Abstract
Background
Studies examining the association of apolipoprotein A5 (APOA5) gene -1131 T>C polymorphism with blood lipids produced inconsistent results. In this meta-analysis encompassing all the relevant studies, we aimed to investigate the association of the -1131 T>C polymorphism with fasting blood lipids.
Methods
We limited our analysis to the following four blood lipid variables: total cholesterol (TC), triglycerides (TG), low density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C). Subjects were confined to adults who were at least 18 years old. A dominant model was used for this meta-analysis. 37 studies with 37859 subjects were included in this meta-analysis.
Results
The results showed that the carriers of -1131C allele have higher blood TC and TG than the non-carriers: standardized mean difference (SMD) = 0.08, 95% confidence interval (CI, 0.05, 0.11), P < 0.00001, Pheterogeneity = 0.42, and SMD = 0.31, 95% CI (0.27, 0.34), P < 0.00001, Pheterogeneity = 0.0003, respectively. Significant association between the -1131 T>C polymorphism and lower blood HDL-C was also detected under the dominant model: SMD = -0.17, 95% CI (-0.21, -0.14), P < 0.00001, Pheterogeneity = 0.003.
Conclusions
Our meta-analysis supports the strong association of the APOA5 -1131 T>C polymorphism with higher levels of TC and TG, and lower levels of HDL-C.
Background
Hyperlipidemia, which is considered to be one of the most important risk factors for coronary heart disease (CHD) and stroke, is characterized by the derangements of one or many of the lipids: elevations of total cholesterol (TC), low density lipoprotein cholesterol (LDL-C) and/or triglycerides (TG), or low levels of high-density lipoprotein cholesterol (HDL-C) [1]. Although a large number of studies have tried to elucidate the pathogenesis of the disease, the exact underlying mechanisms are still not completely understood [2]. In recent years, much has been learned about specific genes that influence hyperlipidemia [3]. However, due to various reasons, including considerable heterogeneity of the disease, the identification of susceptibility genes is difficult and most associations have not been replicated [3].
More recently, apolipoprotein A5 (APOA5) was identified as a strong modulator of blood lipids [4]. The APOA5 is predominantly synthesized in the liver and secreted into the plasma where it plays a central role in regulating TG metabolism [4]. A higher plasma APOA5 would result in lower TG levels [5]. APOA5 knockout mice develop hypertriglyceridemia, whereas transgenic mice overexpressing APOA5 have low TG levels [5]. APOA5 reduces plasma TG by inhibiting very low density lipoprotein-TG production and stimulating lipoprotein lipase-mediated very low density lipoprotein-TG hydrolysis [6]. APOA5 also play important roles in modulating other blood lipid metabolism [6-8]. Several studies have demonstrated that the APOA5 gene polymorphisms are associated with reduced HDL-C levels and decreased low density lipoprotein particle size [6-10]. Given its role in blood lipid metabolism, the APOA5 gene is considered a candidate gene for hyperlipidemia.
The APOA5 gene is located on chromosome 11q23 within the APOA1/C3/A4/A5 gene cluster, and comprises 4 exons encoding 366 amino acids [5,11]. A number of human APOA5 gene nucleotide polymorphisms (SNPs) have been investigated for a possible role in mediating genetic predisposition to hyperlipidemia [11]. The most extensively studied polymorphism is APOA5 -1131 T>C polymorphism (rs662799, SNP3). This polymorphism is located in the promoter region of the APOA5 gene [12]. A number of investigators studied the possible association between this polymorphism and blood lipids, but the results are conflicting and inconclusive [6-42]. In this paper, a meta-analysis was performed on previous reports to investigate the association of the APOA5 -1131 T>C polymorphism with fasting blood lipids.
Methods
Identification and eligibility of relevant studies
We identified all articles published before November 2009 on the APOA5 -1131 T>C polymorphism and its association with blood lipids. A systematic search of the literature was carried out by using PubMed and HugeNavigator. The language was limited to English. The keywords used for this search were "APOAV OR APOA-V OR apolipoprotein A-V OR apolipoprotein AV OR apolipoprotein A5 OR APOA5 OR APO A5" concatenated with "polymorphism OR variant OR SNP OR mutation". We limited our analysis to the following four blood lipid variables: TC, TG, LDL-C, and HDL-C. The selection criteria for studies to be considered for this meta-analysis were as follows: (1) data were reported on at least 1 of the four blood lipid variables; (2) data reported on fasting blood lipid; (3) in case of interventional studies, we used pre-intervention baseline data; (4) we only included studies in which mean blood lipid levels and standard deviations (SD) or standard errors by genotype were available; (5) subjects were confined to adults who were at least 18 years old. All references cited in the studies were also reviewed in order to find other published work that was not indexed by PubMed and HugeNavigator. Animal studies, case reports, review articles, abstracts, reports with incomplete data, and studies based on pedigree data were excluded.
Data extraction
Two investigators independently reviewed the articles to exclude irrelevant and overlapping studies. The results were compared, and disagreements were discussed and resolved by consensus. When overlapping articles were found, we only included the publication that reported the most extensive information. From each study, the following information was extracted: journal, year of publication, first author, demographics, racial background of the study population, fasting status, age, sex, health condition, sample size, mean and SD or standard error by genotypes, genotyping and lipid assay methods, unit of the four lipid variables.
Statistical analysis
All data in this analysis were presented as mean ± SD. When the standard error was reported in the original article, the value of the SD was calculated. Review Manager 5.0 software (The Cochrane Collaboration, Oxford, UK) was used for the meta-analysis. Because the frequencies of the minor allele homozygous were low, to ensure adequate statistical power, we employed a dominant model [(TC+CC) versus TT] for this meta-analysis. When information was reported for more than one subpopulation (for example, male subjects or female subjects, subjects with type 2 diabetes and control subjects, subjects from different geographical areas or different ethnicity) in one study, each subpopulation was treated as a separate comparison in our meta-analysis. In addition, we conducted subgroup analyses by ethnicity, gender, and health condition. Ethnic subgroups were defined as European descendents, East Asian, or populations of other ethnic origins. Considering that the effects of APOA 5 gene on blood lipids may be influenced by lipid-lowering therapy, we also performed a subgroup analysis in subjects without taking lipid-lowering medication. The healthy subjects (normal subjects or healthy subjects) and the subjects who ceased lipid-lowering therapy at least 1 month were also included in this subgroup. For TG, we also conducted a larger study (with sample size more than five hundreds) subgroup analysis. To ensure adequate statistical power, we only performed the meta-analysis on the subgroup with at least four studies.
The existence of heterogeneity between studies was ascertained by Q-statistic. Heterogeneity was considered significant for P < 0.1. A pooled standardized mean difference (SMD), together with 95% confidence interval (CI), was used for this meta-analysis. The SMD was chosen because the blood lipids were measured using different scanners [43]. Because a random effects model considers both between-study and within-study heterogeneity, it provides a more conservative evaluation of the significance of the association than one based on fixed effects [44]. The random effects model was used for this meta-analysis.
All populations described in the studies were tested for Hardy-Weinberg equilibrium (HWE). We repeatedly performed the meta-analysis via excluding the studies not in HWE to test the stability of the results.
Galbraith plot, made by MIX 1.7 software (Kitasato Clinical Research Center, Kitasato University, Japan), was used to detect potential sources of heterogeneity [45,46]. The pooled SMDs were recalculated after removal of the outlier studies identified in the plot.
