Abstract
An integrated HIV-1 genomic DNA leads to an infected cell becoming either an active or a latent virus-producing cell. Upon appropriate activation, a latently infected cell can result in production of progeny viruses that spread the infection to uninfected cells. The host proteins influence several steps of HIV-1 infection including formation of the preintegration complex (PIC), a key nucleoprotein intermediate essential for integration of reverse transcribed viral DNA into the chromosome. Much effort has gone into the identification of host proteins contributing to the assembly of functional PICs. Experimental approaches included the use of yeast two-hybrid system, co-immunoprecipitation, affinity tagged HIV-1 viral proteins and in vitro reconstitution of salt-stripped PIC activity. Several host proteins identified using these approaches have been shown to affect HIV-1 replication in cells and influence catalytic activities of recombinant IN in vitro. However, the comprehensive identification and characterization of host proteins associated with HIV-1 PICs of infected cells have been hindered in part by the technical limitation in acquiring sufficient amount of catalytically active PICs. To efficiently identify additional host factors associated with PICs in infected cells, we have developed the following novel approach. The catalytically active PICs from HIV-1-infected CD4+ cells were isolated using biotinylated target DNA, and the proteins selectively co-purifying with PICs have been analyzed by mass spectrometry. This technology enabled us to reveal at least 19 host proteins that are associated with HIV-1 PICs, of which 18 proteins have not been described previously with respect to HIV-1 integration. Physiological functions of the identified proteins range from chromatin organization to protein transport. A detailed characterization of these host proteins could provide new insights into the mechanism of HIV-1 integration and uncover new antiviral targets to block HIV-1 integration.
Findings
Human immunodeficiency virus type 1 (HIV-1) integrase (IN) is a 288 amino-acid protein with three functional domains: N-terminal domain (NTD), catalytic core domain (CCD) and C-terminal domain (CTD). The NTD contains a zinc binding motif, the CCD has three acidic residues, D64, D116 and E152, which co-ordinate the catalytic divalent metal ions; and the CTD is suggested to nonspecifically bind the DNA substrate [1]. IN catalyzes two endonucleolytic reactions - 3' processing: the removal of two deoxynucleotides from viral DNA ends; and DNA strand transfer: the covalent ligation of viral DNA 3' ends to host chromosomal DNA. While a recombinant IN can catalyze 3' processing and strand transfer reactions [2], the activity of HIV-1 integrase in the context of preintegration complex (PIC) is assisted and modulated by several host factors during proviral DNA formation. The PIC is thought to be derived from the reverse transcription complex and consists of the full length viral DNA and both viral and host proteins that participate in generation of the proviral DNA [3,4].
The PIC formed following reverse transcription is in limiting amounts to permit biochemical purification of the pure complexes and identification of constituent proteins [5]. Previous studies to identify IN-interacting host proteins have primarily used yeast two-hybrid system and co-immunoprecipitations involving ectopically expressed viral and host proteins (Table 1). Another approach has been the in vitro reconstitution of salt-stripped PIC activity (PICs treated with high salt result in integration-defective complexes) using purified or recombinant host proteins. These approaches have helped to identify host proteins that physically interact with HIV-1 IN or stimulate HIV-1 IN catalytic activity. A recent study involving use of a biotinylated IN as a tool to detect interacting host proteins concluded that activity of the modified IN was adversely affected [6].
Table 1.
Summary of previously characterized host proteins interacting with HIV-1 IN
| Host proteins | Methods | References |
|---|---|---|
| BAF | SS | [11] |
| Gemin2 | IP | [14] |
| HAT p300 | IP | [26] |
| HMGA1 | SS | [8] |
| HSP 60 | PD | [27] |
| Human EED protein | THS; PD | [28] |
| Importin 7 | IP | [13] |
| Integrase interactor 1 | THS | [29] |
| LEDGF/p75 | IP | [30] |
| UNG2 | PD | [31] |
A list of host proteins interacting with HIV-1 integrase and the experimental procedure used to identify the protein-protein interaction. THS: two hybrid system; PD: Pull down; SS: reconstitution of salt stripped PIC activity; IP: immunoprecipitation.
