Abstract
Background
MUTYH-associated polyposis (MAP) is a disorder caused by bi-allelic germline MUTYH mutation, characterized by multiple colorectal adenomas. In order to identify mutations in MUTYH gene we applied High Resolution Melting (HRM) genotyping. HRM analysis is extensively employed as a scanning method for the detection of heterozygous mutations. Therefore, we applied HRM to show effectiveness in detecting homozygous mutations for these clinically important and frequent patients.
Methods
In this study, we analyzed phenotype and genotype data from 82 patients, with multiple (>= 10) synchronous (19/82) or metachronous (63/82) adenomas and negative APC study (except one case). Analysis was performed by HRM-PCR and direct sequencing, in order to identify mutations in MUTYH exons 7, 12 and 13, where the most prevalent mutations are located. In monoallelic mutation carriers, we evaluated entire MUTYH gene in search of another possible alteration. HRM-PCR was performed with strict conditions in several rounds: the first one to discriminate the heteroduplex patterns and homoduplex patterns and the next ones, in order to refine and confirm parameters. The genotypes obtained were correlated to phenotypic features (number of adenomas (synchronous or metachronous), colorectal cancer (CRC) and family history).
Results
MUTYH germline mutations were found in 15.8% (13/82) of patients. The hot spots, Y179C (exon 7) and G396D (exon 13), were readily identified and other mutations were also detected. Each mutation had a reproducible melting profile by HRM, both heterozygous mutations and homozygous mutations. In our study of 82 patients, biallelic mutation is associated with being a carrier of ≥10 synchronous polyps (p = 0.05) and there is no association between biallelic mutation and CRC (p = 0.39) nor family history (p = 0.63). G338H non-pathogenic polymorphism (exon 12) was found in 23.1% (19/82) of patients. In all cases there was concordance between HRM (first and subsequent rounds) and sequencing data.
Conclusions
Here, we describe a screening method, HRM, for the detection of both heterozygous and homozygous mutations in the gene encoding MUTYH in selected samples of patients with phenotype of MAP. We refine the capabilities of HRM-PCR and apply it to a gene not yet analyzed by this tool. As clinical decisions will increasingly rely on molecular medicine, the power of identifying germline mutations must be continuously evaluated and improved.
Background
MAP is a disorder caused by bi-allelic germline MUTYH mutations, characterized by multiple colorectal adenomas. Mutations are distributed over the MUTYH locus, most of the changes found are missense mutations, of which Y179C and G396D, located in MUTYH exons 7 and 13 respectively, pose approximately 73% of mutations found in western populations [1,2]. Analysis of 82 selected patients was performed by HRM-PCR and direct sequencing, in order to identify mutations in MUTYH exons 7, 12 and 13. We also analyzed the prevalence of a non-pathogenic polymorphism, located in exon 12: G338H [3]. We evaluated the ability of HRM [2] for genotype at specific positions and also unknown mutations. HRM has become an alternative for screening for molecular diagnosis. In normal application HRM-PCR, discriminates heteroduplexes, which melt at lower temperatures than homoduplexes [2]. However, mutant homozygotes have been reported on the germline MUTYH gene [1]. The importance of identifying these mutations is based on the fact that MAP represents a syndrome predisposing colorectal cancer with an autosomal recessive pattern. In fact, these mutations are responsible for as many as 40% of cases with attenuated familial adenomatous polyposis without mutations in the APC gene. In this regard, downstream assays are limited in clinical decision-making and outcome [4-6]. Thus, this application provides a platform for detection of germline mutations in MUTYH gene. HRM-PCR was performed in several rounds with specific primers: the first one discriminating heteroduplex and homoduplex patterns and the next ones, in order to refine the tight conditions. Biallelic MUTYH mutations have also been found to be appended with 93-fold excess risk of colorectal cancer, with practically complete penetrance by 60 years of age [1].
Methods
Patients
This study was a collaboration amongst 3 research Departments from 12 De Octubre Hospital: Gastroenterology, Oncology and Molecular Biology. In this study, we analyzed phenotype and genotype data from 82 patients, with multiple (>= 10) synchronous (19/82) or metachronous (63/82) adenomas and negative APC study (except one case). The patients, listed in Table 1, were collected and analyzed for the presence of MUTYH mutations. Included were average ages at presentation of polyps (ranging from 17 to 88), type of polyps (≥10) metachronous (63/82) or synchronous (19/82), family history and, if any, location, stage and age at presentation of colorectal cancer (CRC). In this case, the occurrence of CRC staging was classified according to the modified Astler-Coller guide and immunohistochemistry was performed to rule out deficient mismatch repair (MMR) proteins: MLH1, MSH2, MSH6 and PMS2. All subjects signed informed consent forms after collection of blood specimens and this study was approved by the Internal Ethics Committee of 12 De Octubre University Hospital.
