Abstract
Chemokines are important immune proteins, carrying out their function by binding to glycosaminoglycans (GAGs) on the endothelial surface, and to cell surface chemokine receptors. A unique viral chemokine analog, viral macrophage inflammatory protein-II (vMIP-II), encoded by human herpesvirus-8, has garnered interest because of its ability to bind to multiple chemokine receptors including both HIV coreceptors. In addition, vMIP-II binds to cell surface GAGs much more tightly than most human chemokines, which may be the key to its anti-inflammatory function in vivo. The goal of the present work was to determine the mechanism of GAG binding by vMIP-II. The interaction of vMIP-II with heparin-derived disaccharide was characterized using NMR. Important binding sites were further analyzed by mutagenesis studies, in which corresponding vMIP-II mutants were tested for GAG binding ability using heparin chromatography and NMR. It was found that despite having many more basic residues than some chemokines, vMIP-II shares a characteristic binding site similar to its human analogs, utilizing basic residues R18, R46 and R48. Interestingly, a particular mutation (Leu13Phe) caused vMIP-II to forms a pH dependent CC-chemokine-type dimer as determined by analytical ultracentrifugation and NMR. To our knowledge, this is the first example of engineering a naturally predominantly monomeric chemokine into a dissociable dimer by a single mutation. This dimeric vMIP-II mutant binds to heparin much more tightly than wild type vMIP-II, and provides a new model to study the relationship between chemokine quaternary structure and various aspects of function. Structural differences between monomeric and dimeric vMIP-II upon GAG binding were characterized by NMR and molecular docking.
Keywords: vMIP-II, chemokine, glycosaminoglycans, NMR, analytical ultracentrifugation, dimer, molecular docking
vMIP-II1 (also called vCCL2) is a viral chemokine analog that is produced by human herpesvirus-8. This protein is unique because it binds to a wide range of chemokine receptors even across different subfamilies: it binds to CCR1, CCR2, CCR5, CXCR4 as an antagonist, and binds to CCR3 and CCR8 as an agonist (1–3). The ability to selectively block or activate multiple receptors enables vMIP-II to preferentially inhibit acute Th1-associated inflammation, but up-regulate Th2 associated immune response to help the virus evade the host immune system (2–3). Due to its potent anti-inflammatory properties, vMIP-II has been successfully used to protect rat brain after ischemic brain injury and spinal cord injury (4–5), as well as increase tolerance of cardiac and corneal allograft transplant in mice (6–7). More recently, vMIP-II has been shown to be pro-angiogenic in both mature and progenitor endothelial cells, suggesting the possibility that this protein could be useful in organ transplantation (8). vMIP-II is also a valuable candidate as an HIV inhibitor, since it blocks HIV cell entry through both CCR5 and CXCR4 coreceptors and preferentially inhibits inflammation of monocytes and Th1 type T cells which are major targets for HIV-1 (1–3). vMIP-II therefore is biologically relevant in many diverse fields.
While studies have addressed the ability of vMIP-II to bind cell surface receptors (1–3), very little has been reported about its ability to bind glycosaminoglycans (GAGs) on the extracellular surface of the endothelial cells. GAG binding plays an important role in chemokine function. While GAG binding is not essential for receptor binding in vitro (9–12), several studies have demonstrated that the ability of a chemokine to bind GAGs is critical for in vivo function (13–15). GAGs control chemokine function in many ways: GAGs help immobilize chemokines on the endothelial cell surface (16–17), where natural chemokines presumably form a concentration gradient (17–22), and these chemokines recruit leukocytes to move along the gradient by activating chemokine receptors on the leukocyte surface (23). GAG binding also protects chemokines from proteolysis and induce chemokine oligomerization (24–25). Overall, GAG binding helps chemokines to increase their local concentrations and enhance receptor signaling (9, 11, 26–27). The major role of vMIP-II is regulating immune responses, and as with human chemokines, GAG binding may be essential for vMIP-II to perform its natural function in vivo. vMIP-II may build up its own concentration gradient at the infection site and disrupt leukocyte recruitment, possibly by competing with human chemokines for binding to many chemokine receptors on the leukocyte surface, and/or by displacing chemokines from cell surface GAGs to disrupt the normal human chemokine gradient. Therefore, it is important to understand the GAG binding mechanism of vMIP-II.
Several structural studies suggested that chemokines’ GAG binding is mediated by basic residues on the chemokine surface, since cell surface GAGs are negatively charged polysaccharides. Previous studies on MIP-1β and RANTES confirmed that positively charged residues like R18, and the 40’s loop BBXB domain residues (named for Basic-Basic-Any-Basic amino acids) are critical for GAG binding (10, 28–29). For MCP-1, basic residue R18, K19, R24, and K49 forms the primary GAG binding site (30). Quaternary structure of the chemokine also plays an important role in GAG binding. The currently accepted hypothesis is that the dimer form is more capable of binding GAGs, while the monomeric form of the chemokine is responsible for binding the receptors (31). It is likely that chemokines bind GAGs in the dimer or oligomeric form and then dissociate into monomers to bind to the receptors (10, 31–32).
The structure of vMIP-II has been solved by both NMR (33–35) and X-ray crystallography (36–37). vMIP-II is a highly basic protein with 8 Lys and 5 Arg out of 71 total residues, and is predominantly a monomer under most conditions (33–35, 37), although under some crystallization conditions it has been observed as a dimer (36). In the present report, a high affinity vMIP-II dimeric variant has been designed. We present full backbone assignments of the dimeric vMIP-II variant by NMR, and demonstrate that vMIP-II provides an excellent opportunity to study the structural and functional differences between the two quaternary structural forms of chemokines at the atomic level. Mutagenesis, NMR, chromatography, and molecular docking are described to characterize the GAG binding mechanism of both quaternary structural forms of vMIP-II. Results show that vMIP-II uses a very similar mechanism as human chemokines to bind GAGs despite having many more basic residues than its close homolog, human MIP-1β, and also show an interesting role for chemokine dimerization in GAG binding.
EXPERIMENTAL PROCEDURES
Protein Preparation
The gene for vMIP-II was synthesized using overlapping oligonucleotides. vMIP-II mutations were made using the QuikChange Site-Directed Mutagenesis method (Stratagene, La Jolla, CA). vMIP-II wild type and variants were expressed in the pET-32a(+) expression vector along with a thioredoxin fusion tag (Novagen, Madison, WI). The vectors were transformed into BL21(DE3), and grown in 1 liter 15N minimal medium using 15NH4Cl as the only nitrogen source. For NMR chemical shift assignment experiments, 13C-labeled glucose was used as the only carbon source to produce the 13C-15N-labeled L13F variant, while for other mutants unlabeled glucose was used. Protein production was induced with 1 mM IPTG. The cells were harvested by centrifugation 4 hours after induction. The cell pellet was resuspended in refolding buffer (5 M Guanidinium/HCl, 3 mM EDTA, 50 mM Tris, 50 mM NaCl, pH 8.0) with 10 mM benzamidine, and French pressed twice at 16,000 psi. The solution was incubated at room temperature for 2 hours with stirring followed by a centrifugation at 20,000 ×g for 30 minutes. The supernatant containing the denatured protein was passed through a Ni chelating column and eluted with imidazole in 5 M guanidinium chloride, 50 mM Tris (pH 8.0), 500 mM NaCl. The purified proteins were slowly stirred overnight with the addition of 10 mM β-mercaptoethanol, and refolding was carried out by dialysis against 50 mM NaCl, 2 mM CaCl2, 20 mM Tris-HCl (pH 8.0). To remove the fusion tag, recombinant enterokinase (Novagen, Madison, WI) was added and the solution was incubated 3–7 days at room temperature. Precipitated matter was removed by centrifugation at 20,000 ×g for 30 minutes and the cut protein was purified on a C4 reversed phase chromatography column (Vydac, Hesperia, CA) on an Akta purification system (GE Healthcare), and lyophilized by the Labconco freeze dry system (Labconco Corporation, Kansas City, MO).