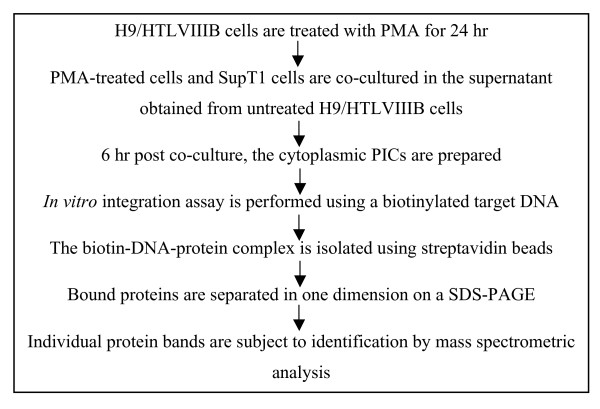
In the current study, a novel approach to identify the host proteins associated with PIC is presented. The protocol involves using a biotinylated target DNA in the standard in vitro PIC reaction assay, and the isolation of the protein complex covalently attached to target DNA using streptavidin beads (Figure 1). As a stable complex that is imported into the nucleus for integration into host chromosome, it is possible that the proteins associated with the HIV-1 DNA remain bound even after catalysis of the integration into a biotinylated target DNA. This assumption is the basis of the approach described here. A well-established protocol has been used to isolate the cytoplasmic PICs (which is a cytoplasmic extract of HIV-1-infected cells) and perform an in vitro integration assay [6]. The H9/HTLVIIIB cell line is a chronically HIV-1 infected H9-derived CD4+ cell line that releases infectious HIV-1 into the culture supernatant [7]. Stimulation of the H9/HTLVIIIB cell line with phorbol 12-myristate 13-acetate (PMA) increases viral production several fold and also increases the cell-to-cell transmission of the virus in co-culture experiments [6]. The parental H9 cells that do not produce HIV-1 were used as a negative control. Co-culture of HIV-1 producing H9/HTLVIIIB cells with HIV-1 susceptible cells such as CD4+ SupT1 cells typically leads to a high proportion of infected cells. SupT1 cells (2.5 × 109) were co-cultured for 6 hours with PMA-treated H9/HTLVIIIB or H9 cells (2.5 × 108) in the supernatant (600 ml) obtained from a 24 hour culture of H9/HTLVIIIB or H9 cells, respectively. (All cells were grown to a density of 1-1.5 × 106 per ml prior to co-culture). The PICs isolated from such co-cultured cells were demonstrated to exhibit high integration activity into naked plasmid DNA [8]. The cytoplasmic PICs generated here (isolated in 50 ml of digitonin-containing lysis buffer) have been used for in vitro integration into a ~1.5 kb biotinylated target DNA (100 μg, prepared by PCR amplification of non-viral DNA in pNL4-3 plasmid using the following primer pair: Biotin - 5' CAA AGT GCT GGG ACA ACC GGG 3' and 5' GCG CTC GGC CCT TCC GGC TGG C 3'). At the end of the integration assay, the biotinylated target DNA-PIC complex was bound to streptavidin magnetic beads (MyOne Streptavidin T1 beads, Invitrogen) at room temperature for 30 minutes (Figure 2A). To efficiently remove the majority of non-specific proteins bound to the streptavidin beads, 10 washes of 15 ml each were performed using the assay buffer [5]. The use of a biotinylated target DNA to isolate the PIC precludes the requirement of a tagged HIV-1 protein, and the potential alteration of protein-protein interactions or activity caused by a tagged-protein. The isolation of the DNA-protein complex based on the catalytic activity of PIC makes it possible to identify physiologically relevant viral-host protein interactions.
Figure 1.
Experimental design for the identification of host proteins associated with HIV-1 PIC. The PICs were generated following a protocol described previously [5]. PICs covalently bound to biotinylated DNA were isolated using streptavidin beads. The proteins in the isolated complex were identified by mass spectrometric analysis. PMA: phorbol 12-myristate 13-acetate.
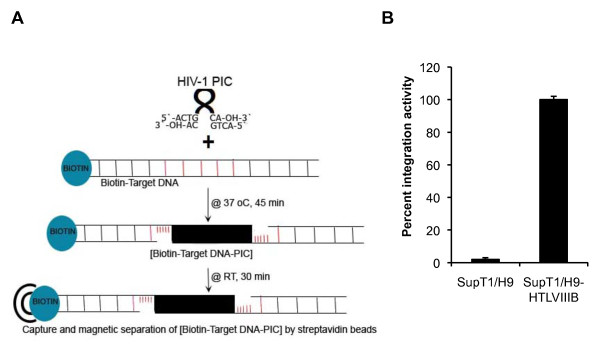
Figure 2.