Table 1.
Phenotypic features of 82 patients
| Type of Adenomas | CRC | Family history | |||
|---|---|---|---|---|---|
| Patients with multiple ≥ 10) polyps synchronous | Patients with multiple (≥ 10) polyps metachronous | Yes | No | Yes | No |
| 23.2% 19/82 |
76.8% 63/82 |
48.8% 40/82 |
51.2% 42/82 |
34.1% 28/82 |
65.9% 54/82 |
| Mean age at presentation 63.7 years | Mean age at presentation 61.0 years | Mean age at presentation 57.1 years |
DNA samples
We extracted genomic DNA from peripheral blood of 82 selected patients from the Endoscopy Department of the 12 De Octubre Hospital. We isolated genomic DNA with the automatic Blood Extraction Kit based on magnetic bed technology according to manufacturer protocol (Masswell Promega, Madison, WI USA). We measured DNA concentration using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) and we diluted the samples to a final concentration of 10 ng/μl.
Primers used for HRM-PCR, identification of the assay conditions
We evaluated 3 amplicons corresponding to MUTYH exons 7, 12 and 13. The primers used for amplification of MUTYH gene segments via HRM-PCR are listed in Table 2, along with the amplicon size. The primers were designed to be annealed at 60°C using Primer Express software (Applied Biosystems, Foster City, CA) to calculate melting temperature (Tm). The primers used for amplification of the remaining MUTYH gene are listed in Table 3.
Table 2.
MUTYH HRM and sequencing primer sequences
| Exon | Primer name | Sequence | Amplicon size (base pairs) |
|---|---|---|---|
| 7 | 7F | 5'-GGGACTGACGGGTGATCTCT-3' | 186 bp |
| 7R | 5'-TTGGAGTGCAAGACTCAAGATT-3' | ||
| 12 | 12F | 5'-AGCCCTCTTGGCTTGAGTA-3' | 297 bp |
| 12R | 5'-TGCCGATTCCCTCCATTCT-3' | ||
| 13 | 13F | 5'-AGGGCAGTGGCATGAGTAAC-3' | 296 bp |
| 13R | 5'-GGGTCAAGGGGTTCAAATAG-3' | ||
Table 3.
MUTYH sequencing primer sequences
| Exon | Primer name | Sequence | Amplicon size (base pairs) |
|---|---|---|---|
| 1 | 1F | 5'-GCGGTGTACAACGGAACTTG-3' | 292 bp |
| 1R | 5'-ATCCCCGACTGCCTGAACC-3' | ||
| 2 | 2F | 5'-CTGCTTTGGCTGGGTCTTT-3' | 262 bp |
| 2R | 5'-CGCACCTGGCCCTTAGTAAG-3' | ||
| 3 | 3F | 5'-CTGCTGTGTCCCAAGACC-3' | 299 bp |
| 3R | 5'-CAACCCCAGATGAGGAGTTAGG-3' | ||
| 4 | 4F | 5'-GACCTACCATGGAGAAGACG-3' | 252 bp |
| 4R | 5'-GGGTTGGCATGAGGACACTG-3' | ||
| 5 | 5F | 5'-GGGCAGGTCAGCAGTGTC-3' | 189 bp |
| 5R | 5'-TACACCCACCCCAAAGTAGA-3' | ||
| 6 | 6F | 5'-TACTTTGGGGTGGGTGTAGA-3' | 185 bp |
| 6R | 5'-AAGAGATCACCCGTCAGTCC-3' | ||
| 8 | 8F | 5'-CCAGGAGTCTTGGGTGTCTT-3' | 240 bp |
| 8R | 5'-AGAGGGGCCAAAGAGTTAGC-3' | ||
| 9 | 9F | 5'-AACTCTTTGGCCCCTCTGTG-3 | 196 bp |
| 9R | 5'-GAAGGGAACACTGCTGTGAAG-3' | ||
| 10 | 10F | 5'-GTGCTTCAGGGGTGTCTGC-3' | 262 bp |
| 10R | 5'-TGTCATAGGGCAGAGTCACTCC-3' | ||
| 11 | 11F | 5'-TAAGGAGTGACTCTGCCCTATG-3' | 251 bp |
| 11R | 5'-GCCAAGAGGGCTTTAGGG-3' | ||
| 14 | 14F | 5'-TTGGCTTTTGAGGCTATATCC-3' | 256 bp |
| 14R | 5'-CATGTAGGAAACACAAGGAAGTA-3' | ||
| 15 | 15F | 5'-TGAAGTTAAGGGCAGAACACC-3' | 207 bp |
| 15R | 5'-GTTCACCCAGACATTCGTTAGT-3' | ||
| 16 | 16F | 5'-AGGACAAGGAGAGGATTCTCTG-3' | 298 bp |
| 16R | 5'-AGACCCCCATCTCAAAAA-3' |
First we designed primers to flank the coding regions of MUTYH exons and analyzed each amplicon in order to ensure that it contained a single melting domain using the Poland program http://www.biophys.uni-duesseldorf.de/local/POLAND/poland.html. We included intronic SNPs, close to the exon boundary, always respecting the condition of amplicons under 300 bp. This was possible as the size of MUTYH exons allowed this design. All primer sequences were analyzed (http://genome.ucsc.edu/cgi-bin/hgPcr) to adjust the likelihood that interferences would not co-amplify with the target sequence melting curves.