Funnel plot was performed to look for evidence of publication bias. The funnel plot should be asymmetric when there is publication bias and symmetric in case of no publication bias. Egger's test, estimated by MIX 1.7 software was performed to measure the funnel plot asymmetry [45-47]. A significance level of 0.1 was used as an indication for the presence of potential publication bias. A P < 0.1 indicates the presence of publication bias.
Results
Selection and characteristics of studies
Additional file 1 (figure legend of additional file 1 is in additional file 2) describes the flow of candidate and eligible papers. Initial search of the literature yielded 220 publications. We excluded 91 irrelevant papers on the basis of title and abstract. The original papers were retrieved and evaluated for compliance with the inclusion criteria. 92 papers were ineligible for the following reasons: 43 papers did not provide complete data for this meta-analysis, 29 papers presented data on other polymorphisms, 8 papers had subjects overlap with other publications, 6 studies were based on pedigree data, 5 papers were review articles, and 1 paper involved subjects younger than 18 years.
The selected study characteristics were summarized in Table S1 of additional file 3. A total of 37 studies were included in this meta-analysis. Of these, 34, 27, 23, and 30 studies separately presented the data on TG, TC, LDL-C, and HDL-C. In the 37 studies, there were 13 studies of European descendents, 15 studies of East Asians, 5 studies of other ethnic origins. 4 studies reported results on different racial descendent population and each population was treated as a separate comparison in the meta-analysis. In 1 study, all the subjects were female, and in 5 studies, all the subjects were male; while in the remaining 31 studies, the subjects consisted of both male and female; among the 31 studies, 4 studies provided the data on male and female subjects respectively. In the eligible studies, 6 studies involved subjects with type 2 diabetes, 9 studies involved healthy subjects, and 6 studies involved subjects with CHD. 19 studies clearly stated that the overall populations or one/several subpopulations did not take lipid-lowering medication. In all 37 studies, there were 19 studies separately providing the information on more than one subpopulation. Each subpopulation was treated as a separate comparison. Genotype distribution in 3 subpopulations from 3 studies significantly deviated from the expected HWE. The units of blood lipids used in the eligible studies included mg/dl and mmol/L. The complete blood lipids data by genotypes can be found in Table S2 of additional file 3.
Summary statistics
We distinguished 66 comparisons on the basis of categories like ethnicity, gender, and health condition. Of these, 60, 42, 37, and 48 comparisons were included for comparing the difference in blood TG, TC, LDL-C, and HDL-C, respectively (Table S2 of additional file 3, Table 1). Overall, 37859 subjects were enrolled in this meta-analysis. Among them, 24226 subjects (64%) had the genotype TT, and 13633 (36%) were carriers of -1131C allele (Table S2 of additional file 3). 34193, 20961, 19860, and 29588 subjects were included in comparing the difference in blood TG, TC, LDL-C, and HDL-C, respectively (Table S2 of additional file 3).
Table 1.
Meta-analysis of the APOA5 -1131T>C polymorphism and blood lipids association
| Group and subgroups under analysis | Comparisons (n) | Q test P value | SMD (95% CI) | P |
|---|---|---|---|---|
| TC | ||||
| All | 42 | 0.42 | 0.08 (0.05, 0.11) | <0.00001 |
| All in HWE | 39 | 0.30 | 0.08 (0.04, 0.12) | 0.0001 |
| No lipid-lowering medicine | 20 | 0.47 | 0.08 (0.03, 0.14) | 0.003 |
| No lipid-lowering medicine in HWE | 19 | 0.41 | 0.08 (0.02, 0.14) | 0.005 |
| European | 16 | 0.16 | 0.07 (-0.00, 0.15) | 0.05 |
| European in HWE | 14 | 0.09 | 0.07 (-0.02, 0.16) | 0.11 |
| East Asian | 18 | 0.69 | 0.09 (0.05, 0.13) | <0.0001 |
| East Asian in HWE | 17 | 0.63 | 0.09 (0.04, 0.15) | 0.0003 |
| Other | 8 | 0.45 | 0.04 (-0.03, 0.11) | 0.23 |
| Healthy | 8 | 0.45 | 0.04 (-0.04, 0.13) | 0.31 |
| Type 2 diabetes | 7 | 0.64 | 0.11 (-0.01, 0.23) | 0.07 |
| CHD | 6 | 0.71 | 0.06 (-0.03, 0.15) | 0.18 |
| CHD in HWE | 5 | 0.58 | 0.06 (-0.04, 0.15) | 0.22 |
| Male | 6 | 0.29 | 0.02 (-0.09, 0.13) | 0.67 |
| Male in HWE | 5 | 0.20 | 0.03 (-0.12, 0.17) | 0.74 |
| TG | ||||
| All | 60 | 0.0003 | 0.31 (0.27, 0.34) | <0.00001 |
| All in HWE | 58 | 0.004 | 0.31 (0.28, 0.35) | <0.00001 |
| No lipid-lowering medicine | 28 | 0.35 | 0.33 (0.29, 0.38) | <0.00001 |
| No lipid-lowering medicine in HWE | 27 | 0.32 | 0.33 (0.29, 0.38) | <0.00001 |