Isolation of HIV-1 PICs and their activity. (A) Magnetic separation of functional HIV-1 PICs. The PIC integrates HIV-1 DNA into the ~1.5 kb biotinylated non-viral DNA from pNL4-3 plasmid that serves as a target DNA in the in vitro assay. The biotin-target DNA-protein complex is then isolated using streptavidin magnetic beads after incubation at room temperature (RT) for 30 minutes. (B) Integration activity of HIV-1 PICs. The integration activity of the PICs isolated from SupT1/H9-HTLVIIIB cell co-cultures (HIV-1-infected, set to 100 percent) and control cytoplasmic extract from SupT1/H9 cell co-cultures (control cells), using biotin-target DNA, is shown. The analysis was performed as described previously [5].
The integration activity of the isolated complex was confirmed by real-time PCR analysis using primers specific to the target and viral DNA [5]. As expected, no activity was detected either in the absence of a target DNA or with the cytoplasmic extract from the SupT1-H9 co-culture as compared to the activity of PICs isolated from the SupT1-H9/HTLVIIIB co-culture (set to 100%) (Figure 2B). Proteins from complex mixtures such as cell lysates have been identified successfully by using mass spectrometric (MS) techniques [9]. The proteins from the complexes bound to streptavidin beads were eluted by boiling the beads in 30 μl of the SDS-PAGE running buffer at 95°C for 5 minutes. The eluted-boiled proteins were loaded into a single well of a 4-15% gradient SDS-PAGE gel (Bio-Rad), and the proteins were separated in one dimension. Differences between the SupT1-H9/HTLVIIIB and SupT1-H9 co-culture samples could not be readily delineated from visual inspection of Coomassie Blue stained gels. This is not surprising considering the minute amounts of PIC proteins in an infected cell and the several cellular proteins that can bind non-specifically to DNA or biotin or streptavidin [10]. To identify the proteins associated specifically with PIC, the protein bands ranging in size from 10 kDa to 250 kDa were sliced into ~ 25 individual gel pieces and subjected to semi-quantitative MS analysis. The false discovery rate as determined using Peptide and Protein Prophet methods was less than 0.6% for all proteins identified. Given the reduced sample complexity, undersampling was not observed.
The output from the MS analysis of two independent experiments of SupT1-H9/HTLVIIIB co-culture infections was compared against that of the SupT1-H9 co-culture control. The list of proteins present in SupT1-H9 co-culture experiment serves to eliminate the non-specifically binding cytoplasmic proteins from those identified in the SupT1-H9/HTLVIIIB co-culture samples. For a more sensitive and accurate analysis of the proteins associated with HIV-1 PICs, the following criteria have been employed: (a) identification of at least two peptides from each protein, and (b) identification of the protein in two independent SupT1-H9/HTLVIIIB co-culture samples. A total of 19 host proteins (~ 6% of the total proteins revealed by the MS analysis) were identified to be specifically associated with the HIV-1 PICs (Table 2). While barrier-to-autointegration factor (BAF) is the only host protein that was characterized previously [11,12], the identification of 18 new host proteins associated with HIV-1 PICs reflects the uniqueness of our approach. Two previously characterized proteins, Importin 7 [13] and Gemin2 [14] were detected in one of the two SupT1-H9/HTLVIIIB co-culture samples, cautioning that some of the characterized and uncharacterized proteins associated with PICs might not have been identified due to detection limits of MS. Lamina-associated polypeptide 2 isoform alpha (LAP2α) protein [15] was identified in the SupT1-H9/HTLVIIIB co-culture samples; however, its presence in SupT1-H9 samples suggests a non-specific interaction with biotinylated DNA. The integrase interacting protein, LEDGF/p75 (lens epithelium-derived growth factor), was not identified in the cytoplasmic PICs. The current analysis is limited to the identification of proteins associated with PIC assembling in the cytoplasm. A similar analysis of nuclear PICs is expected to reveal proteins such as LEDGF/p75 that function at the site of integration in the nucleus [16]. Importantly, the peptides corresponding to two HIV-1 proteins, IN and Rev were identified in SupT1-H9/HTLVIIIB co-culture samples (Figure 3). Recently, Rev has been suggested to regulate HIV-1 integration in infected cells based on its ability to interact with both IN and LEDGF/p75 [17]. Rev could therefore potentially contribute to the formation of PICs. Figure 4 shows a representative immunoblotting of SupT1-H9 and SupT1-H9/HTLVIIIB samples confirming the host proteins that specifically associated with HIV-1 PICs.
Table 2.