In monoallelic mutation carriers, we evaluated by HRM-PCR and sequencing entire MUTYH gene in search of another possible alteration.
HRM-PCR
This was performed and monitored in a Light Cycler 480 machine (Roche Diagnostics, Penzberg, Germany). HRM-PCR was performed in several rounds, the first one discriminating heteroduplex and homoduplex patterns. First round: we amplified DNA fragments (MUTYH exons 7, 12 and 13) from 5-10 ng genomic DNA. Full reactions contained final concentrations of reagents as follows: 2 mM MgCl2, 0.12 μΜ forward and reverse primers listed in Table 2, 2 × HRM Master (containing ResoLight dye) and DNA. Full HRM-PCR cycling and melting conditions were as follows: 95°C,10 min; 40 cycles of (95°C, 10 s; 60°C fluorescence reading, 10 s; 72°C, 15 s) then melting of (95°C, 1 min; 40°C, 1 min; 60°C, 1 s; 95°C 25 acquisitions per °C). We performed several rounds in each exon, using re-extracted DNA samples and the same concentration of arrangements and conditions. For reproducibility, the analyst repeated the procedures on three different days.
HRM analyses were performed on the latest Software (v.1.5) and the results were blinded to the sequencing data. The melting curves were normalized and temperature shifted, to permit samples to be compared. Significant differences in fluorescence from the horizontal baseline, previously selected, were indicative of mutations. For an evaluation of the mutation detection obtained via HRM-PCR, mutation-containing DNA was amplified using the full HRM-PCR program. In order to remove background fluorescence we subjected raw fluorescence data to normalization and temperature shifting. The probability that the observed melting temperature (Tm) separation of alternative homozygotes was assessed using the nonparametric Mann-Whitney test. We started HRM analyses with sensitivity 0.3 and it was adjusted in the following rounds.
In monoallelic mutation carriers, we evaluated entire MUTYH gene in search of another possible alteration, by HRM-PCR: Full reactions contained final concentrations of reagents as follows: 2 mM MgCl2, 0.12 μΜ forward and reverse primers listed in Table 3, 2 × HRM Master (containing ResoLight dye). Full HRM-PCR touch-down cycling and melting conditions were as follows: 95°C,10 min; 40 cycles of (95°C, 10 s; (65°C-58°C step size 0,5°C and step delay 1 cycle, fluorescence reading), 10 s; 72°C, 15 s) then melting of (95°C, 1 min; 40°C, 1 min; 60°C, 1 s; 95°C 25 acquisitions per °C). The analyst repeated the procedures on three different days.
DNA sequencing
We sequenced all samples, to evaluate the detection of MUTYH mutations and correlate these with HRM results. Amplifying DNA fragments from 50-100 ng. Reactions contained final concentrations of reagents as follows: 0.8 mM MgCl2, 0.7 μΜ forward and reverse primers listed in Table 2 and Table 3, 1 × Taq polymerase fast start (Roche Diagnostics, Penzberg, Germany) and DNA. PCR cycling and conditions were: 95°C, 10 min; 35 cycles of (95°C, 30 s; 60°C, 30 s; 72°C, 30 s) and 72°C, 10 min. PCR products were purified with ExosapIT (GE Healthcare) followed by sequencing reaction with Big Dye Terminator v3.1 (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. The sequencing products were purified using AutoSeq G-50 Dye Terminator (GE Healthcare) before running on a 3130 Genetic Analyser (Applied Biosystems, Foster City, CA).
Downstream assays
HRM genotyping of MUTYH gene was performed at the 12 De Octubre Hospital for genomics genotyping Core Facility.