NMR Spectroscopy
All NMR data were acquired at 25 °C on Varian Inova 500 or 600 MHz, or Bruker 600 MHz spectrometers. NMR samples were prepared by adding lyophilized proteins into 20 mM sodium phosphate (pH = 2.5, 5.4, or 7.4, depending on the specific requirement for each experiment). The effect of pH variation on chemokine quaternary structure was also evaluated by NMR. While many CC-chemokines require low pH to avoid aggregation, vMIP-II was soluble throughout the range. The chemical shift was referenced relative to internal DSS (2,2-dimethyl-2-silapentane-5-sulfonic acid) (38). The data were processed using NmrPipe (39) and analyzed using PIPP (40). For 2D HSQC spectra, SW=6982.631 Hz (1H) and 1700.030 Hz (15N), with 512* points in 1H and 128* points in 15N. The 3D HNCACB experiments were acquired with SW=7804.117 Hz (1H), 9599.232 Hz (13C) and 1619.991 (15N), number of points are 512* in 1H, 64* in 13C, and 32* in 15N.
NMR Titration
Titrations of wild type vMIP-II and vMIP-II dimeric variant Leu13Phe with disaccharide I-S (V-labs, Covington, LA) were performed in 20mM Sodium Phosphate buffer at pH 7.4. Aliquots from stock solutions of disaccharide (I-S was also dissolved in 20mM Sodium Phosphate buffer at pH 7.4) were added to samples of 0.1–0.3 mM 15N-labeled protein. At the concentrations used, wild type vMIP-II was observed to be entirely monomeric except upon the addition of 1500 µM disaccharide, and for the L13F variant, only a single set of peaks (dimer) were observed. Maximum concentration of I-S titrated was 1500 µM in wild type vMIP-II, and 900 µM in vMIP-II L13F. One 1H-15N HSQC spectrum was acquired after each titration. The change in pH of the protein sample upon addition of disaccharides was negligible. For every I-S titration experiment, the change in HSQC cross-peaks of the backbone amide groups of the vMIP-II proteins were calculated as in Equation 1 (41), the observed total chemical shift perturbation (Δδobs) for each residue can be calculated as the weighted average chemical shift changes of the 1H (ΔδH) and 15N (ΔδN).
| (Eq. 1) |
In the presence of different concentrations of disaccharides, the chemical shifts for vMIP-II is the population-weighted average of the chemical shifts of free and disaccharide-bound protein. As defined in Equation 2, the observed chemical shift perturbation Δδobs is related to the ratio of disaccharide-bound vMIP-II to total vMIP-II (Vb/Vt),
| (Eq. 2) |
Where Δδmax is the difference of the chemical shifts between the fully bound and free form of vMIP-II, Vb is the concentration of bound vMIP-II, and Vt is the total concentration of vMIP-II. Assuming that one disaccharide binds only one vMIP-II monomer (or one subunit of the dimeric vMIP-II L13F variant), the ratio Vb/Vt can be calculated by Equation 3 (42),
| (Eq. 3) |
Where Dt is the total concentration of disaccharide I-S and Kd is the apparent dissociation constant of the vMIP-II I-S complex. For each titration, the ratio Dt/Vt and the Δδobs for those residues that undergo disaccharide-induced chemical shift changes are fit to Equations 2 and 3 using KaleidaGraph 3.5 (Synergy Software, Reading, PA) to give an apparent dissociation constant Kd for each residue.
Heparin Chromatography
Lyophilized proteins were dissolved in 0.5 mL 50 mM Tris buffer (pH 7.4) and injected into a 1 mL Hi-Trap heparin column (GE Healthcare). The column was rinsed with 5 mL Tris buffer and followed by a NaCl concentration gradient from 0 M to 1 M in the same buffer (50 mM Tris, pH 7.4) with the flow rate of 0.5 mL/min. The absorbance at 280 nm was monitored. Each experiment was done in duplicate to ensure the repeatability of the results. The same experiments were repeated on 1 mL Hi-Trap SP columns (GE Pharmacia) as a control. Essentially no differences in results were observed when 50 mM sodium phosphate buffer was used instead of Tris buffer.
Analytical Ultracentrifugation (AUC)
Sedimentation equilibrium analyses were performed at 25 °C on a Beckman Optima XLA analytical ultracentrifuge, using an AN 60 Ti rotor with multiple speeds and variable protein concentrations to obtain the molecular weight and dimer dissociation constant of the proteins. Wild type vMIP-II and L13F samples of different concentrations were dissolved in 20 mM sodium phosphate buffer containing 150 mM NaCl at pH 2.5, 5.4 and 7.4. Samples were centrifuged at 3,000, 15,000, 35,000, and 42,000 rpm. During each equilibrium experiment, samples were monitored by measuring the absorbance at 280 nm. The data were processed with the software “Origin” (Beckman), and each set of experimental data was fit to single ideal model or monomer–dimer equilibrium model using a nonlinear least squares fit. Wild type vMIP-II at pH 2.5 and 5.4 only fit to single ideal monomer model, and were not able to be fit with monomer-dimer equilibrium. Wild type vMIP-II at pH7.4 and vMIP-II L13F at all pH values fit best to monomer-dimer equilibrium models. Vbar for each protein was estimated using software Sednterp (www.rasmb.org), and the dimer dissociation constant (43) was calculated using the standard method described previously (44–45) using the fit Ka from Origin (Beckman).
NMR constraint-driven molecular docking
Molecular docking of disaccharide Heparin I-S on monomeric and dimeric vMIP-II (protein data bank code: 1VMP, 1CM9 respectively) was performed using the software HADDOCK2.1 (Alexandre Bonvin, Utrecht University). Experimental data such as the chemical shift perturbation data from the NMR titration and mutagenesis data on the heparin column are introduced as Ambiguous Interaction Restraints (AIRs) to drive the HADDOCK docking process.
Residues with a chemical shift perturbation higher than the average chemical shift perturbation plus one standard deviation were selected as “active” residues as defined in HADDOCK instructions. The protein structures were further analyzed using the VADAR server, and residues with less than 50% surface accessibility were excluded. “Passive” residues are defined as the residues that are within or near the “active” residues, but only residues with more than 50% surface accessibility were selected. “Active” and “passive” residues were used to generate the Ambiguous Interaction Restraints (AIRs) (46) to drive the docking.
Optimized parameters for liquid simulation (OPLS) were used, as described in the HADDOCK instruction manual. The geometric coordinate and parameter files for the proteins were parallhdg5.3.pro and topallhdg5.3.pro. For the ligand (H1S from the RANTES disaccharide complex 1U4L), the parameters were calculated and optimized using the PRODRG server (47).
For both monomeric and dimeric vMIP-II, the docking was performed in 1:1 ratio of disaccharide: protein (the dimeric vMIP-II was defined to use only one of the two identical subunits for docking). For each docking experiment, a total of 1000 complex structures were calculated during the rigid body docking, and the best 200 structures in terms of intermolecular energies were further analyzed with semiflexible annealing and water refinement. The root-mean-square deviations (rmsd) of the final set of docked structures were calculated, and the final structures were clustered using the algorithm described in Daura et al. (48). The clusters were scored using a weighted sum of a combination of energy terms.