Host proteins selectively co-purifying with HIV-1 PICs
| Host Proteins | Accession numbers | Molecular Weight | No. of peptides |
|---|---|---|---|
| Chromatin organization | |||
| Barrier-to-autointegration factor | baf_bovin | 10 kDa | 3 |
| Nucleosome assembly protein 1-like 1 | np1l1_bovin | 45 kDa | 5 |
| Histone-binding protein RBBP4 | rbbp4_bovin | 48 kDa | 3 |
| Transcription regulation | |||
| Acidic leucine-rich nuclear phosphoprotein 32 family member A | an32a_bovin | 29 kDa | 4 |
| Acidic leucine-rich nuclear phosphoprotein 32 family member E | an32e_human | 31 kDa | 3 |
| Calreticulin | calr_cerae | 48 kDa | 5 |
| NF-kappa-B essential modulator | nemo_bovin | 49 kDa | 7 |
| Non-POU domain-containing octamer-binding protein | nono_human | 54 kDa | 8 |
| RNA polymerase-associated protein LEO1 | leo1_human | 75 kDa | 6 |
| RNA processing/localization | |||
| ATP-dependent RNA helicase DDX19A | dd19a_bovin | 54 kDa | 4 |
| Double-stranded RNA-binding protein Staufen homolog 1 | stau1_human | 63 kDa | 2 |
| Heterogeneous nuclear ribonucleoprotein H1 | hnrh1_human | 49 kDa | 2 |
| Heterogeneous nuclear ribonucleoprotein H3 | hnrh3_human | 37 kDa | 3 |
| Plasminogen activator inhibitor 1 RNA-binding protein | pairb_human | 45 kDa | 7 |
| Splicing factor 3B subunit 2 | sf3b2_human | 98 kDa | 19 |
| Splicing factor, arginine/serine-rich 3 | sfrs3_bovin | 19 kDa | 3 |
| U4/U6.U5 tri-snRNP-associated protein 1 | snut1_human | 90 kDa | 17 |
| Translation | |||
| Eukaryotic translation initiation factor 4 gamma 1 | if4g1_human | 176 kDa | 3 |
| Cytoplasmic trafficking | |||
| Dynactin subunit 2 | dctn2_human | 44 kDa | 3 |
The host proteins have been broadly categorized based on the known physiological function. The number of peptides identified by mass spectroscopic analysis and assigned to the specific protein in the database is shown.
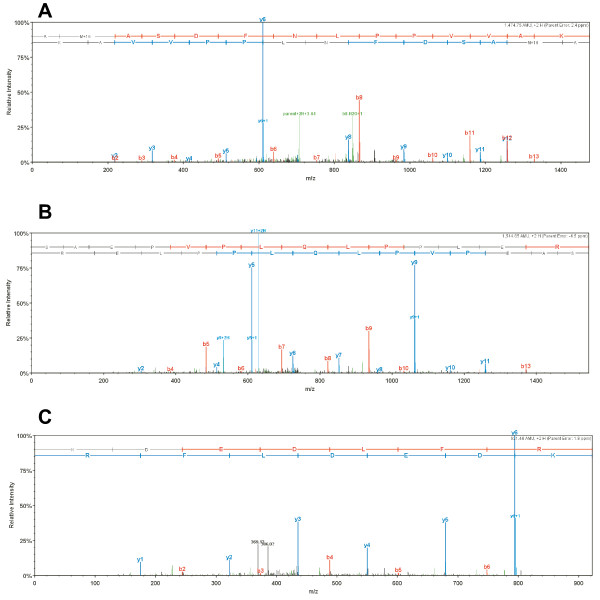
Figure 3.
Representative MS/MS data for HIV-1 and host proteins associated with PICs. (A) HIV-1 integrase peptide AMASDFNLPPVVAK. (B) HIV-1 Rev peptide SAEPVPLQLPPLER. (C) Cellular barrier-to-autointegration factor (BAF) peptide KDEDLFR. The 'b" and "y" ion series derived from the amide bond cleavage during collision induced dissociation of the peptide provide amino acid sequence information. The b-ion series (shown in red) is read from the N-terminus to C-terminus, while the y-ion series (shown in blue) is read from the C-terminus to N-terminus, providing thus complementary sequence information [25]. Other minor fragments resulted from peptide fragmentations at other sites are shown in green.