Statistical analysis
The statistical program used for the analysis was SPSS version 15.0. The association between categorical variables was made by χ2 test and Fisher exact test and odds ratio was used to measure the strength of the association. The association between continuous variables was conducted by comparing means using the T test of Student or Mann-Whitney. The significance level was at <or equal to 0.05.
Results
MUTYH germline mutations were found in 15.8 percent, i.e. 13/82 of the patients. Biallelic MUTYH germline mutations were found in 8.5%, i.e. 7/82 which also showed an attenuated polyposis phenotype. Monoallelic MUTYH germline mutations were found in 7.3% i.e. 6/82 of the patients. In monoallelic mutation carriers, we performed HRM-PCR and sequenced entire MUTYH gene in search of another possible alteration: another mutation was detected in two cases (2/8 entire gene studied cases).
The hot spots: Y179C (exon 7) and G396D (exon 13) were found, as well as the five mutations already reported E410GfsX43 (exon 13), R426C (exon 13), R354GfsX40 (exon 12), V232F (exon 9) and V22M (exon 2).
G338H polymorphism (exon 12) was also analyzed in the cohort of 82 subjects and it was found in 23.1%, i.e. 19/82.
The two most frequent mutations reported to date [7] (Y179C and G396D) were detected in quite a number of the mutated cases, the frequency of these alleles being 61.5%, i.e. 8/13. Amongst the other mutations found [8], the E410GfsX43 (exon 13) accounted for 23.0%, i.e. 3/13, of the mutant alleles reported (Table 4).
Table 4.
MUTYH mutation prevalence for 82 patients with multiple adenomas (≥10), determined via HRM-PCR and via sequencing
| Wild type (%) |
Homozygous (%) |
Heterozygous (%) |
||
|---|---|---|---|---|
| Mutations founded | Y179C (exon 7) | 96.4 (79/82) | 1.2 (1/82) | 2.4 (2/82) |
| G396D (exon 13) | 92.7 (76/82) | 1.2 (1/82) | 6.1 (5/82) | |
| E410GfsX43 (exon 13) | 95.1 (78/82) | 0.0 (0/82) | 4.9 (4/82) | |
| R426C (exon 13) | 97.6 (80/82) | 0.0 (0/82) | 2.4 (2/82) | |
| V232F (exon 9) | 98.8 (81/82) | 0.0 (0/82) | 1.2 (1/82) | |
| V22M (exon 2) | 98.8 (81/82) | 0.0 (0/82) | 1.2 (1/82) | |
| R354GfsX40 (exon 12) | 98.8 (81/82) | 0.0 (0/82) | 1.2 (1/82) | |
| Polymorphism tested | G338H (exon 12) | 76.8 (63/82) | 0.0 (0/82) | 23.2 (19/82) |
Phenotypic and genotypic features in the 7 patients carriers of biallelic MUTYH germline mutations are shown in Table 5, collecting the variables: type of detected mutation, type of adenomas, family history, analysis of G338H non-pathogenic polymorphism and, in the case of colorectal cancer (CRC), stage and age at presentation. In our study biallelic mutation is associated with being a carrier of synchronous polyps (p = 0.05) and there is no association between biallelic mutation and colorectal cancer (p = 0.39) nor family history (p = 0.63).
Table 5.
Phenotypic and genotypic features in the 7 patients carriers of biallelic MUTYH germline mutations
| Patients with biallelic germline mutations | Number of adenomas (≥10) synchronous or metachronous | CRC | Family history | Polymorphism G338H | |
|---|---|---|---|---|---|
| Patient 1 | Y179C (exon 7) homozygous | Polyps synchronous Age 58 | No | No | No |
| Patient 2 | V22M (exon 2) E410GfsX43 (exon 13) double heterozygote | Polyps synchronous Age 42 | No | Yes | No |
| Patient 3 | R426C (exon 13) E410GfsX43 (exon 13) double heterozygote | Polyps synchronous | Yes Age diagnosis Astler-Coller A | Yes 50 | No |
| Patient 4 | R426C (exon 13) R354GfsX40 (exon 12) double heterozygote | Polyps metachronous Age 67 | No | No | Yes |
| Patient 5 | Y179C (exon 7) E410GfsX43 (exon 13) double heterozygote | Polyps synchronous | Yes Age diagnosis Astler-Coller B1 | No 63 | No |
| Patient 6 | G396D (exon 13) V232F (exon 9) double heterozygote | Polyps synchronous Age 82 | No | No | No |
| Patient 7 | G396D (exon 13) homozygous | Polyps synchronous Age 58 | No | No | No |
Phenotypic and genotypic features in the 6 patients carrying monoallelic MUTYH germline mutations show that monoallelic mutations have no association with being a carrier of synchronous polyps (p = 0.66) nor colorectal cancer (p = 0.79) and also no association between monoallelic mutations and family history (p = 0.38) (Table 6).