RESULTS
Design of a vMIP-II dimer in solution
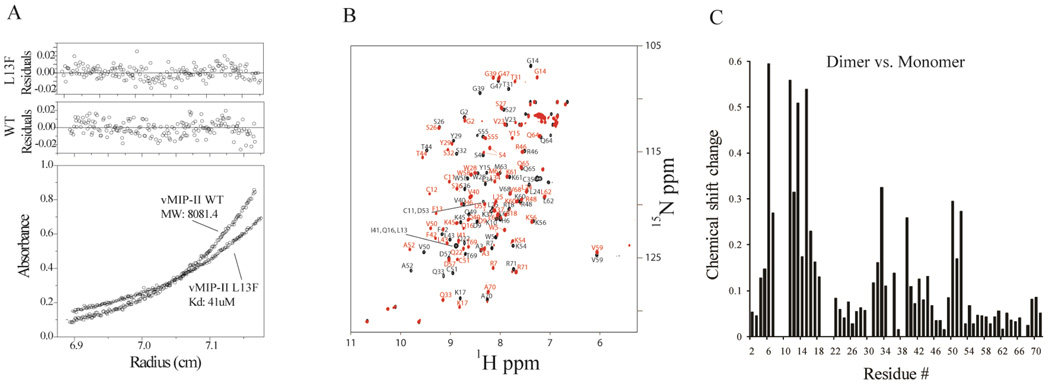
Chemokines can exist in a variety of quaternary states, which are influenced by solution conditions (28, 32, 49). However, the viral chemokine analog vMIP-II is observed to be predominantly monomeric in solution under most conditions, even at millimolar concentrations (Figure 1B) (33–35). To investigate how different quaternary states of vMIP-II interact with disaccharide in solution, we designed a vMIP-II variant which is a high affinity dissociable dimer in solution. Most human CC chemokines have a Phe or Tyr in the 13th position. In MIP-1β, the human analog of vMIP-II, Phe13 is the single most important residue for both of its quaternary structure and function. The mutant Phe13Leu disrupts dimer formation in MIP-1β, greatly reduces its GAG binding affinity (10), and also caused more than a 100 fold drop in CCR5 receptor binding affinity (50). vMIP-II, unlike its human analogs, has a Leu in its 13th position. To further investigate the role of the 13th position in vMIP-II, we mutated Leu13 to Phe and analyzed the structural and functional changes. Analytical ultracentrifugation (AUC) experiments on this vMIP-II variant revealed that vMIP-II L13F is a dimer in solution, with a Kd of 41 µM at pH 5.4, while wild type vMIP-II is a monomer under these conditions (Figure 1A). Moreover, the dimerization of vMIP-II L13F was found to be dependent on pH, with a Kd ranging from 129 µM at pH 2.5; a Kd of 41 µM at pH 5.4; to a Kd 0.74 µM at pH 7.4 (data not shown). This is in contrast to wild type vMIP-II, which is almost universally observed as a monomer, although AUC experiments show that it has a very weak tendency to dimerize at pH 7.4 (data not shown).
Figure 1.
A. Analytical Ultracentrifugation data at pH 5.4. Wild type vMIP-II fits only to a single ideal monomer model, giving a fitted molecular weight that is the same as that calculated from the protein sequence. The vMIP-II L13F mutant fits to a monomer-dimer equilibrium model, with a dissociation constant of 41 µM. B. Overlay of the chemical shift assigned 15N HSQC spectra of wild type vMIP-II (black) and vMIP-II L13F (red), both at pH 5.4 and 1 mM concentration. Peak shifts upon mutation were observed at many residues, not only at the mutation site. C. Chemical perturbation chart showing the chemical shift change of each residue from monomer (wild type) form to the dimer form (vMIP-II L13F).
As shown in Figure 1B, the L13F mutation causes dramatic change in the 15N HSQC spectrum compared to the wild type protein, not only at the 13th position, but also at many other residues. Given the results of the analytical ultracentrifugation, it is likely that the change in spectrum in the mutant is due to dimerization, a possibility that is supported by the known structures of chemokine dimers. Chemokines form two different kinds of dimer, the CC type dimer (which uses the N-terminus and 30’s loop as dimer interface) (51), and the CXC type dimer (which uses β-strand 1 and the C-terminal helix to dimerize) (52–54). Since a crystal structure of wild type vMIP-II shows a CC-type dimer (PDB code 1CM9) (36), it is likely that the dimer form of vMIP-II L13F is also a CC type dimer. To enable further analysis of the dimerization of L13F, an HNCACB experiment was performed. Full backbone chemical shift assignments ( 1H, 15N,13Cα, 13Cβ) of vMIP-II L13F were determined using standard methods, and the assignment is shown in Figure 1B. This sequence assignment enabled a residue-by-residue comparison with the wild type protein, resulting in an estimation of the relative extent of structural changes for each residue. The results (Figure 1C) showed that the greatest chemical shift perturbations occurred in the N-terminus, N-loop, 30’s loop, and residues 50–52 which are the same areas expected to be involved in a CC chemokine dimer, and correspond well to the vMIP-II dimer crystal structure, indicating that vMIP-II L13F indeed very likely forms a CC type dimer.
Wild type vMIP-II and vMIP-II L13F therefore show two distinct forms of quaternary structure in solution, which provide a good model to study the relationship between chemokine quaternary structure and chemokine - GAG binding function.
NMR studies of disaccharide binding by wild type vMIP-II
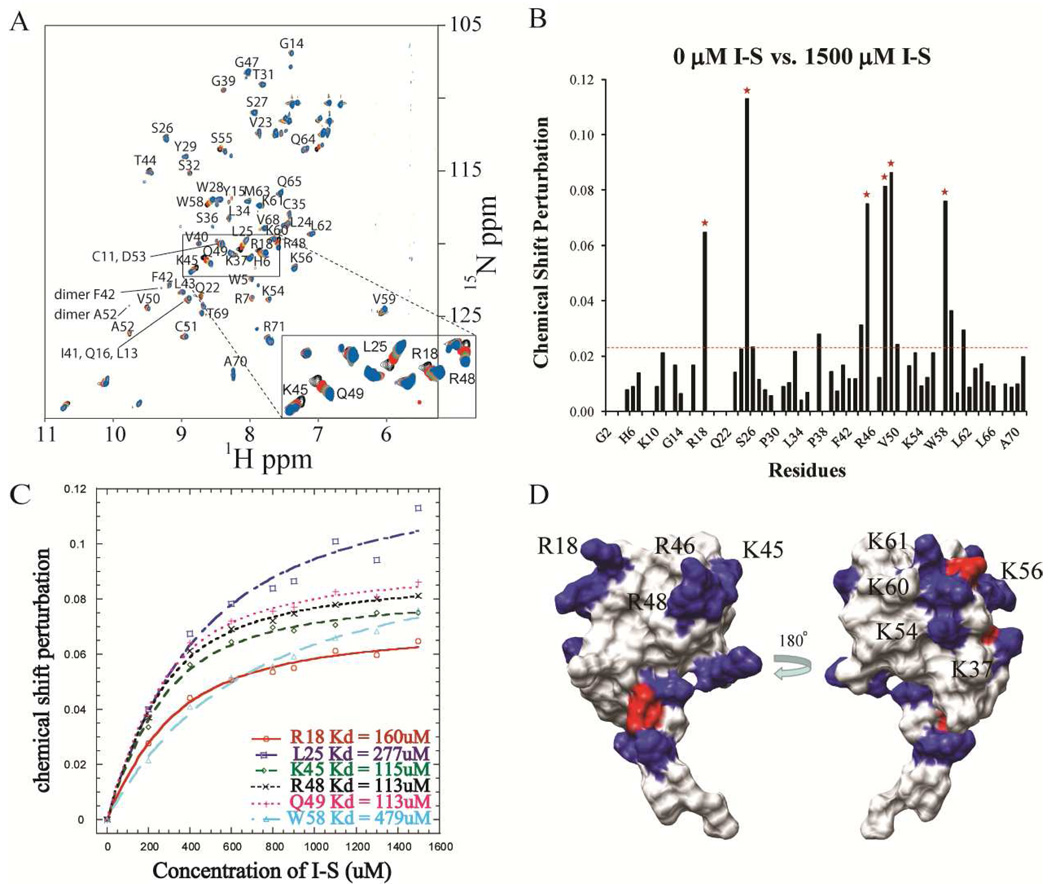
NMR studies were carried out to map the important residues in the vMIP-II - GAG interaction. Polysaccharides would be ideal for studying this interaction, but the presence of either heparin or low molecular weight heparin (which is a mixture of heparins with an average length of about 10 saccharide units) in solution caused dramatic aggregation of the protein. So the disaccharide I-S, which has been used in several chemokine binding studies, was used instead (28–29, 32, 55). Using the known sequential assignments of the wild type vMIP-II, 1H-15N HSQC experiments were carried out to monitor the chemical shift changes of each residue upon titration of the protein with heparin disaccharide I-S. An overlay of the HSQC spectra of wild type vMIP-II containing different concentrations of disaccharide clearly shows changes, indicating binding or conformational change in several areas (Figure 2A). Another notable difference observed after titrating in 1500 µM I-S was the appearance of new peaks. These indicate the formation of a vMIP-II dimer, consistent with previous results that adding high concentration disaccharide does promote dimerization of chemokines (10, 26, 32, 56), even for this “monomeric” chemokine.