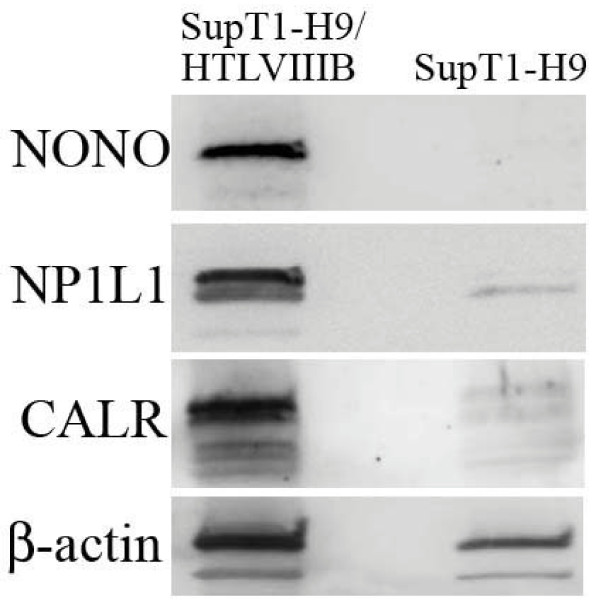
Figure 4.
Immunoblotting for host proteins that are specifically associated with HIV-1 PIC. The proteins bound to the streptavidin magnetic beads after integration assay were probed with specific antibodies. SupT1-H9/HTLVIIIB represents HIV-1 infected cell samples, and SupT1-H9 represents non-infected control samples. The host proteins are indicated by accession names on the left. The 'NONO' is Non-POU domain-containing octamer-binding protein, 'NP1L1' is Nucleosome assembly protein 1-like 1 protein and 'CALR' is Calreticulin for which 8, 5 and 5 peptides were identified by MS analysis respectively. Beta-actin found in both samples is also shown.
Of the host proteins identified here to be specifically associated with HIV-1 PICs, histone-binding protein RBBP4 is known to influences transcription activation by facilitating histone acetylation [18], and non-POU domain-containing octamer-binding protein is characterized to function with respect to double strand DNA break repair [19]. Nucleosome assembly protein 1-like 1 protein has been shown to interact with HIV-1 Tat and promote viral transcription [20], while splicing factor 3B subunit 2 protein interacts with HIV-1 Vpr and activates G2 checkpoint activation [21]. Moreover, the double-stranded RNA-binding protein Staufen homolog 1 is incorporated in HIV-1 and plays a role in viral genomic RNA encapsidation and viral particle assembly [22-24]. It is tempting to speculate that such factors might play a role in the assembly of PICs and assist the formation of proviral DNA similar to that of BAF or LEDGF/p75 and fulfill the roles not attributed to the previously characterized host factors. A detailed characterization of the host proteins identified here is essential to elucidate their role in HIV integration and to verify their potential utility as cellular targets for drug development. In conclusion, the approach described here for the identification of host proteins associated with HIV-1 PIC revealed a number of previously not described host proteins which potentially contribute to HIV-1 integration. In addition, the application of the method depicted here could be used for characterizing nucleoprotein complexes from other retroviruses.
List of abbreviations
IN: integrase; PIC: preintegration complex; HAT: histone acetyltransferase; HMGA1: high mobility group A1; HSP 60: heat shock protein 60; EED: embryonic ectoderm development; LEDGF/p75: lens epithelium-derived growth factor; UNG2: uracil-DNA glycosylase 2.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NKR conceived the study, designed and performed the biochemical experiments and drafted the manuscript. SH and MK designed, and NS and RLJG performed the mass spectrometry and helped in drafting the manuscript. LW coordinated the study, participated in the experimental design and the drafting of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Nidhanapati K Raghavendra, Email: rnidhana@cvm.osu.edu.
Nikolozi Shkriabai, Email: shkriabai.1@osu.edu.
Robert LJ Graham, Email: bobbyg@caltech.edu.
Sonja Hess, Email: shess@caltech.edu.
Mamuka Kvaratskhelia, Email: kvaratskhelia.1@osu.edu.
Li Wu, Email: wu.840@osu.edu.
Acknowledgements
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: H9 and H9/HTLV-IIIB cells from Dr. Robert Gallo. We thank Dr. Kathleen Boris-Lawrie for generous gift of antibodies used in the immunoblotting. This work was supported in part by grants to LW (R01AI068493 and R21AI078762) and to MK (R01AI062520 and P01CA100730) from the NIH.