Table 6.
Phenotypic and genotypic features in the 6 patients carriers of monoallelic MUTYH germline mutations
| Patients with monollelic germline mutations | Number of adenomas (≥ 10) synchronous or metachronous | CRC | Family history | Polymorphism G338H | |
|---|---|---|---|---|---|
| Patient 1 | Y179C (exon 7) heterozygote | Polyps synchronous Age 43 | No | Yes | No |
| Patient 2 | G396D (exon 13) heterozygote | Polyps metachronous Age 42 | Yes Age diagnosis Astler-Coller A | Yes 40 | No |
| Patient 3 | R426C (exon 13) heterozygote | Polyps synchronous Age 74 | No | No | No |
| Patient 4 | G396D (exon 13) heterozygote (MUTYH and APC genes are mutated) In APC gene L126S heterozygous | Polyps synchronous Age 51 | No | Yes | Yes |
| Patient 5 | G396D (exon 13) heterozygote | Polyps metachronous Age 70 | No | Yes | No |
| Patient 6 | E410GfsX43 (exon 13) heterozygote | Polyps metachronous Age 39 | No | Yes | No |
In relation to the 19 carriers of G338H polymorphism [9] we found no association with any of the following variables: synchronous adenomas (p = 0,76), CRC (p = 0,79) and family history (p = 0,11) in this selected group. (Table 7).
Table 7.
Phenotypic and genotypic features in the 19 patients carriers of polymorphism G338H (exon 12)
| Type of adenomas | CRC | Family history In 9 cases no information was available |
|||
|---|---|---|---|---|---|
| Patients with multiple (≥ 10) polyps synchronous | Patients with multiple (≥ 10) polyps metachronous | Yes | No | Yes | No |
| 31,6% 6/19 |
68,4% 13/19 |
52,6% 10/19 |
47,4% 9/19 |
26,3% 5/19 |
26,3% 5/19 |
To assess the ability to differentiate between alleles, by high resolution melting technique [10-12], PCR products (first and subsequent rounds) were processed by latest Software (v.1.5) and the results were blinded to the sequencing data [13-15]. First round HRM-PCR (exon 7 with sensitivity of 0.3 and exon 13 with sensitivity of 0.45) discriminated between heteroduplex, wild homoduplex and mutant homoduplex patterns. Several rounds were carried out, as a confirmatory method and to set up the appropriate sensitivity of each exon. For reproducibility, the analyst repeated the procedures on three different days, with equivalent results. We did not have any false calls.
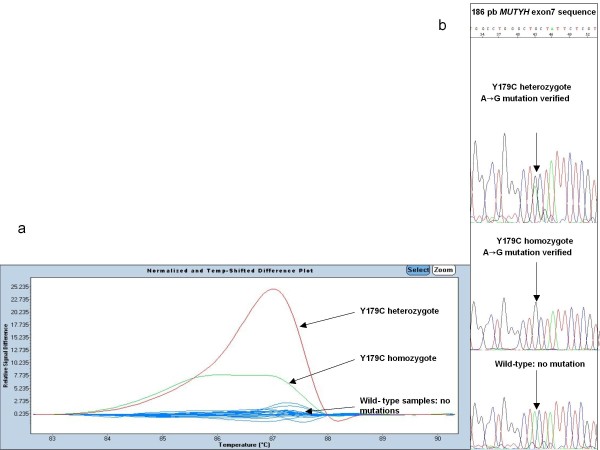
In relation to MUTYH exon 7, the results are listed in Figure 1: mutations were clearly distinct from the wild type controls in the first round. Melt curves of each mutation heterozygote, homozygote, were plotted against the wild types.
Figure 1.
Different plot and sequence traces for MUTYH exon 7 mutations. (a) The melting profile with sensitivity 0,3. The figure shows that mutations were clearly distinct from the wild type controls. Melt curves of each mutation (red: Y176C heterozygote, green: Y179C homozygote were plotted against the wild types (blue). (b) Sequencing electropherograms show a Y179C heterozygous mutation, a Y179C homozygous mutation and a wild type.