Figure 2.
A. 15N HSQC spectral overlay of wild type vMIP-II (150 µM, pH 7.4) titrated with heparin I-S disaccharide with peak assignment. Selected peaks are labeled. A separate color is shown for the spectrum at each titration amount: Black, 0 µM I-S; Red, 200 µM I-S; Green, 400 µM I-S; yellow, 600 µM I-S; Purple, 800 µM I-S; Brown, 900 µM I-S; Turquoise, 1100 µM I-S; Salmon, 1300 µM I-S; Blue, 1500 µM I-S. B. Chemical shift perturbation of wild type vMIP-II after titration with 1500 mM disaccharide I-S. The red dashed line indicates the average value of the chemical shift perturbation; the red stars indicate the residues that undergo the significant chemical shift change and do not overlap with other residues in the spectra. C. Heparin I-S binding affinity calculation for the 6 residues labeled by red stars in B. D. Surface charge figure of vMIP-II. Residues with positive charge and negative charge are colored blue and red, respectively. Left panel, the "front" surface, containing R18, R46, and R48; Right panel, the "back" surface of vMIP-II.
The chemical shift perturbation of each residue upon titration with heparin I-S disaccharide is shown in Figure 2B, where red stars indicate the regions that undergo the most change in chemical shift during the titration. Residue R18, L25, K45, R48, Q49 and W58 undergo the most significant chemical shift change, and the apparent affinity of I-S binding to these residues is calculated by plotting the chemical shift perturbations at different I-S concentrations and fitting to a saturation curve (Figure 2C). The maximum concentration of disaccharide titrated was 1500 µM, since higher concentration caused severe precipitation, so the fully saturated concentration may not have been achieved. This residue-specific data allows an estimation of the binding affinity of the heparin I-S to vMIP-II. The data of the 3 adjacent 40’s loop residues K45, R48 and Q49 all can be fit with a similar Kd around 113 µM, and another positively charged residue R18 also has a similar apparent Kd of 160 µM. These residues possibly form a binding groove for the disaccharide, which is in agreement with findings on its human chemokine analogs that R18 and the 40’s loop BBXB domain are critical GAG binding sites. The other basic residue, R46 in vMIP-II’s BBXB domain, was not visible on these HSQC spectra at pH 7.4, so we were unable to assess the importance of this residue on disaccharide binding with this NMR experiment. Interestingly, a comparison of the structure of vMIP-II with the chemical shift perturbation shows that the residues undergoing the most significant chemical shift changes are all located on one face of the protein. In contrast, residues from the other surface of the protein do not show significant changes, even the positively charged residues K37, K54, K56, K60 and K61 (Figure 2D).
Heparin Chromatography of vMIP-II variants
The small size of heparin I-S (MW = 665) makes it in several ways unlike a physiological GAG, which has many more saccharide units that likely confer higher affinity binding due to avidity and possibly structural arrangement. As mentioned above, NMR experiments in the presence of the longer “low molecular weight heparin” resulted in precipitation and no usable results (data not shown). Therefore, to confirm our findings from the NMR experiments, and to provide data on the interaction of vMIP-II with longer chain GAGs, heparin chromatography was carried out on vMIP-II and its variants to determine whether the residues mapped by NMR in the presence of disaccharide are also important in binding polysaccharides.
Several variants of vMIP-II were constructed and purified (Table 1). The HSQC NMR spectrum of each variant was measured, and all mutants showed similar spectral attributes as the wild type vMIP-II (data not shown), indicating the mutations did not affect the overall structure of the protein.
Table 1.
Heparin binding affinities for vMIP-II variants as measured by NaCl elution concentrations
| vMIP-II Mutant | Heparin NaCl (mM) |
SP NaCl (mM) |
ΔNaClHep (mM) |
ΔNaClSP (mM) |
|---|---|---|---|---|
| WT | 630±1.5 | 623±1 | / | / |
| R18A | 473±1/585±1.5 | 487±0.5 | 157/45 | 136 |
| K45A | 593±0.5 | 545±0.5 | 37 | 78 |
| R46A | 535±0.5 | 503±1.5 | 95 | 120 |
| R48A | 514±2 | 512±0.5 | 116 | 111 |
| R46AR48A | 449±0.5 | 419±1 | 181 | 204 |
| K45AR46AR48A | 438±1 | 390±1 | 192 | 233 |
| K54A | 611±1.5 | 617±1 | 19 | 6 |
| L13F | 753±1 | 653±2 | −123 | −30 |
| P8AL13F | 561±1 | 626±1 | 69 | −3 |
| L13A | 564±2 | 626±0.5 | 66 | −3 |
Every experiment was done in duplicate.
ΔNaCl indicates change in NaCl elution concentration from the wild type value.
All elution profiles are similar in peak shape and height, and no unbound flow-through peaks were observed.
For R18A, 2 peaks of about equal size were observed.
Commercially prepared immobilized heparin is a highly sulfated polysaccharide with 20–25 repeated units of disaccharides. Proteins are loaded onto the heparin column, and then eluted using NaCl buffer. The elution concentration of NaCl indicates the strength of the interaction between the protein and heparin. As a control, the same experiments are repeated on an SP cation exchange column, which contains only the negatively charged sulfate but not the polysaccharides, to estimate the extent of specific binding to heparin versus the electrostatic binding to the sulfate groups of the SP column.
As shown in Table 1, wild type vMIP-II binds tightly to the heparin column, requiring 630 mM NaCl for elution. As a comparison, human chemokine MIP-1β has been shown to elute earlier, at 500 mM NaCl (10, 31); and the chemokine RANTES, a more basic protein than MIP-1β and also a dimer, has been shown to elute at 800 mM (57). Single point mutation in vMIP-II at R18 and each of the basic residues of the BBXB motif to Ala significantly decreases the binding affinity of vMIP-II to heparin (Table 1). Among the 40's loop point mutants, R46 and R48 show a significant effect, and K45A shows the least effect. Double mutant R46A/R48A and triple mutant K45A/R46A/R48A on the 40’s loop BBXB domain were also made, and these variants showed the greatest loss of heparin binding affinity among all mutants (Table 1). As a control, each mutant was also bound to a sulfate-containing cation-exchange SP column (Table 1). As expected, all the mutants with a loss of a positively charge residue exhibited reduced affinity for the negatively charged SP column.
Unlike the basic residues in the putative binding site formed by R18, K45, R46 and R48, positively charged residues on the other face of vMIP-II are not very important for GAG binding. For instance, residue K54 is located in the middle of the “back” surface. Heparin and SP chromatography results showed that the K54A mutation did not affect binding to GAGs (Table 1).
For these variants with mutations on the positive charged residues, the heparin and SP chromatography profiles showed that the vMIP-II - GAG interaction is site-specific, and the nature of this interaction is almost purely electrostatic between the positively charged residues of the binding site and the negatively charged sulfate groups on GAGs.
Dimeric vMIP-II variant binds GAGs more tightly than the wild type protein
The vMIP-II L13F dimeric variant requires 753 mM NaCl for elution from the heparin column, which is much more than for the wild type protein (Table 1). As a control for the presence of Phe at the 13th position (as opposed to dimer formation), a mutation at a different site was made to vMIP-II L13F to abrogate the dimer. The mutation from proline to alanine at the 8th position has been shown for several chemokines to disrupt the wild type dimer (50, 58). We made a P8A mutation in addition to the L13F mutation on vMIP-II, and this double mutant was indeed a folded monomer in solution as judged by NMR (data not shown). The P8AL13F vMIP-II did bind to heparin column with much lower affinity than L13F vMIP-II (Table 1), indicating that quaternary state is indeed a key mediator of GAG binding. In a comparison with wild type vMIP-II, the P8AL13F variant showed slightly reduced binding affinity to the heparin column. To further test the importance of the 13th position in vMIP-II, we made an L13A mutation which would reduce the size of the wild type side chain. As judged by HSQC spectrum, L13A was also observed to be a monomer in solution as expected (data not shown). L13A not only showed a large decrease in the elution concentration of NaCl from the dimeric L13F, but also a moderate decrease compared to the wild type protein (Table 1).