| European | 21 | 0.29 | 0.36 (0.29, 0.43) | <0.00001 |
| European in HWE | 20 | 0.26 | 0.36 (0.29, 0.43) | <0.00001 |
| East Asian | 23 | 0.001 | 0.34 (0.29, 0.39) | <0.00001 |
| East Asian in HWE | 22 | 0.10 | 0.35 (0.30, 0.39) | <0.00001 |
| Other | 16 | 0.27 | 0.21 (0.16, 0.26) | <0.00001 |
| Healthy | 8 | 0.89 | 0.36 (0.27, 0.44) | <0.00001 |
| Type 2 diabetes | 7 | 0.77 | 0.40 (0.29, 0.52) | <0.00001 |
| CHD | 6 | 0.88 | 0.39 (0.29, 0.48) | <0.00001 |
| CHD in HWE | 5 | 0.81 | 0.38 (0.28, 0.48) | <0.00001 |
| Male | 10 | 0.39 | 0.26 (0.19, 0.33) | <0.00001 |
| Female | 7 | 0.13 | 0.21 (0.11, 0.31) | <0.0001 |
| Larger | 25 | 0.009 | 0.26 (0.22, 0.30) | <0.00001 |
| Larger in HWE | 24 | 0.05 | 0.27 (0.23, 0.31) | <0.00001 |
| LDL-C | ||||
| All | 37 | 0.010 | 0.03 (-0.02, 0.07) | 0.22 |
| All in HWE | 34 | 0.009 | 0.02 (-0.02, 0.07) | 0.34 |
| No lipid-lowering medicine | 16 | 0.32 | 0.02 (-0.05, 0.08) | 0.61 |
| No lipid-lowering medicine in HWE | 15 | 0.26 | 0.02 (-0.05, 0.09) | 0.58 |
| European | 11 | 0.01 | -0.00 (-0.10, 0.10) | 0.99 |
| European in HWE | 9 | 0.005 | -0.01 (-0.14, 0.12) | 0.93 |
| East Asian | 20 | 0.04 | 0.04 (-0.01, 0.10) | 0.14 |
| East Asian in HWE | 19 | 0.05 | 0.04 (-0.03, 0.10) | 0.25 |
| Other | 6 | 0.44 | 0.02 (-0.06, 0.09) | 0.68 |
| Healthy | 8 | 0.03 | 0.05 (-0.09, 0.18) | 0.50 |
| Type 2 diabetes | 8 | 0.11 | 0.04 (-0.09, 0.16) | 0.55 |
| CHD | 5 | 0.73 | -0.03 (-0.12, 0.07) | 0.55 |
| CHD in HWE | 4 | 0.57 | -0.03 (-0.13, 0.07) | 0.56 |
| Male | 5 | 0.02 | -0.06 (-0.25, 0.13) | 0.51 |
| Male in HWE | 4 | 0.02 | -0.09 (-0.34, 0.15) | 0.47 |
| HDL-C | ||||
| All | 48 | 0.003 | -0.17 (-0.21, -0.14) | <0.00001 |
| All in HWE | 45 | 0.003 | -0.17 (-0.21, -0.14) | <0.00001 |
| No lipid-lowering medicine | 24 | 0.003 | -0.20 (-0.27, -0.13) | <0.00001 |
| No lipid-lowering medicine in HWE | 23 | 0.002 | -0.20 (-0.28, -0.13) | <0.00001 |
| European | 13 | 0.03 | -0.15 (-0.24, -0.06) | 0.002 |
| European in HWE | 11 | 0.01 | -0.16 (-0.27, -0.05) | 0.006 |
| East Asian | 25 | 0.07 | -0.20 (-0.24, -0.16) | <0.00001 |
| East Asian in HWE | 24 | 0.07 | -0.19 (-0.24, -0.15) | <0.00001 |
| Other | 10 | 0.13 | -0.14 (-0.22, -0.06) | 0.0008 |
| Healthy | 8 | 0.29 | -0.17 (-0.26, -0.07) | 0.0007 |
| Type 2 diabetes | 9 | 0.36 | -0.18 (-0.28, -0.09) | 0.0002 |
| CHD | 5 | 0.003 | -0.21 (-0.42, -0.01) | 0.06 |
| CHD in HWE | 4 | 0.002 | -0.24 (-0.48, -0.01) | 0.06 |
| Male | 6 | 0.55 | -0.11 (-0.20, -0.02) | 0.02 |
| Male in HWE | 5 | 0.41 | -0.11 (-0.21, -0.00) | 0.05 |
Association between the APOA5 -1131 T>C polymorphism and TC
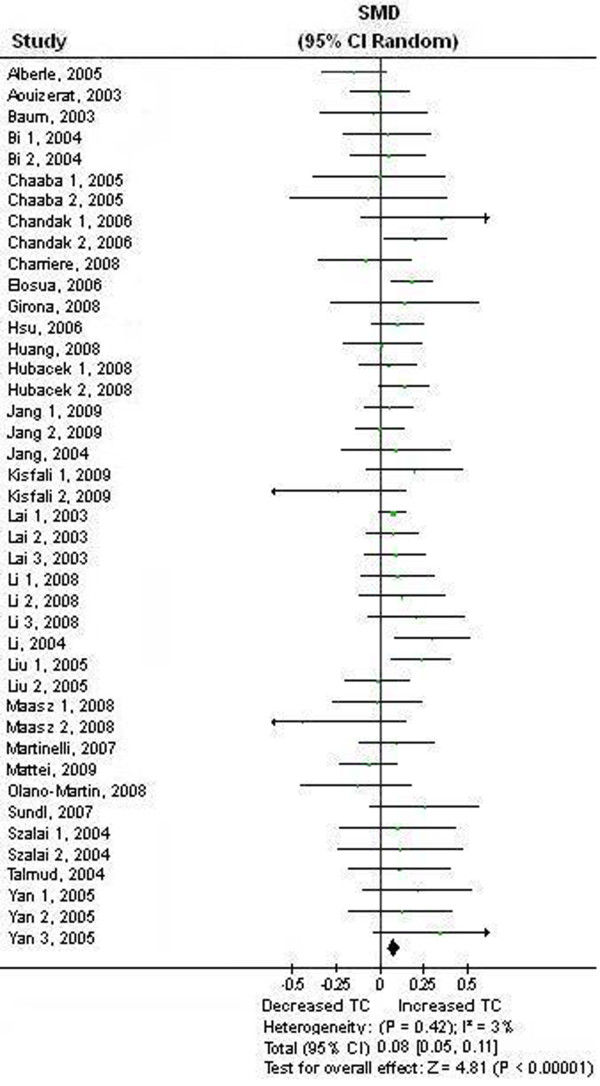
We first performed the meta-analysis on all 42 comparisons. The result showed that the carriers of -1131C allele have higher blood TC than the non-carriers: SMD = 0.08, 95% CI (0.05, 0.11), P < 0.00001, Pheterogeneity = 0.42 (Table 1, Figure 1). We then performed the subgroup analyses stratified by the characteristics of the subjects. The significant association between the APOA5 -1131 T>C polymorphism and blood TC in subjects without taking lipid-lowering medication was also detected under the dominant models: SMD = 0.08, 95% CI (0.03, 0.14), P = 0.003, Pheterogeneity = 0.47 (Table 1). The association between the APOA5 -1131 T>C polymorphism and higher levels of TC under dominant model was significant in East Asians, and marginally significant in European descendents: SMD = 0.09, 95% CI (0.05, 0.13), P < 0.0001, Pheterogeneity = 0.69, and SMD = 0.07, 95% CI (-0.00, 0.15), P = 0.05, respectively (Table 1). The other pooled SMDs from the subgroup analyses were not significant (P>0.05) (Table 1).
Figure 1.
Forest plot of the APOA5 -1131 T>C polymorphism and TC association. Result from the analysis on all 42 comparisons.
We next conducted the analyses excluding the studies not in HWE (Table 1). The association between the APOA5 -1131 T>C polymorphism and higher levels of TC under dominant model was not significant in European descendents: SMD = 0.07, 95% CI (-0.02, 0.16), P = 0.11, Pheterogeneity = 0.09. The other results were similar to those when the studies not in HWE were included.
Association between the APOA5 -1131 T>C polymorphism and TG
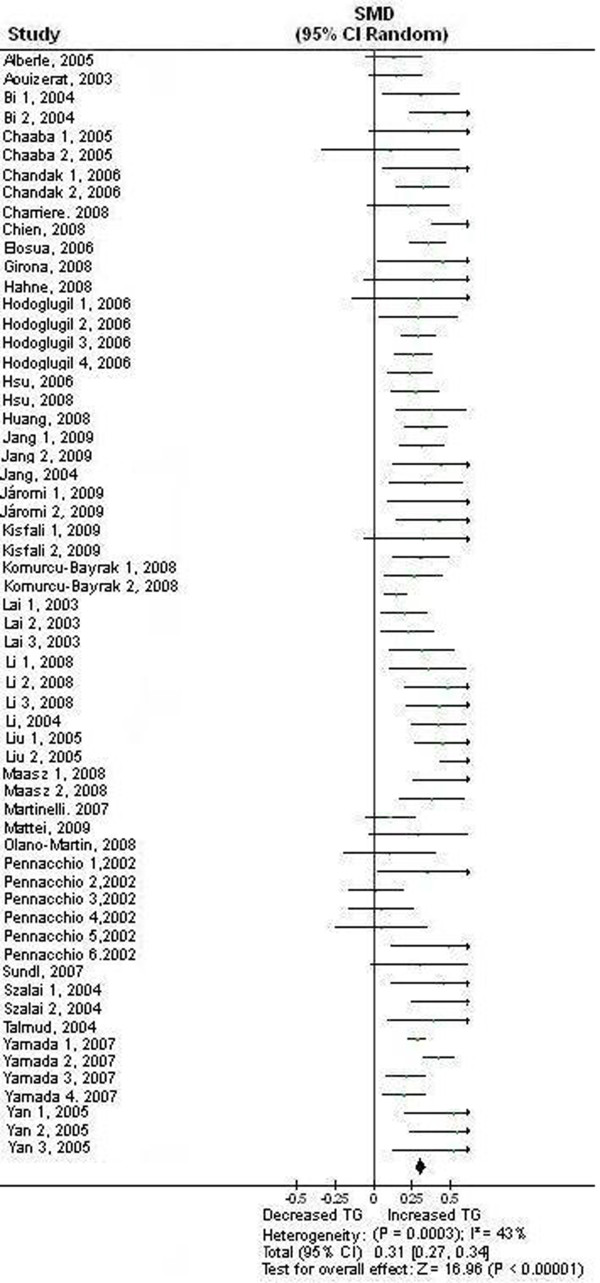
The result from the meta-analysis on all 60 comparisons showed that the carriers of -1131C allele have higher blood TG than the non-carriers in overall population: SMD = 0.31, 95% CI (0.27, 0.34), P < 0.00001, Pheterogeneity = 0.0003 (Table 1, Figure 2). The association between the APOA5 -1131 T>C polymorphism and higher levels of TG under dominant model was also significant across all subpopulations (P < 0.0001) (Table 1).