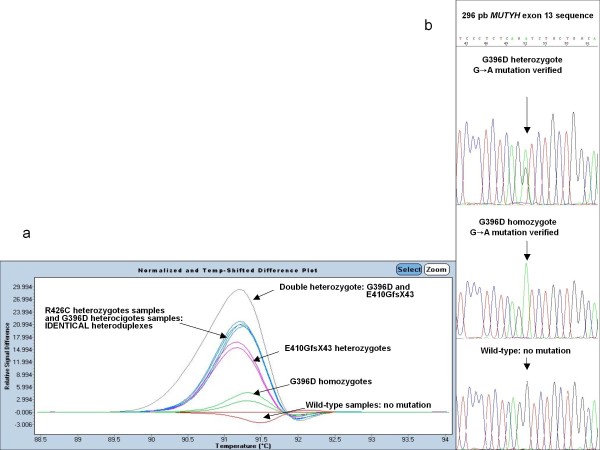
Similar to the previous analysis, Figure 2 includes different plot and sequence traces for MUTYH exon 13; First round: mutations clearly distinct from the wild type controls; but R426C heterozygote and G396C heterozygote have identical heteroduplex melting patterns (this differed in the subsequent sequencing).
Figure 2.
Difference plot and sequence traces for MUTYH exon 13 mutations. (a) Difference plot and sequence traces for MUTYH exon 13 mutations. (a) The melting profile with sensitivity 0,45. The figure shows that mutations were clearly distinct from the wild type controls. R426C heterozygote and G396D heterozygote have identical heteroduplex melting patterns. Melt curves of each mutation (green: G396D homozygotes, pink: E410GfsX43 heterozygotes, blue: R426C heterozygotes and G396D heterozygotes, grey: double heterozygote G396D and E410GfsX43 against the wild type (red). (b) Sequencing electropherograms show a G396D heterozygous mutation, a G396D homozygous mutation and a wild type.
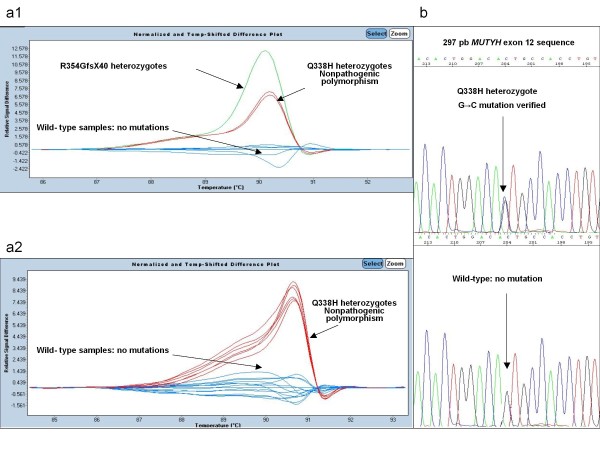
In Figure 3 different plot and sequence traces are indicated for MUTYH exon 12 mutations.
Figure 3.
Different plot and sequence traces are indicated for MUTYH exon 12 mutations. (a1) (a2) The melting profile with sensitivity 0,3. The figures shows that mutations were clearly distinct from the wild type controls. Melt curves of each mutation, green: R354GfsX40 heterozygotes, red: G338H non-pathogenic heterozygotes polymorphism against the wild type (blue). (b) Sequencing electropherograms show a G338H heterozygous polymorphism and a wild type.
In all cases there was concordance between HRM and sequencing data. (Figures 1, 2 and 3).
Discussion
We found MUTYH germline mutations in 15.8 percent, i.e. 13/82 of the patients with multiple adenomas. This percentage is obtained from the analysis of three exons, 7,12 and 13 in the 82 cases and the posterior analysis of entire MUTYH gene in monoallelic mutation carriers. We believe it feasible that this percentage would be slightly increased if we analyze the entire gene in the 82 cases[1]. We gathered information from the most prevalent hot spots at that moment in Spain.
The average age at diagnosis of CRC in studied families was 57 years (ranging from 24 to 86).
In contrast, classical FAP (familial adenomatous polyposis) patients, as is described in the literature [16-18], show a CRC onset 10 years earlier than MAP (mean age at presentation 39 versus 53 respectively)[16].
It appears that disease symptoms in the 7 MAP patients are not as severe as those observed in APC driven FAP. The mutations found along the MUTYH gene were as follows: the hot spots Y179C and G396D; and as well as the five mutations already reported. The highest prevalence of mutations corresponds to the hot spots mentioned above.
In the study of G338H non-pathogenic polymorphism (exon 12) our data indicate no association with attenuated familial adenomatous polyposis (MAP) in this selected group.