Chromatography experiments on an SP sepharose column were carried out as a control for electrostatic interactions versus specific binding to GAGs (Table. 1). Since no change in charge was involved in these mutations, little difference between L13 mutants and wild type protein was observed for SP column elution, as expected, unlike the large difference observed for the heparin column. Similarly, the P8AL13F and L13A mutations, which gave a moderate decrease in affinity for heparin column, had almost no effect on binding the cation-exchange SP column. This indicates that the overall structure (i.e. dimer) of the L13F mutant is responsible for the tighter binding to heparin, rather than simply an increase in local positive charges.
NMR studies of disaccharide binding by the dimeric vMIP-II L13F
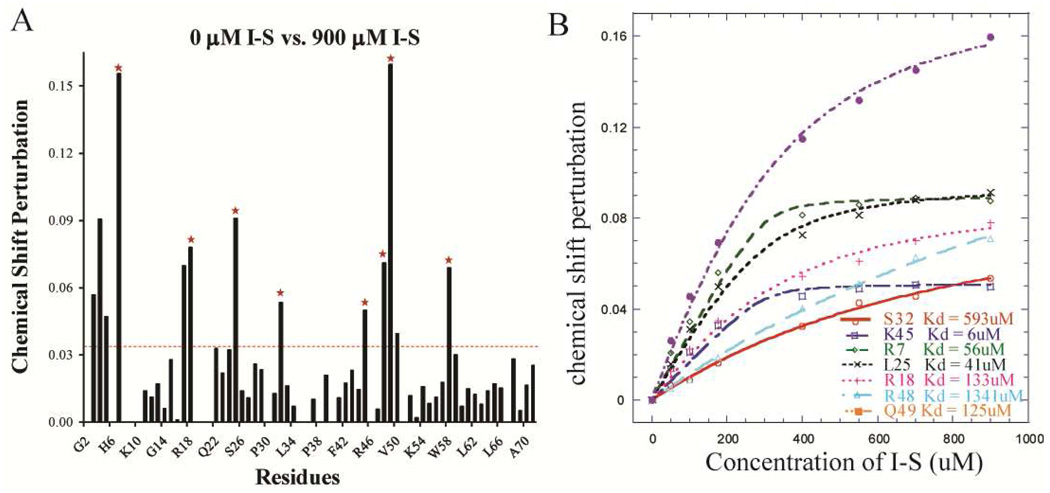
Since different quaternary states of vMIP-II clearly have different GAG binding ability, a NMR titration experiment was carried out to study the disaccharide binding properties of the vMIP-II L13F dimer. Heparin I-S was titrated to the vMIP-L13F protein solution, up to 900 µM (higher concentrations of I-S caused aggregation). The resulting chemical shift perturbation upon titration of heparin I-S with dimeric L13F is shown in Figure 3A. The backbone sequence assignment allows a residue-by-residue comparison between the wild type and dimeric variant upon GAG binding. Similar to the wild type protein, the important residues for heparin I-S binding on the L13F variant are also apparently located at R18 and the 40’s loop. The residues L25, V59, basic residues of the N-loop K17, R18, basic residues of the 40’s loop K45, R46, and R48 undergo the most spectral change upon heparin I-S binding. Large differences in chemical shift are also apparent at the N-terminus of the L13F mutant upon titration with disaccharide (Figure 3A), unlike the wild type protein, where the N-terminal peaks are either not significantly perturbed or not visible in the spectrum.
Figure 3.
A. Chemical shift perturbation chart of vMIP-II L13F titrated with heparin disaccharide I-S. The red dashed line indicates the average value of the chemical shift perturbation; the red stars indicate the residues that undergo the most significant chemical shift change and that do not overlap with other residues on the spectra. B. Heparin I-S binding affinity calculation for the residues labeled by red stars in A.
Modeling of disaccharide interaction with monomeric and dimeric vMIP-II
To further characterize and compare the GAG binding mechanisms of monomeric and dimeric vMIP-II, we used molecular docking calculations, using HADDOCK2.1 (59), a well-established method which uses experimental data as well as energetic and shape complementarity factors to drive molecular docking (60). Chemical-shift data were used to generate the ambiguous interaction restraints (AIRs) for the docking. HADDOCK calculations produced clusters of complex structures, and revealed interesting insights into the likely GAG binding mechanism of vMIP-II.
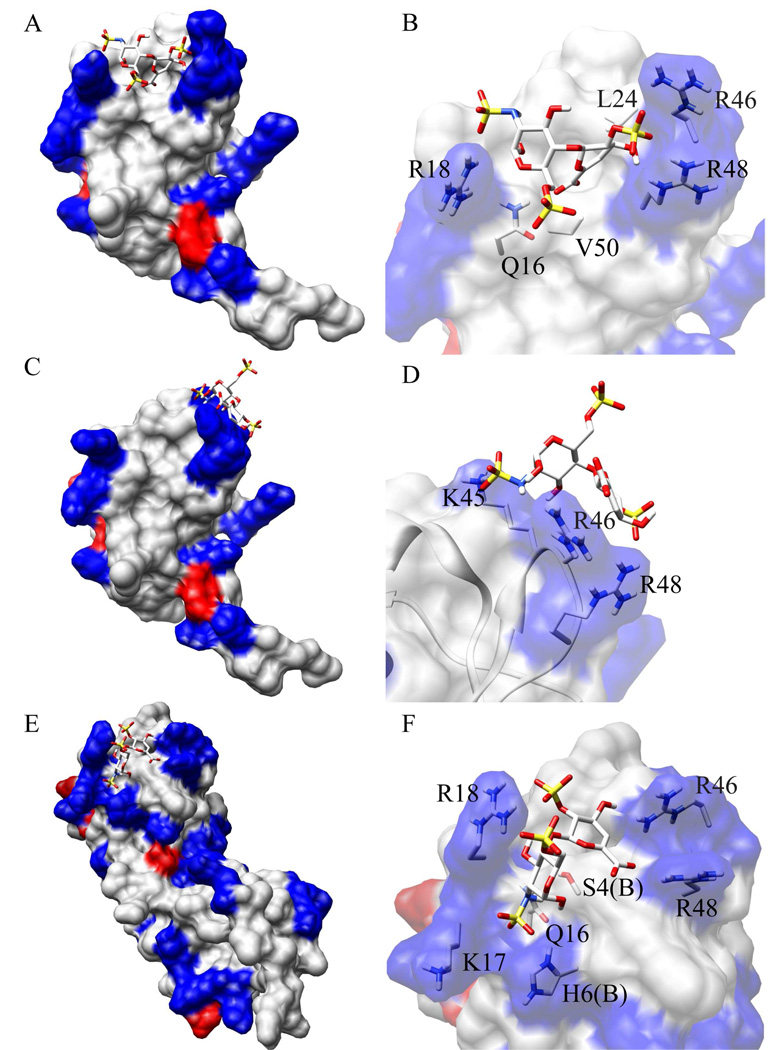
A representative structure of I-S binding to wild type monomeric vMIP-II (the best structure of the best ranking cluster, in terms of intermolecular binding energy, Table S1) is shown in Figure 4A and 4B, where the disaccharide fits in the binding pocket formed mainly by R46, R48 and R18. An overlay of 8 structures from the same cluster showed a similar binding site with slight variations on the orientation of I-S (Figure S1A). The binding site correlates very well with the NMR titration data (Figure S1C). Figure 4B shows the residues of vMIP-II that possibly contribute to I-S binding. As expected, electrostatic interactions between the negatively charged sulfate groups and the positively charged Arg residues play an important role. While NMR showed a large chemical shift change for L25, which is located in the middle between R46, R48 and R18, this residue is not exposed on the surface, so the significant chemical shift change is probably due to structural perturbations, not direct saccharide binding.
Figure 4.