Figure 2.
Forest plot of the APOA5 -1131 T>C polymorphism and TG association. Result from the analysis on all 60 comparisons.
There was significant heterogeneity among the available studies. However, the heterogeneity was effectively decreased or removed in the subgroups stratified by the characteristics of the subjects (Table 1).
Association between the APOA5 -1131 T>C polymorphism and LDL-C
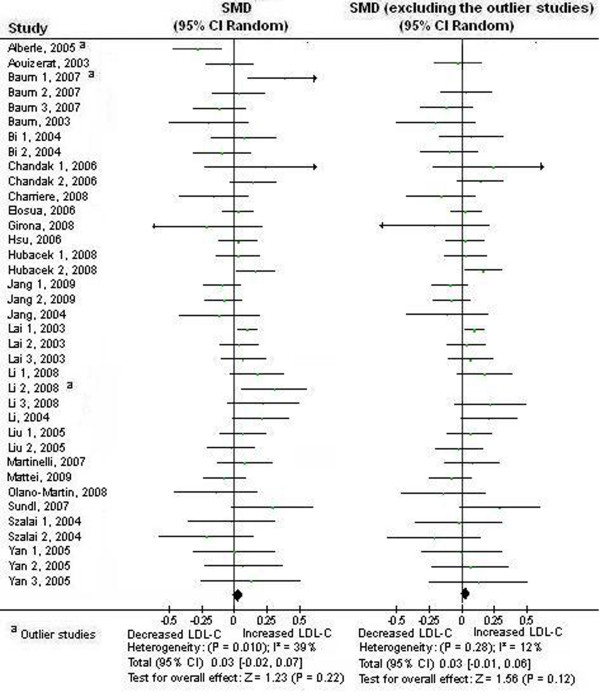
The results from our meta-analysis did not suggest a significant association between the -1131 C>T polymorphism and higher levels of LDL-C under dominant model (P>0.05) (Table 1, Table 2, Figure 3).
Table 2.
Meta-analysis of the APOA5 -1131T>C polymorphism and blood lipids association (excluding the outlier studies)
| Group and subgroups under analysis | Comparisons (n) | Q test P value | SMD (95% CI) | P |
|---|---|---|---|---|
| LDL-C | ||||
| All | 34 | 0.28 | 0.03 (-0.01, 0.06) | 0.12 |
| All in HWE | 31 | 0.28 | 0.02 (-0.02, 0.06) | 0.34 |
| No lipid-lowering medicine | 15 | 0.76 | -0.00 (-0.06, 0.06) | 0.94 |
| No lipid-lowering medicine in HWE | 14 | 0.69 | -0.00 (-0.06, 0.06) | 0.96 |
| European | 10 | 0.26 | 0.05 (-0.04, 0.13) | 0.27 |
| European in HWE | 8 | 0.14 | 0.05 (-0.06, 0.16) | 0.38 |
| East Asian | 18 | 0.24 | 0.02 (-0.03, 0.07) | 0.37 |
| East Asian in HWE | 17 | 0.40 | 0.00 (-0.05, 0.06) | 0.87 |
| Healthy | 7 | 0.18 | -0.00 (-0.11, 0.11) | 0.98 |
| Type 2 diabetes | 7 | 0.36 | -0.01 (-0.11, 0.10) | 0.89 |
| Male | 4 | 0.14 | 0.01 (-0.17, 0.19) | 0.94 |
| Male in HWE | 3 | 0.07 | -0.01 (-0.30, 0.29) | 0.96 |
| HDL-C | ||||
| All | 44 | 0.55 | -0.15 (-0.18, -0.13) | <0.00001 |
| All in HWE | 41 | 0.66 | -0.15 (-0.17, -0.12) | <0.00001 |
| No lipid-lowering medicine | 21 | 0.66 | -0.15 (-0.20, -0.10 | <0.00001 |
| No lipid-lowering medicine in HWE | 20 | 0.61 | -0.15 (-0.20, -0.10) | <0.00001 |
| European | 12 | 0.46 | -0.11 (-0.17, -0.05) | 0.0005 |
| European in HWE | 10 | 0.29 | -0.12 (-0.20, -0.04) | 0.005 |
| East Asian | 23 | 0.71 | -0.18 (-0.21, -0.15) | <0.00001 |
| East Asian in HWE | 22 | 0.80 | -0.17 (-0.20, -0.13) | <0.00001 |
| Other | 9 | 0.74 | -0.10 (-0.16, -0.03) | 0.004 |
| CHD | 4 | 0.61 | -0.12 (-0.23, -0.01) | 0.04 |
| CHD in HWE | 3 | 0.42 | -0.12 (-0.24, -0.01) | 0.04 |
Figure 3.
Forest plot of the APOA5 -1131 T>C polymorphism and LDL-C association. Result from the analysis on all 37 comparisons. There was significant heterogeneity among the studies. The pooled SMD was recalculated after removal of the outlier studies.
There was significant heterogeneity among the available studies. Significant heterogeneity was also observed in European subgroup, East Asian subgroup, healthy subject subgroup, and male subject subgroup (Table 1). 3 studies (Alberle, 2005, Baum 1, 2007, and Li 2, 2008) were identified as the main contributors of heterogeneity by using the Galbraith plot [Table S1 of additional file 3, additional file 4A (Figure legend is in additional file 2)] [20,35]. The heterogeneity was effectively removed or decreased after exclusion of these outlier studies (Table 2).
Association between the APOA5 -1131 T>C polymorphism and HDL-C
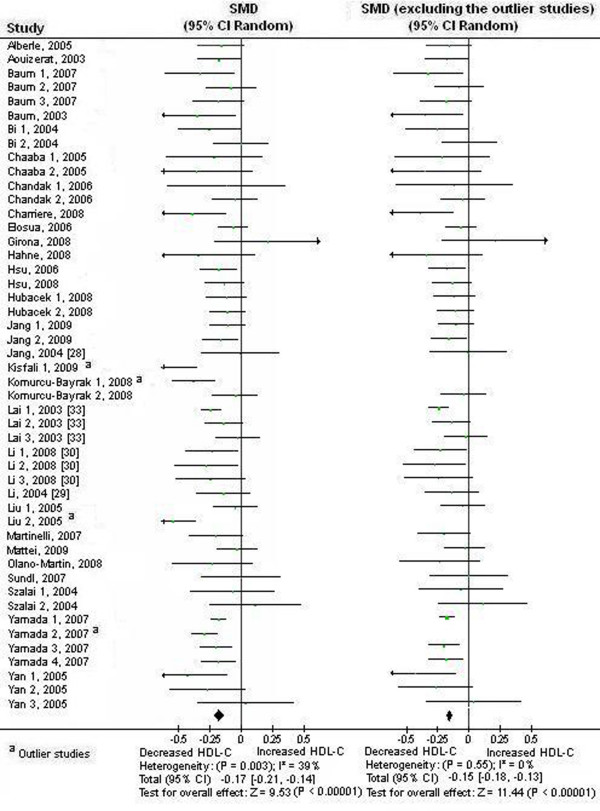
We first performed the meta-analysis on all 48 comparisons. The result showed that the carriers of -1131C allele have lower blood HDL-C than the non-carriers: SMD = -0.17, 95% CI (-0.21, -0.14), P < 0.00001, Pheterogeneity = 0.003 (Table 1, Figure 4). We then performed the subgroup analyses stratified by the characteristics of the subjects. All findings from these analyses were significant (P < 0.05) except that the association between the APOA5 -1131 T>C polymorphism and lower blood HDL-C under dominant model was marginally significant in CHD subjects: SMD = -0.21, 95% CI (-0.42, -0.01), P = 0.06, Pheterogeneity = 0.003 (Table 1).