By discriminating between distinctive individuals based on their featured high-resolution melting curves[11], one of the most challenging tasks is the discrimination of amplicons differing by homoduplexes. HRM is a rapid and informative toll, but requires specific primers and refines the sensitivity post-PCR analysis in case of differing homoduplexes. We demonstrate that this tool, HRM, can also be used to detect homozygous mutations in MAP patients. In this way, homozygous mutations show a mutated homoduplex by HRM which is perfectly discriminated against wild type homoduplex and against mutated heteroduplex. (Figures 1, 2 and 3).
A HRM pre-screening assay of exons 7, 12 and 13 could be integrated into the laboratory routine, so that a large number of samples can be screened for identification of samples of interest.
We estimate that by using HRM as a screening method, the number of sequencing reactions requiring MUTYH mutation detection can be reduced by up to 80% thus resulting in substantial time and cost savings[13]. Sequencing is reduced, because once HRM-PCR is optimized for this gene, only abnormal patterns are sequenced. This shows the high resolving power of HRM, compared with direct sequencing[14]. Hence HRM can be used to combat unknown mutation-detection technologies. We refine the capabilities of HRM-PCR and apply it to a gene not yet analyzed by this method[15].
The combination of real-time PCR and high-resolution melting curve analysis provides an approach to successfully scan exons 7, 12 and 13 of MUTYH gene for these clinically important and frequent mutations.
Conclusions
In conclusion, we describe a screening method HRM, to detect heterozygous and homozygous mutations in the gene encoding MUTYH in selected samples of patients with phenotype of MAP. As clinical decisions will increasingly rely on molecular medicine, the power of identifying germline mutations must be continuously evaluated and improved.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
ILV, JW, RA, EL, SG and JML experimental design, project setup and manuscript preparation; JDM, JCM, JDT and LR clinical considerations and rationale. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Isabel López-Villar, Email: isabellv2004@yahoo.es.
Rosa Ayala, Email: rayaladiaz@yahoo.es.
Jan Wesselink, Email: jjwesselink@cbm.uam.es.
Juan Diego Morillas, Email: jdmorillas@hotmail.com.
Elena López, Email: elopezv@farm.ucm.es.
José Carlos Marín, Email: jmarin.hdoc@salud.madrid.org.
José Díaz-Tasende, Email: josediaztasende@hotmail.com.
Sara González, Email: sgonzalez@neiconcologia.net.
Luis Robles, Email: lrobles.hdoc@salud.madrid.org.
Joaquín Martínez-López, Email: jmartinezlo@hematologia12octubre.com.
Acknowledgements
We gratefully acknowledge the assistance of E. Gil and E. Rueda at the 12 De Octubre Hospital Partners for Genetics and Genomics High Throughput Genotyping Facility.
This work was backed by a training grant from Laín Entralgo Agency.
References
- Gómez-Fernández N, Castellví-Bel S, Fernández-Rozadilla C, Balaguer F, Muñoz J, Madrigal I, Milà M, Graña B, Vega A, Castells A, Carracedo A, Ruiz-Ponte C. Molecular analysis of the APC and MUTYH genes in Galician and Catalonian FAP families: a different spectrum of mutations. BMC Med Genet. 2009;10:57–68. doi: 10.1186/1471-2350-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF. Mutation scanning using high-resolution melting. Biochem Soc Tran. 2009;37:433–7. doi: 10.1042/BST0370433. [DOI] [PubMed] [Google Scholar]
- Tao H, Shinmura K, Suzuki M, Kono S, Mibu R, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, Yasunami Y, Maekawa T, Takenaka K, Ichimiya H, Imaizumi N, Sugimura H. Association between genetic polymorphisms of the base excision repair gene MUTYH and increased colorectal cancer risk in a Japanese population. Cancer Sci. 2008;99:355–60. doi: 10.1111/j.1349-7006.2007.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbe SJ, Di Bernardo MC, Chandler IP, Houlston RS. Clinical implications of the colorectal cancer risk associated with MUTYH mutation. J Clin Onco. 2009;24:3975–80. doi: 10.1200/JCO.2008.21.6853. (2009) [DOI] [PubMed] [Google Scholar]