HADDOCK model of the interaction of vMIP-II in complex with heparin disaccharide I-S. Positively charged and negatively charged surface regions of vMIP-II are shown in blue and red, respectively. For both vMIP-II and disaccharide, yellow, red, and blue colors represent sulfur atoms, oxygen atoms, and nitrogen atoms, respectively. A. A model of the interaction of monomeric vMIP-II (PDB code 1VMP) in complex with disaccharide I-S (from PDB 1U4L). A representative complex structure from Cluster 1 is shown. B. A close-up view of the binding site of A, with side chains of the important residues shown. C. An energetically less favorable model calculated by HADDOCK which represents a possible alternative mode for monomeric vMIP-II – disaccharide interaction. D. A close-up view of the binding site of C, with side chains of the important residues shown. E. HADDOCK model of interaction of dimeric vMIP-II (PDB code 1CM9) in complex with disaccharide I-S (from PDB 1U4L). F. A close-up view of the binding site of E, with side chains of the important residues shown.
The docking calculation also generated an energetically less favorable cluster, Cluster 2. A representative structure (Figure 4C, 4D) and the structure overlay (Figure S1B) shows that in this case IS binds on a different, nearby site of the protein, making major contact with K45, R46 and R48 (Figure 4D). The presence of this Cluster 2 may suggest a possible secondary binding site that may be used when multiple disaccharides or a long chain polysaccharide binds to vMIP-II.
When calculating disaccharide binding of the dimeric form of vMIP-II, the structures were closely clustered and generated only one binding model. Dimer clusters 1, 2, and 3 all showed very similar structures within 1 Å rmsd to each other, and in all structures the I-S binds in the middle of the pocket formed by R46, R48 and R18 of one subunit and the N-terminus of the other subunit (Figure 4E, 4F and S2A). The results also correlated well with the NMR data (Figure S2B). Figure 4F shows that in the bound dimer of vMIP-II, the N-terminus of the other subunit inserts into the binding cleft, and is clearly involved in I-S binding. The much lower energy terms compared to the monomer binding calculations also indicates that disaccharide binding to the dimer is more stable and tight, which is consistent with our experimental results such as heparin chromatography.
DISCUSSION
The importance of chemokine quaternary structure in GAG binding has been shown in many chemokine studies (10, 28, 31–32). The quaternary state of chemokines is determined by many factors. For instance, it has been shown that concentration, pH, and the presence of certain ions such as phosphate can affect the monomer-dimer equilibrium for chemokines (49, 61). It has also been shown that GAG binding promotes dimer formation of chemokines (10, 26, 32, 49, 56).
The viral chemokine homolog vMIP-II is observed as a monomer in solution under most conditions, including millimolar concentration, in the presence of phosphate, and at a variety of pH values, although under certain crystallization conditions, it has been observed as a CC-type dimer (33–37). In the present experiments, wild type vMIP-II is predominantly observed as a monomer in solution. But the dimeric form of vMIP-II may also play a role in its function as evidenced by its weak tendency to dimerize in analytical ultracentrifugation experiments at physiological pH, and also by the appearance of a small proportion of dimer peaks in the HSQC spectrum in the presence of 1500 µM of Heparin I-S disaccharide. To assist in understanding the GAG binding mechanism of vMIP-II, the ability to manipulate its quaternary state is important.
The point mutant L13F apparently transforms vMIP-II from a largely monomeric protein to a CC chemokine-type dimer. The NMR chemical shift changes upon L13F mutation are wholly consistent with expected changes based on the dimeric crystal structure of vMIP-II (36). In particular, residues undergoing the most significant chemical shift change include the N-terminus and N-loop, which are a critical part of the CC chemokine dimer interface, making contact with residues 50–52 and the 30's loop of the other subunit (respectively), all of which also exhibit chemical shift changes.
Many studies have been performed to drastically increase the dissociation of dimeric chemokines to form largely monomeric variants (28, 50, 58, 62–64), but vMIP-II L13F is the first dimer made from a predominantly monomeric chemokine by a single point mutation. With wild type vMIP-II being predominantly monomeric and vMIP-II L13F being predominantly dimeric throughout the NMR titration, we have an excellent model to study the role of dimerization in chemokine disaccharide interaction.
The NMR titration results provide an indication of which region is used by wild type vMIP-II to interact with GAGs (Figure 2B). Basic residues, in particular the N-loop residue R18 and the 40’s loop residues, play an important role in the interaction with the negatively charged sugar. Two hydrophobic residues, L25 and W58, also have resonances that shift significantly upon binding I-S, but it is likely that the chemical shift change is due to overall structural change propagation rather than being involved in GAG binding directly, since L25 is an internal residue located in the middle of the groove formed by R46, R48 and R18, and W58 is located outside of this groove with the bulky hydrophobic side chain pointing near the backbone of R18.
To investigate how vMIP-II interacts with longer polysaccharides, heparin chromatography was used. Variants R18A, R46A, and R48A showed the most significant decrease in heparin binding affinity which is consistent with NMR titration results indicating these residues form a heparin binding site, similar to human chemokines MIP-1β and RANTES (10, 29, 57). Perhaps surprisingly, the vMIP-II variant K45A did not show a great change in heparin binding affinity, although the residue is part of the so-called BBXB GAG binding domain, and significant change was observed for the nearby single mutants R46A and R48A. Further, the double mutant R46A/R48A showed almost the same loss of affinity for the heparin column as the triple mutant K45A/R46A/R48A, again indicating that K45A is not particularly important in GAG binding. This was not expected since the NMR titration in the presence of the disaccharide heparin I-S, showed large movement of this residue. In similar studies with MIP-1β, it was found that K45A, and R46A mutations caused significant decrease in NaCl elution concentration in heparin column binding, while in that case K48A mutation had no effect (10). While this may suggest a different binding mode between vMIP-II and MIP-1β, examination of both of their structures clarifies the issue. Both proteins have basic residues at positions 18, 45, 46, and 48 (the latter three being the BBXB motif). However, the surface charge figure of vMIP-II (Figure 2D) shows that R18, R46 and R48 form a clear binding surface while the residue K45 is located away from the surface. In contrast, the dimeric MIP-1β has a clear binding surface comprised of R18, K45 and R46 (28, 65). These findings indicate that GAG binding on chemokines is site-specific, and vMIP-II likely binds GAGs using its positively charged surface formed by R18, R46, and R48, which is shifted from the binding site of MIP-1β which primarily uses R18, K45 and R46.
Interestingly, as opposed to the patch of basic residues encompassed by R18, R46 and R48, the other face of vMIP-II, which also has many basic residues (Figure 2D), does not appear to be involved in high affinity GAG binding. Basic residues K37, K54, K56, K60 and K61 do not show significant chemical shift change upon titration with I-S, suggesting that these basic residues may not be directly involved in disaccharide binding. Further, when mutation was made to K54, this variant showed similar GAG binding affinity as wild type vMIP-II in heparin chromatography (Table 1). This indicates that vMIP-II binds GAGs fairly specifically at one site, encompassed by residues R18, R46, and R48, despite being overall a highly basic protein with many other positive charges throughout its surface. Basic residues on the “back” side of vMIP-II do not appear to contribute to high affinity GAG binding as much as the residues from the binding site do, but overall they may help maintain a basal level of electrostatic attraction to the GAG target. Mutating the whole BBXB domain in vMIP-II with the triple mutant K45A/R46A/R48A causes a significant drop in heparin binding affinity, but does not totally abolish GAG binding as the analogous mutant does in MIP-1β (10). This may be similar to the very basic chemokine RANTES, where mutating the BBXB domain significantly disrupts binding to heparin column but does not abolish binding completely (57).
Molecular docking based on our experimental data gave us a more comprehensive understanding of the disaccharide binding mechanism of vMIP-II. As is shown in Figure 4A and 4B, the docking model predicts that the groove encompassed by positively charged R46, R48 and R18 is the energetically favored binding site for disaccharide. Another set of less favorable complex structures (Cluster 2, Figure 4C and 4D) suggested a possible secondary binding site for disaccharide when multiple disaccharides or a long chain polysaccharide binds to vMIP-II. This is supported by the heparin chromatography data of R18A variant which showed 2 elution peaks. When R18 is mutated to alanine, the original binding pocket may become less favorable, and a saccharide could then bind to multiple sites. No calculated structure shows binding to the positively charged residues on the “back” side of vMIP-II, indicating that this is not a favorable disaccharide binding site, which is also consistent with our NMR and mutagenesis data. Possibly, the “back” side is unfavorable because of the interference of the negatively charged D57 residue located in the middle of the patch, or perhaps the shape of the pocket is not energetically favorable.