Figure 4.
Forest plot of the APOA5 -1131 T>C polymorphism and HDL-C association. Result from the analysis on all 48 comparisons. There was significant heterogeneity among the studies. The pooled SMD was recalculated after removal of the outlier studies.
We next conducted the analyses excluding the studies not in HWE (Table 1). The association between the APOA5 -1131 T>C polymorphism and lower levels of HDL-C under dominant model was marginally significant in male subjects: SMD = -0.11, 95% CI (-0.21, -0.00), P = 0.05, Pheterogeneity = 0.41. The remaining results from these analyses were similar to those when the studies not in HWE were included.
There was significant heterogeneity among the available studies. Significant heterogeneity was also observed in European subgroup, East Asian subgroup, subgroup of subjects with CHD, and subgroup of subjects without taking lipid-lowering medicine (Table 1). 4 studies (Kisfali 1, 2009, Komurcu-Bayrak 1, 2008, Liu 2, 2005, and Yamada 2, 2007) were identified as the main contributors of heterogeneity by using the Galbraith plot [Table S1 of additional file 3, additional file 4B (Figure legend is in additional file 2)] [20,35]. The heterogeneity was effectively removed or decreased after exclusion of these outlier studies (Table 2). The association between the APOA5 -1131 T>C polymorphism and lower blood HDL-C under dominant model was significant in CHD subjects: for the analysis including all studies on subjects with CHD: SMD = -0.12, 95% CI (-0.23, -0.01), P = 0.04, Pheterogeneity = 0.61, and for the analysis excluding the studies not in HWE: SMD = -0.12, 95% CI (-0.24, -0.01), P = 0.04, Pheterogeneity = 0.42 (Table 2). The other results from these analyses were similar to those when the outlier studies were included (Table 2, Figure 4).
Publication bias
Publication bias was assayed by visual funnel plot inspection and Egger's test. The funnel plots comparing the differences in TC, LDL-C, and HDL-C were all basically symmetric [Additional file 5A, 5B, 5C (Figure legend is in additional file 2)] and Egger's test did not indicate asymmetry of the plots: Intercept = -0.1374, 95% CI (-0.9238, 0.6491), P = 0.7259, Intercept = -0.2542, 95% CI (-1.397, 0.8886), P = 0.6544, and Intercept = 0.0825, 95% CI (-0.7668, 0.9319), P = 0.8458, respectively. The funnel plot comparing the difference in TG showed asymmetry that suggested the existence of publication bias [Additional file 5D (Figure legend is in additional file 2)], and Egger's test did not indicate symmetry of the plots: Intercept = 1.0584, 95% CI (0.3716, 1.7451), P = 0.0031. To further evaluate the relationship between the APOA5 -1131 T>C polymorphism and blood TG, we carried the subgroup analysis only including the studies with sample size more than five hundreds. The funnel plot for these studies was basically symmetric [Additional file 5E (Figure legend is in additional file 2)] and Egger's test did not indicate asymmetry of the plot: Intercept = -0.0596, 95% CI (-1.7364, 1.6173), P = 0.9421.
Discussion
The APOA5 -1131 T>C polymorphism has been suggested as a possible genetic factor associated with hyperlipidemia [4,5]. However, the results from the published studies on the association of this polymorphism with hyperlipidemia are conflicting and inconclusive [6-42]. The lack of concordance across many of these studies reflects limitation in the studies, such as small sample sizes, ethnic difference and research methodology.
In this meta-analysis, we investigated the association of the APOA5 -1131 T>C polymorphism with fasting blood lipids. Because the frequencies of the minor allele homozygous were low, most of the studies included in this meta-analysis only provided mean blood lipid levels of all allele C carriers and did not respectively provide mean blood lipid levels of homozygote CC and heterozygote CT. To ensure adequate statistical power, we employed a dominant model for this meta-analysis. The results suggest that the APOA5 -1131 T>C polymorphism under dominant model is significantly associated with fasting blood lipids. The carriers of -1131C allele have higher levels of TC and TG, and lower levels of HDL-C than the non-carriers. In the eligible studies, some studies clearly stated that the overall populations or one/several subpopulations did not take lipid-lowering medication. Because the effects of APOA5 gene on blood lipids may be influenced by lipid-lowering therapy, we performed the subgroup analyses in subjects without taking lipid-lowering medication. The similar association between the -1131 T>C polymorphism and the three blood lipid variables was also detected in these subjects. Since gender, ethnicity, and health condition probably were important variables in determining associative risk with hyperlipidemia, we performed subgroup analyses of gender, ethnicity, and health condition. The effect of this polymorphism on TC especially exists in East Asians, and the effect on TG exists in all subpopulations included in this meta-analysis. For the association of the -1131 T>C polymorphism with lower levels of HDL-C, all findings from this meta-analysis were significant except that the association was marginally significant in male subjects. More studies should be conducted to further examine the association of this polymorphism with HDL-C in male subjects. The association of the -1131 T>C polymorphism with blood HDL-C, TG, and TC was very robust, which did not vary materially when the analyses that removed the studies not in HWE were performed. The significant association of the APOA5 -1131 T>C polymorphism with blood HDL-C was also supported by the results from the meta-analysis performed after removal of the outlier studies. However, our meta-analysis does not suggest the significant association of the APOA5 -1131 T>C polymorphism with blood LDL-C under dominant model. Genome-wide association studies have suggested that the polymorphisms in or near the APOA5 gene are among the strongest known genetic determinants of triglyceride concentration [48,49]. Our findings also are consistent the results from a meta-analysis by Triglyceride Coronary Disease Genetics Consortium and Emerging Risk Factors Collaboration, in which they reported that the APOA5 -1131 T>C polymorphism is associated with increased blood TG and lower HDL-C, and is not associated with increased LDL-C [50]. The different effects of the -1131 T>C polymorphism on TC, TG, HDL-C, and LDL-C suggest that pathogenesis may different in different lipid fractions and the -1131 T>C polymorphism may exert varying effect.
Significant heterogeneity was respectively found across the studies for TG, LDL-C, and HDL-C. Prominent sources of heterogeneity include: differences in ancestry, different study design, gender difference, and healthy status of the subjects included in the meta-analysis, etc. To explore the potential source of the observed heterogeneity, we performed subgroup analyses stratified by the characteristics of the subjects. For TG, the heterogeneity was effectively decreased or removed in the subgroup analyses. For LDL-C and HDL-C, significant heterogeneity was still observed in some subgroups. The sources of heterogeneity were further evaluated by Galbraith plot. Outlier studies were identified as the main contributors of heterogeneity by using the plot. The heterogeneity was effectively removed or decreased after exclusion of these outlier studies.