References
- Poeschla EM. Integrase, LEDGF/p75 and HIV replication. Cell Mol Life Sci. 2008;65:1403–1424. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandgenett DP, Bera S, Pandey KK, Vora AC, Zahm J, Sinha S. Biochemical and biophysical analyses of concerted (U5/U3) integration. Methods. 2009;47:229–236. doi: 10.1016/j.ymeth.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CW, Engelman A. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J Virol. 2003;77:5030–5036. doi: 10.1128/JVI.77.8.5030-5036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka AM, Katz RA. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 2005;12(Suppl 1):971–978. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- Engelman A, Oztop I, Vandegraaff N, Raghavendra NK. Quantitative analysis of HIV-1 preintegration complexes. Methods. 2009;47:283–290. doi: 10.1016/j.ymeth.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshan M, Schweitzer CJ, Donnellan MR, Lu R, Engelman A. In vivo biotinylation and capture of HIV-1 matrix and integrase proteins. J Virol Methods. 2009;159:178–184. doi: 10.1016/j.jviromet.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M, Sarngadharan MG, Read E, Gallo RC. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984;224:497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- Farnet CM, Bushman FD. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/S0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- Leblond-Francillard M, Dreyfus M, Rougeon F. Isolation of DNA-protein complexes based on streptavidin and biotin interaction. Eur J Biochem. 1987;166:351–355. doi: 10.1111/j.1432-1033.1987.tb13522.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Z, Huang G, Yao H, Xu Z, Labine M, Cochrane AW, Yao X. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J Biol Chem. 2007;282:13456–13467. doi: 10.1074/jbc.M610546200. [DOI] [PubMed] [Google Scholar]
- Hamamoto S, Nishitsuji H, Amagasa T, Kannagi M, Masuda T. Identification of a novel human immunodeficiency virus type 1 integrase interactor, Gemin2, that facilitates efficient viral cDNA synthesis in vivo. J Virol. 2006;80:5670–5677. doi: 10.1128/JVI.02471-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulky A, Cohen TV, Kozlov SV, Korbei B, Foisner R, Stewart CL, KewalRamani VN. The LEM domain proteins emerin and LAP2alpha are dispensable for human immunodeficiency virus type 1 and murine leukemia virus infections. J Virol. 2008;82:5860–5868. doi: 10.1128/JVI.00076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin A, Rosenbluh J, Hayouka Z, Friedler A, Loyter A. Integration of HIV-1 DNA is regulated by interplay between viral rev and cellular LEDGF/p75 proteins. Mol Med. 2010;16:34–44. doi: 10.2119/molmed.2009.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Vo N, Goodman RH. Histone binding protein RbAp48 interacts with a complex of CREB binding protein and phosphorylated CREB. Mol Cell Biol. 2000;20:4970–4978. doi: 10.1128/MCB.20.14.4970-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bladen CL, Udayakumar D, Takeda Y, Dynan WS. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J Biol Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- Vardabasso C, Manganaro L, Lusic M, Marcello A, Giacca M. The histone chaperone protein Nucleosome Assembly Protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology. 2008;5:8. doi: 10.1186/1742-4690-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y, Yasuda Y. Human immunodeficiency virus type 1 Vpr induces G2 checkpoint activation by interacting with the splicing factor SAP145. Mol Cell Biol. 2006;26:8149–8158. doi: 10.1128/MCB.01170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/JVI.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev MP, Brown CM, Mouland AJ. Live cell visualization of the interactions between HIV-1 Gag and the cellular RNA-binding protein Staufen1. Retrovirology. 2010;7:41. doi: 10.1186/1742-4690-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen H, Mann M. The ABC's (and XYZ's) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, Lusic M, Marcello A, Giacca M. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 2005;24:3070–3081. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parissi V, Calmels C, De Soultrait VR, Caumont A, Fournier M, Chaignepain S, Litvak S. Functional interactions of human immunodeficiency virus type 1 integrase with human and yeast HSP60. J Virol. 2001;75:11344–11353. doi: 10.1128/JVI.75.23.11344-11353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violot S, Hong SS, Rakotobe D, Petit C, Gay B, Moreau K, Billaud G, Priet S, Sire J, Schwartz O, Mouscadet JF, Boulanger P. The human polycomb group EED protein interacts with the integrase of human immunodeficiency virus type 1. J Virol. 2003;77:12507–12522. doi: 10.1128/JVI.77.23.12507-12522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- Willetts KE, Rey F, Agostini I, Navarro JM, Baudat Y, Vigne R, Sire J. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J Virol. 1999;73:1682–1688. doi: 10.1128/jvi.73.2.1682-1688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]