With the NMR sequence assignment of both the monomeric and dimeric forms of vMIP-II, NMR titration allows a residue-specific comparison between their mechanisms of GAG binding. The chemical shift perturbation charts show that while many features of the overall binding site appear to be the same for the monomer and dimer forms of vMIP-II, differences are observed, particularly at the N-terminal region. In the dimeric L13F variant, the N-terminal residues undergo much larger chemical shift changes, indicating that the N-terminal residues are involved in disaccharide binding in the dimeric form of the protein. In the CC-type chemokine dimer structure, the N-terminus of one subunit is in close proximity to the conserved basic residues on other subunit, likely forming part of the saccharide binding site. A calculated docking model (Figure 4E and 4F) confirmed this hypothesis, predicting that disaccharide binds to dimeric vMIP-II (PDB 1CM9) at the new binding pocket encompassed by basic residues R46, R48, R18, and several N-terminal residues from the other subunit. The biggest difference compared to the monomer model is the involvement of the N-terminus from the other subunit which is consistent with our NMR data. Compared to the monomer disaccharide binding model which has two major binding conformation and higher energy terms, the dimer vMIP-II – disaccharide interaction is more specific and energetically more favorable. This is consistent with the heparin chromatography data showing that vMIP-II L13F binds to the heparin column more tightly than wild type vMIP-II.
Previous GAG binding studies on vMIP-II’s human analogs MIP-1β and RANTES have shown similar binding mechanisms to what we have shown for vMIP-II. For MIP-1β, basic residues of the BBXB domain and R18 form the principal binding pocket, and N-terminal residues are also shown to be involved in GAG binding for this dimeric chemokine (28). For RANTES, positively charged residues of the BBXB domain, but not the ones on the other side of the protein, are responsible for GAG binding (57). An X-ray crystal structure of RANTES in complex with disaccharide confirmed the role of the BBXB motif in GAG binding, but also showed unexpected disaccharide binding in the N-loop and 30s loop which was not easily explainable from previous studies (29). Generally, we can conclude that the viral chemokine vMIP-II binds GAGs using a similar mechanism as its human CC chemokines analogs. This could provide an explanation for its natural function: This viral chemokine that antagonizes many chemokine receptors could build up its own concentration gradient upon viral infection (as the human chemokines do), by binding to endothelial surface GAGs. vMIP-II could be protected from proteolysis and could interact with human cell surface receptors more efficiently, and carry out its anti-inflammatory function. Since vMIP-II tends to bind to GAGs more tightly than many human chemokines, it might also disrupt the concentration gradient of human chemokines by displacing them from GAGs on the endothelial surface.
The present study also raises certain questions: assuming the viral chemokine vMIP-II evolved from a human chemokine ancestor, why did it adopt a Leu at the 13th position which resulted in weaker dimerization and GAG binding, instead of Phe, as its human analogs do? A functional comparison between vMIP-II and human CC-chemokine analog MIP-1β suggests a possible explanation. Unlike MIP-1β, which requires a Phe in the 13th position for both receptor binding and GAG binding (10, 50), our preliminary results (manuscript in preparation) show that although the L13F mutation does significantly enhance the heparin binding ability, it does not change the affinity of vMIP-II for cell surface receptors CCR5 or CXCR4. So for receptor binding, L13 and F13 are equivalent for vMIP-II, and it has apparently achieved promiscuous receptor binding without Phe13. Regarding GAG binding, although the lack of Phe residue at the 13th position leads to a predominantly monomeric form, vMIP-II is still able to tightly bind GAGs, more tightly than the dimeric human chemokine MIP-1β. Considering vMIP-II’s natural function, which is apparently to subvert the human immune system and antagonize multiple chemokine receptors, it is possible that binding GAGs more tightly (as does the Phe13-containing dimer form) would hinder its anti-inflammatory function, perhaps by decreasing its mobility.
In conclusion, NMR, mutagenesis and molecular docking were carried out to elucidate the structural features of GAG binding for the anti-inflammatory viral chemokine homolog vMIP-II. These experiments show that vMIP-II tightly binds glycosaminoglycans using residues distributed along only one face of the protein, such as R18, R46 and R48, even though the protein is highly basic throughout its surface. This study also provides a valuable model to study the role of chemokine quaternary state in GAG binding. With the wild type vMIP-II being predominantly monomeric, and the point mutant vMIP-II L13F being predominantly dimeric throughout the NMR titration experiments, we were able to analyze the difference in GAG binding mechanisms between the two quaternary states at the atomic level. NMR studies reveal a shift in the GAG binding site between monomer and dimer, where the N-terminus is involved in GAG binding for the dimeric form. Docking models confirmed this finding and suggested that GAG binding to the dimer form of vMIP-II is energetically more favorable. The tighter GAG binding of the vMIP-II L13F dimer also reinforces past results suggesting that the major physiologic role of the chemokine dimer may be to bind to GAGs. The results presented here clarify several aspects of the biochemistry of vMIP-II, suggest possibilities for how this viral chemokine analog may function, and shed light into understanding the mechanism of how quaternary structure affects GAG binding.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr. Amanda Jacks and Michael Pirics for synthesizing the vMIP-II gene; Dr. Yongguang Gao, Dr. Hongjun Jin, Dr. Giannis Kagiampakis, and Dr. Xiangming Kong for technical assistance.
Funding was provided by the American Heart Association Texas Affiliate 0655070Y, and NIH R21AI079777.
Abbreviations
- vMIP
viral macrophage inflammatory protein. The international standardized name of vMIP-II is vCCL2
- IPTG
isopropyl R-D-thiogalactopyranoside
- CCR
CC chemokine receptor
- CXCR
CXC chemokine receptor
- GAG
glycosaminoglycan
- NMR
nuclear magnetic resonance
- HSQC
heteronuclear single-quantum coherence
- DSS
4,4-dimethyl-4-silapentane-1-sulfonate
- MW
molecular weight
- WT
wild type
- BBXB
a part of the 40's loop of chemokines, for "Basic-Basic-Any-Basic" amino acid
- PSI
pounds per square inch
- SW
sweep width
- AIR
ambiguous interaction restraint
- HADDOCK
High ambiguity driven biomolecular docking
- RMSD
root-mean-square deviation
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental materials may be accessed free of charge at online at http://pubs.acs.org. Please refer to the separate supplemental file.