Significant publication bias was present in the studies on TG. Any meta-analysis carries the risk of publication bias caused by the fact that small studies with positive results are more likely to be published than those with negative results [51]. Therefore, a meta-analysis of larger studies will generally yield more conservative results than a meta-analysis of all published studies [51]. To further evaluate the relationship between the APOA5 -1131 T>C polymorphism and blood TG, we carried subgroup analysis only including the studies with sample size more than five hundreds. There was no evidence of publication bias for this subgroup analysis. The results from this analysis also suggest a significant association of the -1131 T>C polymorphism with lower levels of TG.
The association of the APOA5 -1131 T>C polymorphism with blood TC, TG, and HDL-C is not likely to be due to type I errors (false-positive results). First, the random effects model was used in our meta-analysis. Because the random effects model provides a more conservative evaluation of the significance of the association than one based on fixed effects, the results from our meta-analysis are based on a more conservative evaluation, which was expected to avoid false-positive results. Second, 37 studies with 37859 subjects were included in this meta-analysis. Among the subjects, 36% of them were carriers of -1131C allele. If the incidence of the Thr 54 carriers was sufficiently high, this may have prevented the type I error.
The possible mechanism by which -1131 T>C polymorphism modulates lipids metabolism has not yet been investigated in detail. Study has shown that APOA5 knockout mice develop hypertriglyceridemia, whereas transgenic mice overexpressing APOA5 have low triglyceride levels [5]. Since the -1131 T>C polymorphism is located within the promoter region of the APOA5 gene and might potentially result in a decreased rate of APOA5 mRNA translation, which could lead to lower plasma APOA5 levels, and then cause to higher levels of TG [32]. It is well known that plasma levels of TG and HDL-C are inversely related. A higher blood TG commonly associates with a lower blood HDL-C. A hypothesis for this association is that the delayed TG hydrolysis, due APOA5 deficiency, reduces the availability of surface components of TG-rich lipoproteins (which contribute to HDL-C formation), thereby leading to a decreased formation of HDL-C [52]. Another hypothesis for the association of the APOA5 -1131 T>C polymorphism with lower HDL-C is that the absence of APOA5, which is a minor protein component of HDL-C, renders HDL-C more unstable and more easily removed from the circulation [52]. Our meta-analysis also suggests the significant association of the -1131 T>C polymorphism with lower TC. It is more likely that the -1131 T>C polymorphism is in linkage disequilibrium with other causative mutations or nearby genes involved in the metabolism of TC [19]. APOA5 gene was identified as part of the APOA1/C3/A4/A5 gene cluster which is key components in modulating lipid metabolism [5,11]. Several SNPs in the APOA1/C3/A4/A5 gene cluster have been reported to significantly affect blood lipid levels [23,24]. Studies have reported linkage disequilibrium of the -1131 T>C polymorphism with other variants located in the APOA1/C3/A4/A5 gene cluster [19].
The current meta-analysis has several limitations which should be noted. First, hyperlipidemia originates from the interactions of multiple genes, environmental factors, and behavior. Lacking of the original data of the included studies limited our further evaluation of potential interactions because the interactions among gene-gene, gene-environment and even different polymorphic loci of the same gene may modulate blood lipid levels. For example, we did not perform the stratification analyses by environmental factors such as diet, exercise, and smoking. A more precise analysis could be conducted if more detailed individual data were available, which would allow for an adjusted estimate. Second, we did not perform a meta-analysis for haplotype analysis of the APOA5 gene, because a very limited number of studies were available. Third, for LDL-C, a relative small number of subjects were included in this meta-analysis, which may lead to lower statistic power and type II errors (false- negative results). More studies with larger population will be required to further examine the association. Fourth, because it was very difficult to get the full papers published in various languages, we only included the studies published in English. Thus the limitations mentioned may affect our final conclusions.
Conclusions
In conclusion, our meta-analysis supports the strong association of the APOA5 -1131 T>C polymorphism with higher levels of TC and TG, and lower levels of HDL-C.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TFZ participated in the design, data acquisition and analysis, interpretation of results, and manuscript writing. JPZ participated in the data acquisition and analysis. Both authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
A figure describing the flow of candidate and eligible papers.
Supplementary tables. Two supplementary tables including: Table S1. Characteristics of individual studies included in the meta-analysis. Table S2. Blood lipid levels by genotypes of individual studies included in the meta-analysis.
Galbraith plot detecting potential sources of heterogeneity.
Funnel plot detecting potential publication bias.
Contributor Information
Tongfeng Zhao, Email: zhaotongfeng@yahoo.com.cn.
Jiangpei Zhao, Email: zhaojiangpei1234@yahoo.com.cn.
Acknowledgements
This work was supported by research grant from the National Natural Science foundation of China (No: 30671007) and the grant from the Natural Science foundation of Zhejiang Province, China (No: Y2081111).
References
- Menown IB, Murtagh G, Maher V, Cooney MT, Graham IM, Tomkin G. Dyslipidemia therapy update: the importance of full lipid profile assessment. Adv Ther. 2009;7:711–718. doi: 10.1007/s12325-009-0052-3. [DOI] [PubMed] [Google Scholar]
- Garg A, Simha V. Update on dyslipidemia. J Clin Endocrinol Metab. 2007;5:581–589. doi: 10.1210/jc.2007-0275. [DOI] [PubMed] [Google Scholar]
- Bosse Y, Chagnon YC, Despres JP, Rice T, Rao DC, Bouchard C, Pérusse L, Vohl MC. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Quebec Family Study. J Lipid Res. 2004;3:419–426. doi: 10.1194/jlr.M300401-JLR200. [DOI] [PubMed] [Google Scholar]
