez G. A single domestication for maize shown by multilocus microsatellite genotyping. Proceedings of the National Academy of Sciences of the USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minch E, Ruiz-Linares A, Goldstein D, Feldman M, Cavalli-Sforza LL. Microsat (version 1·5): a computer program for calculating various statistics on microsatellite allele data. Stanford, CA: Stanford University Medical Center; 1996. [Google Scholar]
- Peakall R, Smouse P. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray JD, Kilen TC, Abel CA, Paris RL. Soybean natural cross-pollination rates under field conditions. Environmental Biosafety Research. 2003;2:133–138. doi: 10.1051/ebr:2003005. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Rosenberg NA, Woolf E, Pritchard JK, et al. Distinctive genetic signatures in the Libyan Jews. Proceedings of the National Academy of Sciences of the USA. 2001;98:858–863. doi: 10.1073/pnas.98.3.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Salamini F, Ozkan H, Brandolini A, Schafer-Pregl R, Martin W. Genetics and geography of wild cereal domestication in the Near East. Nature Reviews Genetics. 2002;3:429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- Shimamoto Y, Fukushi H, Abe J, et al. RFLPs of chloroplast and mitochondrial DNA in wild soybean, Glycine soja, growing in China. Genetic Resources and Crop Evolution. 1998;45:433–439. [Google Scholar]
- Shimamoto Y, Abe J, Gao Z, Gai JY, Thseng FS. Characterizing the cytoplasmic diversity and phyletic relationship of Chinese landraces of soybean, Glycine max, based on RFLPs of chloroplast and mitochondrial DNA. Genetic Resources and Crop Evolution. 2000;47:611–617. [Google Scholar]
- Singh RJ, Hymowitz T. Soybean genetic resources and crop improvement. Genome. 1999;42:605–616. [Google Scholar]
- Smith BD. Eastern North America as an independent center of plant domestication. Proceedings of the National Academy of Sciences of the USA. 2006;103:12223–12228. doi: 10.1073/pnas.0604335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QJ, Marek LF, Shoemaker RC, et al. A new integrated genetic linkage map of the soybean. Theoretical and Applied Genetics. 2004;109:122–128. doi: 10.1007/s00122-004-1602-3. [DOI] [PubMed] [Google Scholar]
- Spooner DM, McLean K, Ramsay G, Waugh R, Bryan GJ. A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proceedings of the National Academy of Sciences of the USA. 2005;102:14694–14699. doi: 10.1073/pnas.0507400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley SD, McCouch SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;277:1063–1666. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- Tanno K, Willcox G. How fast was wild wheat domesticated? Science. 2006;311:1886. doi: 10.1126/science.1124635. [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, U'Ren J, Tenaillon O, Gaut BS. Selection versus demography: a multilocus investigation of the domestication process in maize. Molecular Biology and Evolution. 2004;21:1214–1225. doi: 10.1093/molbev/msh102. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van K, Hwang EY, Kim MY, Park HJ, Lee SH, Cregan PB. Discovery of SNPs in soybean genotypes frequently used as the parents of mapping populations in the United States and Korea. Journal of Heredity. 2005;96:529–535. doi: 10.1093/jhered/esi069. [DOI] [PubMed] [Google Scholar]
- Vavilov NI. Origin and geography of cultivated plants. Cambridge: Cambridge University Press; 1992. [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, et al. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Wu FN, Mueller LA, Crouzillat D, Pétiard V, Tanksley SD. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics. 2006;174:1407–1420. doi: 10.1534/genetics.106.062455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DH, Gai JY. Genetic diversity of wild and cultivated soybeans growing in China revealed by RAPD analysis. Plant Breeding. 2003;122:503–506. [Google Scholar]
- Xu DH, Abe J, Gai JY, Shimamoto Y. Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: evidence for multiple origins of cultivated soybean. Theoretical and Applied Genetics. 2002;105:645–653. doi: 10.1007/s00122-002-0972-7. [DOI] [PubMed] [Google Scholar]
- Xu DH, Gao Z, Tian QZ, et al. Genetic diversity of the annual wild soybean (Glycine soja) in China. Chinese Journal of Applied & Environmental Biology. 1999;5:439–443. [Google Scholar]
- Zhao TJ, Gai JY. The origin and evolution of cultivated soybeans [Glycine max (L.) Merr.] Scientia Agricultura Sinica. 2004;37:954–962. [Google Scholar]
- Zhu QH, Zheng XM, Luo JC, Gaut BS, Ge S. Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Molecular Biology and Evolution. 2007;24:875–888. doi: 10.1093/molbev/msm005. [DOI] [PubMed] [Google Scholar]
- Zhu YL, Song QJ, Hyten DL, et al. Single-nucleotide polymorphisms in soybean. Genetics. 2003;163:1123–1134. doi: 10.1093/genetics/163.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang BC, Hui DW, Wang YM, Gu J, Xu B, Chen SY. RAPD analysis of cultivated and wild soybean from different areas in China. Chinese Science Bulletin. 1994;39:2178–2180. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.