- Fuster O, Barragán E, Bolufer P, Cervera J, Larráyoz MJ, Jiménez-Velasco A, Martínez-López J, Valencia A, Moscardó F, Sanz MA. Rapid detection of KIT mutations in core-binding factor acute myeloid leukaemia using high-resolution melting analysis. J Mol Diagn. 2009;5:458–63. doi: 10.2353/jmoldx.2009.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson JR, Jones N. MUTYH-associated polyposis. Best Pract Res Clin Gastroenterol. 2009;23:209–18. doi: 10.1016/j.bpg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Bouguen G, Manfredi S, Blayau M, Dugast C, Buecher B, Bonneau D, Siproudhis L, David V, Bretagne JF. Colorectal adenomatous polyposis Associated with MYH mutations: genotype and phenotype characteristics. Dis Colon Rectum. 2007;50:1612–7. doi: 10.1007/s10350-007-9027-0. [DOI] [PubMed] [Google Scholar]
- Isidro G, Laranjeira F, Pires A, Leite J, Regateiro F, Castro e Sousa F, Soares J, Castro C, Giria J, Brito MJ, Medeira A, Teixeira R, Morna H, Gaspar I, Marinho C, Jorge R, Brehm A, Ramos JS, Boavida MG. Germline MUTYH (MYH) mutations in Portuguese individuals with multiple colorectal adenomas. Hum Mutat. 2004;24:353. doi: 10.1002/humu.9282. [DOI] [PubMed] [Google Scholar]
- Yanaru-Fujisawa R, Matsumoto T, Ushijima Y, Esaki M, Hirahashi M, Gushima M, Yao T, Nakabeppu Y, Iida M. Genomic and functional analyses of MUTYH in Japanese patients with adenomatous polyposis. Clin Genet. 2008;73:545–53. doi: 10.1111/j.1399-0004.2008.00998.x. [DOI] [PubMed] [Google Scholar]
- Wittwer CT. High-resolution DNA melting analysis: advancements and limitations. Hum Mutat. 2009;30:857–9. doi: 10.1002/humu.20951. [DOI] [PubMed] [Google Scholar]
- Whitehall V, Tran K, Umapathy A, Grieu F, Hewitt C, Evans TJ, Ismail T, Li WQ, Collins P, Ravetto P, Leggett B, Salto-Tellez M, Soong R, Fox S, Scott RJ, Dobrovic A, Iacopetta B. A multicenter blinded study to evaluate KRAS mutation testing methodologies in the clinical setting. J Mol Diagn. 2009;11:543–52. doi: 10.2353/jmoldx.2009.090057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ES, Wong CL, Law FB, Chan WK, Siu D. Detection of KRAS mutations in colorectal cancer by high resolution melting analysis. J Clin Pathol. 2009;10:886–91. doi: 10.1136/jcp.2008.063677. [DOI] [PubMed] [Google Scholar]
- Heideman DA. A panel of high resolution melting (HRM) technology-based assays with direct sequencing possibility for effective mutation screening of EGFR and K-ras genes. Cell Oncol. 2009;31:329–33. doi: 10.3233/CLO-2009-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez Ide J, Esteban Cardeñosa E, Palanca Suela S, González EB, Bolufer Gilabert P. Advantages of the high resolution melting in the detection of BRCA1 or BRCA2 mutation carriers. Clin Biochem. 2009;42:1572–6. doi: 10.1016/j.clinbiochem.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Doi Y. High-resolution melting analysis for a reliable and two-step scanning of mutations in the tyrosine kinasine domain of the chimerical bcr-abl gene. Int J Hematol. 2009;90:37–43. doi: 10.1007/s12185-009-0337-y. [DOI] [PubMed] [Google Scholar]
- Jenkins MA, Croitoru ME, Monga N, Cleary SP, Cotterchio M, Hopper JL, Gallinger S. Risk of colorectal cancer in monoallelic and biallelic carriers of MYH mutations: a population-based case-family study. Cancer Epidemiol Biomarkers Prev. 2006;15:312–4. doi: 10.1158/1055-9965.EPI-05-0793. [DOI] [PubMed] [Google Scholar]
- Filipe B, Baltazar C, Albuquerque C, Fragoso S, Lage P, Vitoriano I, Mão de Ferro S, Claro I, Rodrigues P, Fidalgo P, Chaves P, Cravo M, Nobre Leitão C. APC or MUTYH mutations account for the majority of clinically well-characterized families with FAP and AFAP phenotype and patients with more than 30 adenomas. Clin Genet. 2009;76:242–55. doi: 10.1111/j.1399-0004.2009.01241.x. [DOI] [PubMed] [Google Scholar]
- Sinicrope FA, Rego RL, Halling KC, Foster N, Sargent DJ, La Plant B, French AJ, Laurie JA, Goldberg RM, Thibodeau SN, Witzig TE. Prognostic impact of microsatellite instability and DNA ploidy in human colon carcinoma patients. Gastroenterology. 2006;131:729–37. doi: 10.1053/j.gastro.2006.06.005. [DOI] [PubMed] [Google Scholar]