- Tai ES, Ordovas JM. Clinical significance of apolipoprotein A5. Curr Opin Lipidol. 2008;4:349–354. doi: 10.1097/MOL.0b013e328304b681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Olivier M, Hubacek JA, Cohen JC, Cox DR, Fruchart JC, Krauss RM, Rubin EM. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- Elosua R, Ordovas JM, Cupples LA, Lai CQ, Demissie S, Fox CS, Polak JF, Wolf PA, D'Agostino RB Sr, O'Donnell CJ. Variants at the APOA5 locus, association with carotid atherosclerosis, and modification by obesity: the Framingham Study. J Lipid Res. 2006;5:990–996. doi: 10.1194/jlr.M500446-JLR200. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang S, Lin J, Li H, Huang A, Xiao C, Li X, Su Z, Wang C, Nebert DW, Zhou B, Zheng K, Shi J, Li G, Huang D. Association between DNA variant sites in the apolipoprotein A5 gene and coronary heart disease in Chinese. Metabolism. 2005;5:568–572. doi: 10.1016/j.metabol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Komurcu-Bayrak E, Onat A, Poda M, Humphries SE, Palmen J, Guclu F, Can G, Erginel-Unaltuna N. Gender-modulated impact of apolipoprotein A5 gene (APOA5) -1131T>C and c.56C>G polymorphisms on lipids, dyslipidemia and metabolic syndrome in Turkish adults. Clin Chem Lab Med. 2008;6:778–784. doi: 10.1515/CCLM.2008.161. [DOI] [PubMed] [Google Scholar]
- Hahne P, Krempler F, Schaap FG, Soyal SM, Höffinger H, Miller K, Oberkofler H, Strobl W, Patsch W. Determinants of plasma apolipoprotein A-V and APOA5 gene transcripts in humans. J Intern Med. 2008;5: 452–462. doi: 10.1111/j.1365-2796.2008.01987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y, Paik JK, Hyun YJ, Chae JS, Kim JY, Choi JR, Lee SH, Shin DJ, Ordovas JM, Lee JH. The apolipoprotein A5 -1131T>C promoter polymorphism in Koreans: association with plasma APOA5 and serum triglyceride concentrations, LDL particle size and coronary artery disease. Clin Chim Acta. 2009;1-2:83–87. doi: 10.1016/j.cca.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Járomi L, Csöngei V, Polgár N, Szolnoki Z, Maász A, Horvatovich K, Faragó B, Sipeky C, Sáfrány E, Magyari L, Kisfali P, Mohás M, Janicsek I, Lakner L, Melegh B. Functional Variants of Glucokinase Regulatory Protein and Apolipoprotein A5 Genes in Ischemic Stroke. J Mol Neurosci. in press . [DOI] [PubMed]
- Sundl I, Guardiola M, Khoschsorur G, Solà R, Vallvé JC, Godàs G, Masana L, Maritschnegg M, Meinitzer A, Cardinault N, Roob JM, Rock E, Winklhofer-Roob BM, Ribalta J. Increased concentrations of circulating vitamin E in carriers of the apolipoprotein A5 gene - 1131T>C variant and associations with plasma lipids and lipid peroxidation. J Lipid Res. 2007;11:2506–2513. doi: 10.1194/jlr.M700285-JLR200. [DOI] [PubMed] [Google Scholar]
- Maasz A, Kisfali P, Jaromi L, Horvatovich K, Szolnoki Z, Csongei V, Safrany E, Sipeky C, Hadarits F, Melegh B. Apolipoprotein A5 gene IVS3+G476A allelic variant confers susceptibility for development of ischemic stroke. Circ J. 2008;7:1065–1070. doi: 10.1253/circj.72.1065. [DOI] [PubMed] [Google Scholar]
- Kisfali P, Mohás M, Maász A, Polgár N, Hadarits F, Markó L, Brasnyó P, Horvatovich K, Oroszlán T, Bagosi Z, Bujtor Z, Gasztonyi B, Rinfel J, Wittmann I, Melegh B. Haplotype analysis of the apolipoprotein A5 gene in patients with the metabolic syndrome. Nutr Metab Cardiovasc Dis. in press . [DOI] [PubMed]
- Charriere S, Bernard S, Aqallal M, Merlin M, Billon S, Perrot L, Le Coquil E, Sassolas A, Moulin P, Marcais C. Association of APOA5 -1131T>C and S19W gene polymorphisms with both mild hypertriglyceridemia and hyperchylomicronemia in type 2 diabetic patients. Clin Chim Acta. 2008;1-2:99–103. doi: 10.1016/j.cca.2008.04.013. [DOI] [PubMed] [Google Scholar]
- Chandak GR, Ward KJ, Yajnik CS, Pandit AN, Bavdekar A, Joglekar CV, Fall CH, Mohankrishna P, Wilkin TJ, Metcalf BS, Weedon MN, Frayling TM, Hattersley AT. Triglyceride associated polymorphisms of the APOA5 gene have very different allele frequencies in Pune, India compared to Europeans. BMC Med Genet. 2006;7:76. doi: 10.1186/1471-2350-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli N, Trabetti E, Bassi A, Girelli D, Friso S, Pizzolo F, Sandri M, Malerba G, Pignatti PF, Corrocher R, Olivieri O. The -1131 T>C and S19W APOA5 gene polymorphisms are associated with high levels of triglycerides and apolipoprotein C-III, but not with coronary artery disease: an angiographic study. Atherosclerosis. 2007;2:409–417. doi: 10.1016/j.atherosclerosis.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Hodoglugil U, Tanyolaç S, Williamson DW, Huang Y, Mahley RW. Apolipoprotein A-V: a potential modulator of plasma triglyceride levels in Turks. J Lipid Res. 2006;1:144–153. doi: 10.1194/jlr.M500343-JLR200. [DOI] [PubMed] [Google Scholar]
- Szalai C, Keszei M, Duba J, Prohászka Z, Kozma GT, Császár A, Balogh S, Almássy Z, Fust G, Czinner A. Polymorphism in the promoter region of the apolipoprotein A5 gene is associated with an increased susceptibility for coronary artery disease. Atherosclerosis. 2004;1:109–114. doi: 10.1016/j.atherosclerosis.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Talmud PJ, Martin S, Taskinen MR, Frick MH, Nieminen MS, Kesäniemi YA, Pasternack A, Humphries SE, Syvänne M. APOA5 gene variants, lipoprotein particle distribution, and progression of coronary heart disease: results from the LOCAT study. J Lipid Res. 2004;4:750–756. doi: 10.1194/jlr.M300458-JLR200. [DOI] [PubMed] [Google Scholar]
- Girona J, Guardiola M, Cabré A, Manzanares JM, Heras M, Ribalta J, Masana L. The apolipoprotein A5 gene -1131T-->C polymorphism affects vitamin E plasma concentrations in type 2 diabetic patients. Clin Chem Lab Med. 2008;4:453–457. doi: 10.1515/CCLM.2008.110. [DOI] [PubMed] [Google Scholar]
- Huang MC, Wang TN, Wang HS, Sung YC, Ko YC, Chiang HC. The -1131T>C polymorphism in the apolipoprotein A5 gene is related to hypertriglyceridemia in Taiwanese aborigines. Kaohsiung J Med Sci. 2008;4:171–179. doi: 10.1016/S1607-551X(08)70114-1. [DOI] [PubMed] [Google Scholar]
- Chien KL, Fang WH, Wen HC, Lin HP, Lin YL, Lin SW, Wu JH, Kao JT. APOA1/C3/A5 haplotype and risk of hypertriglyceridemia in Taiwanese. Clin Chim Acta. 2008;1-2:56–62. doi: 10.1016/j.cca.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Matsuo H, Warita S, Watanabe S, Kato K, Oguri M, Yokoi K, Metoki N, Yoshida H, Satoh K, Ichihara S, Aoyagi Y, Yasunaga A, Park H, Tanaka M, Nozawa Y. Prediction of genetic risk for dyslipidemia. Genomics. 2007;5:551–558. doi: 10.1016/j.ygeno.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Kato K, Hibino T, Yokoi K, Matsuo H, Segawa T, Watanabe S, Ichihara S, Yoshida H, Satoh K, Nozawa Y. Prediction of genetic risk for metabolic syndrome. Atherosclerosis. 2007;2:298–304. doi: 10.1016/j.atherosclerosis.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Hsu LA, Ko YL, Chang CJ, Hu CF, Wu S, Teng MS, Wang CL, Ho WJ, Ko YS, Hsu TS, Lee YS. Genetic variations of apolipoprotein A5 gene is associated with the risk of coronary artery disease among Chinese in Taiwan. Atherosclerosis. 2006;1:143–149. doi: 10.1016/j.atherosclerosis.2005.05.031. [DOI] [PubMed] [Google Scholar]
- Bi N, Yan SK, Li GP, Yin ZN, Chen BS. A single nucleotide polymorphism -1131T>C in the apolipoprotein A5 gene is associated with an increased risk of coronary artery disease and alters triglyceride metabolism in Chinese. Mol Genet Metab. 2004;3:280–286. doi: 10.1016/j.ymgme.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Jang Y, Kim JY, Kim OY, Lee JE, Cho H, Ordovas JM, Lee JH. The -1131T-->C polymorphism in the apolipoprotein A5 gene is associated with postprandial hypertriacylglycerolemia; elevated small, dense LDL concentrations; and oxidative stress in nonobese Korean men. Am J Clin Nutr. 2004;4:832–840. doi: 10.1093/ajcn/80.4.832. [DOI] [PubMed] [Google Scholar]
- Li GP, Wang JY, Yan SK, Chen BS, Xue H, Wu G. Genetic effect of two polymorphisms in the apolipoprotein A5 gene and apolipoprotein C3 gene on serum lipids and lipoproteins levels in a Chinese population. Clin Genet. 2004;6:470–476. doi: 10.1111/j.1399-0004.2004.00251.x. [DOI] [PubMed] [Google Scholar]
- Li X, Xu Y, Ding Y, Qin C, Dai Z, Niu L. Polymorphism of apolipoprotein A5 is a risk factor for cerebral infarction in type 2 diabetes. J Huazhong Univ Sci Technolog Med Sci. 2008;6: 653–656. doi: 10.1007/s11596-008-0608-5. [DOI] [PubMed] [Google Scholar]
- Hsu LA, Ko YL, Chang CJ, Teng MS, Wu S, Hu CF. Apolipoprotein A5 gene -1131T/C polymorphism is associated with the risk of metabolic syndrome in ethnic Chinese in Taiwan. Clin Chem Lab Med. 2008;12:1714–1719. doi: 10.1515/CCLM.2008.352. [DOI] [PubMed] [Google Scholar]
- Yan SK, Cheng XQ, Song YH, Xiao XH, Bi N, Chen BS. Apolipoprotein A5 gene polymorphism -1131T-->C: association with plasma lipids and type 2 diabetes mellitus with coronary heart disease in Chinese. Clin Chem Lab Med. 2005;6:607–612. doi: 10.1515/CCLM.2005.105. [DOI] [PubMed] [Google Scholar]
- Lai CQ, Tai ES, Tan CE, Cutter J, Chew SK, Zhu YP, Adiconis X, Ordovas JM. The APOA5 locus is a strong determinant of plasma triglyceride concentrations across ethnic groups in Singapore. J Lipid Res. 2003;12:2365–273. doi: 10.1194/jlr.M300251-JLR200. [DOI] [PubMed] [Google Scholar]
- Aouizerat BE, Kulkarni M, Heilbron D, Drown D, Raskin S, Pullinger CR, Malloy MJ, Kane JP. Genetic analysis of a polymorphism in the human apoA-V gene: effect on plasma lipids. J Lipid Res. 2003;6:1167–1173. doi: 10.1194/jlr.M200480-JLR200. [DOI] [PubMed] [Google Scholar]
- Aberle J, Evans D, Beil FU, Seedorf U. A polymorphism in the apolipoprotein A5 gene is associated with weight loss after short-term diet. Clin Genet. 2005;2:152–154. doi: 10.1111/j.1399-0004.2005.00463.x. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Olivier M, Hubacek JA, Krauss RM, Rubin EM, Cohen JC. Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum Mol Genet. 2002;24:3031–3038. doi: 10.1093/hmg/11.24.3031. [DOI] [PubMed] [Google Scholar]
- Olano-Martin E, Abraham EC, Gill-Garrison R, Valdes AM, Grimaldi K, Tang F, Jackson KG, Williams CM, Minihane AM. Influence of apoA-V gene variants on postprandial triglyceride metabolism: impact of gender. J Lipid Res. 2008;5:945–953. doi: 10.1194/jlr.M700112-JLR200. [DOI] [PubMed] [Google Scholar]
- Chaaba R, Attia N, Hammami S, Smaoui M, Mahjoub S, Hammami M, Masmoudi AS. Association of SNP3 polymorphism in the apolipoprotein A-V gene with plasma triglyceride level in Tunisian type 2 diabetes. Lipids Health Dis. 2005;4:1. doi: 10.1186/1476-511X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei J, Demissie S, Tucker KL, Ordovas JM. Apolipoprotein A5 polymorphisms interact with total dietary fat intake in association with markers of metabolic syndrome in Puerto Rican older adults. J Nutr. 2009;12:2301–2308. doi: 10.3945/jn.109.109900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubacek JA, Skodová Z, Lánská V, Adámková V. Apolipoprotein A-V variant (T-1131>C) affects plasma levels of non-high-density lipoprotein cholesterol in Caucasians. Exp Clin Cardiol. 2008;3:129–132. [PMC free article] [PubMed] [Google Scholar]
- Baum L, Tomlinson B, Thomas GN. APOA5-1131T>C polymorphism is associated with triglyceride levels in Chinese men. Clin Genet. 2003;5:377–379. doi: 10.1034/j.1399-0004.2003.00063.x. [DOI] [PubMed] [Google Scholar]
- Baum L, Ng MC, So WY, Poon E, Wang Y, Lam VK, Tomlinson B, Chan JC. A case-control study of apoA5 -1131T-->C polymorphism that examines the role of triglyceride levels in diabetic nephropathy. J Diabetes Complications. 2007;3:158–163. doi: 10.1016/j.jdiacomp.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Welton NJ, Caldwell DM, Adamopoulos E, Vedhara K. Mixed treatment comparison meta-analysis of complex interventions: psychological interventions in coronary heart disease. Am J Epidemiol. 2009;9:1158–1165. doi: 10.1093/aje/kwp014. [DOI] [PubMed] [Google Scholar]
- Fleiss JL. The statistical basis of meta-analysis. Stat Methods Med Res. 1993;2:121–145. doi: 10.1177/096228029300200202. [DOI] [PubMed] [Google Scholar]
- Bax L, Yu LM, Ikeda N, Tsuruta H, Moons KGM. Development and validation of MIX: comprehensive free software for meta-analysis of causal research data. BMC Med Res Methodol. 2006;6:50. doi: 10.1186/1471-2288-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax L, Yu LM, Ikeda N, Tsuruta H, Moons KGM. MIX: comprehensive free software for meta-analysis of causal research data. Version 1.7. 2008. http://mix-for-meta-analysis.info [DOI] [PMC free article] [PubMed]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simples, graphical test. BMJ. 1997;315:629-634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;2:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;2:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglyceride Coronary Disease Genetics Consortium and Emerging Risk Factors Collaboration. Sarwar N, Sandhu MS, Ricketts SL, Butterworth AS, Di Angelantonio E, Boekholdt SM, Ouwehand W, Watkins H, Samani NJ, Saleheen D, Lawlor D, Reilly MP, Hingorani AD, Talmud PJ, Danesh J. Triglyceride-mediated pathways and coronary disease: collaborative analysis of 101 studies. Lancet. 2010;9726:1634–1639. doi: 10.1016/S0140-6736(10)60545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Trikalinos TA, Ntzani EN, Contopoulos-Ioannidis DG. Genetic associations in large versus small studies: an empirical assessment. Lancet. 2003;361:567-571. doi: 10.1016/S0140-6736(03)12516-0. [DOI] [PubMed] [Google Scholar]
- Priore Oliva C, Carubbi F, Schaap FG, Bertolini S, Calandra S. Hypertriglyceridaemia and low plasma HDL in a patient with apolipoprotein A-V deficiency due to a novel mutation in the APOA5 gene. J Intern Med. 2008;4:450–458. doi: 10.1111/j.1365-2796.2007.01912.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A figure describing the flow of candidate and eligible papers.
Supplementary tables. Two supplementary tables including: Table S1. Characteristics of individual studies included in the meta-analysis. Table S2. Blood lipid levels by genotypes of individual studies included in the meta-analysis.
Galbraith plot detecting potential sources of heterogeneity.
Funnel plot detecting potential publication bias.