REFERENCES
- 1.Kledal TN, Rosenkilde MM, Coulin F, Simmons G, Johnsen AH, Alouani S, Power CA, Luttichau HR, Gerstoft J, Clapham PR, Clark-Lewis I, Wells TNC, Schwartz TW. A broad-spectrum chemokine antagonist encoded by Kaposi's Sarcoma-associated herpesvirus. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 2.Sozzani S, Luini W, Bianchi G, Allavena P, Wells TN, Napolitano M, Bernardini G, Vecchi A, D'Ambrosio D, Mazzeo D, Sinigaglia F, Santoni A, Maggi E, Romagnani S, Mantovani A. The viral chemokine macrophage inflammatory protein-II is a selective Th2 chemoattractant. Blood. 1998;92:4036–4039. [PubMed] [Google Scholar]
- 3.Weber KS, Grone HJ, Rocken M, Klier C, Gu S, Wank R, Proudfoot AE, Nelson PJ, Weber C. Selective recruitment of Th2-type cells and evasion from a cytotoxic immune response mediated by viral macrophage inhibitory protein-II. European Journal of Immunology. 2001;31:2458–2466. doi: 10.1002/1521-4141(200108)31:8<2458::aid-immu2458>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 4.Takami S, Minami M, Nagata I, Namura S, Satoh M. Chemokine receptor antagonist peptide, viral MIP-II, protects the brain against focal cerebral ischemia in mice. Journal of Cerebral Blood Flow & Metabolism. 2001;21:1430–1435. doi: 10.1097/00004647-200112000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Ghirnikar RS, Lee YL, Eng LF. Chemokine antagonist infusion promotes axonal sparing after spinal cord contusion injury in rat. Journal of Neuroscience Research. 2001;64:582–589. doi: 10.1002/jnr.1110. [DOI] [PubMed] [Google Scholar]
- 6.DeBruyne LA, Li K, Bishop DK, Bromberg JS. Gene transfer of virally encoded chemokine antagonists vMIP-II and MC148 prolongs cardiac allograft survival and inhibits donor-specific immunity. Gene Therapy. 2000;7:575–582. doi: 10.1038/sj.gt.3301128. [DOI] [PubMed] [Google Scholar]
- 7.Pillai RG, Beutelspacher SC, Larkin DFP, George AJT. Expression of the chemokine antagonist vMIP II using a non-viral vector can prolong corneal allograft survival. Tranplantation. 2008;85:1640–1647. doi: 10.1097/TP.0b013e318172813f. [DOI] [PubMed] [Google Scholar]
- 8.Cherqui SK, Kenneth M, Thorpe Camille, Kurian Sunil M, Salomon Daniel R. Lentiviral Gene Delivery of vMIP-II to Transplanted Endothelial Cells and Endothelial Progenitors Is Proangiogenic In Vivo. Molecular Therapy. 2007;15:1264–1272. doi: 10.1038/sj.mt.6300183. [DOI] [PubMed] [Google Scholar]
- 9.Ali S, Palmer ACV, Banerjee B, Fritchley SJ, Kirby JA. Examination of the function of RANTES, MIP-1α, and MIP-1β following interaction with heparin-like glycosaminoglycans. J. Biol. Chem. 2000;275:11721–11727. doi: 10.1074/jbc.275.16.11721. [DOI] [PubMed] [Google Scholar]
- 10.Laurence JS, Blanpain C, De Leener A, Parmentier M, LiWang PJ. The Importance of Basic Residues and Quaternary Structure in the Function of MIP-1β: CCR5 Binding and Cell Surface Sugar Interactions. Biochemistry. 2001;40:4990–4999. doi: 10.1021/bi002593w. [DOI] [PubMed] [Google Scholar]
- 11.Valenzuela-Fernandez A, Palanche T, Amara A, Magerus A, Altmeyer R, Delaunay T, Virelizier JL, Baleux F, Galzi JL, Arenzana-Seisdedos F. Optimal inhibition of X4 HIV isolates by the CXC chemokine stromal cell-derived factor 1 alpha requires interaction with cell surface heparan sulfate proteoglycans. Journal of Biological Chemistry. 2001;276:26550–26558. doi: 10.1074/jbc.M100411200. [DOI] [PubMed] [Google Scholar]
- 12.Amara A LO, Valenzuela A, Magerus A, Thelen M, Montes M, Virelizier JL, Delepierre M, Baleux F, Lortat-Jacob H, Arenzana-Seisdedos F. Stromal cell-derived factor-1alpha associates with heparan sulfates through the first beta-strand of the chemokine. J Biol Chem. 2001;274:23916–23925. doi: 10.1074/jbc.274.34.23916. [DOI] [PubMed] [Google Scholar]
- 13.Proudfoot AE, Handel TM, Johnson Z, Lau EK, LiWang P, Clark-Lewis I, Borlat F, Wells TN, Kosco-Vilbois MH. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc. Natl. Acad. Sci. USA. 2003;100:1885–1890. doi: 10.1073/pnas.0334864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuinstra RL, Peterson FC, Kutlesa S, Elgin ES, Kron MA, Volkman BF. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc Natl Acad Sci U S A. 2008;105:5057–5062. doi: 10.1073/pnas.0709518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 16.Ley K. Arrest Chemokines. Microcirculation. 2003;10:289–295. doi: 10.1038/sj.mn.7800194. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat. Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 18.Lau EK, Allen S, Hsu AR, Handel TM. Chemokine-receptor interactions: GPCRs, glycosaminoglycans and viral chemokine binding proteins. Advances in Protein Chemistry. 2004;68:351–391. doi: 10.1016/S0065-3233(04)68010-7. [DOI] [PubMed] [Google Scholar]
- 19.Yoshie O, Imai T, Nomiyama H. Chemokines in immunity. Adv. Immunol. 2001;78:57–110. doi: 10.1016/s0065-2776(01)78002-9. [DOI] [PubMed] [Google Scholar]
- 20.Rot A. Neutrophil attractant/activation protein-1 (interleukin-8) induces in vitro neutrophil migration by haptotactic mechanism. Eur. J. Immunol. 1993;23:303–306. doi: 10.1002/eji.1830230150. [DOI] [PubMed] [Google Scholar]
- 21.Wiedermann CJ, Kowald E, Reinisch N, Kaehler CM, von Luettichau I, Pattison JM, Huie P, Sibley RK, Nelson PJ, Krensky AM. Monocyte haptotaxis induced by the RANTES chemokine. Curr. Biol. 1993;3:735–739. doi: 10.1016/0960-9822(93)90020-o. [DOI] [PubMed] [Google Scholar]
- 22.Middleton J, Patterson AM, Gardner L, Schmutz C, Ashton BA. Leukocyte extravasation: chemokine transport and presentation by the endothelium. Blood. 2002;100:3853–3860. doi: 10.1182/blood.V100.12.3853. [DOI] [PubMed] [Google Scholar]
- 23.Parish CR. The role of heparan sulphate in inflammation. Nat. Rev. Immunol. 2006;6:633–643. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- 24.Vives RR, Imberty A, Sattentau QJ, Lortat-Jacob H. Heparan sulfate targets the HIV-1 envelope glycoprotein gp120 coreceptor binding site. Journal of Biological Chemistry. 2005;280:21353–21357. doi: 10.1074/jbc.M500911200. [DOI] [PubMed] [Google Scholar]
- 25.Webb LM, Ehrengruber MU, Clark-Lewis I, Baggiolini M, Rot A. Binding to heparan sulfate or heparin enhances neutrophil responses to interleukin-8. Proc. Natl. Acad. Sci. USA. 1993;90:7158–7162. doi: 10.1073/pnas.90.15.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoogewerf AJ, Kuschert GS, Proudfoot AE, Bortat F, Clark-Lewis I, Power CA, Wells TN. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 27.Park PW, Reizes O, Bernfield M. Cell surface heparan sulfate proteoglycans: selective regulators of ligand-receptor encounters. J Biol Chem. 2000;275:29923–29926. doi: 10.1074/jbc.R000008200. [DOI] [PubMed] [Google Scholar]
- 28.McCornack MA, Cassidy CK, LiWang PJ. The binding surface and affinity of monomeric and dimeric chemokine MIP-1β for various glycosaminoglycan disaccharides. Journal of Biological Chemistry. 2003;278:1946–1956. doi: 10.1074/jbc.M207440200. [DOI] [PubMed] [Google Scholar]
- 29.Shaw JP, Johnson Z, Borlat F, Zwahlen C, Kungl A, Roulin K, Harrenga A, Wells TN, Proudfoot AE. The X-ray structure of RANTES: heparin-derived disaccharides allows the rational design of chemokine inhibitors. Structure. 2004;12:2081–2093. doi: 10.1016/j.str.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 30.Lau EK, Paavola CD, Johnson Z, Gaudry JP, Geretti E, Borlat F, Kungl AJ, Proudfoot AE, Handel TM. Identification of the glycosaminoglycan binding site of the CC chemokine, MCP-1: implications for structure and function in vivo. Journal of Biological Chemistry. 2004;279:22294–22305. doi: 10.1074/jbc.M311224200. [DOI] [PubMed] [Google Scholar]
- 31.Jin H, Shen X, Baggett B, Kong X, LiWang P. The human CC chemokine MIP-1b dimer is not competent to bind the CCR5 receptor. Journal of Biological Chemistry. 2007;282:27976–27983. doi: 10.1074/jbc.M702654200. [DOI] [PubMed] [Google Scholar]
- 32.McCornack MA, Boren DM, LiWang PJ. Glycosaminoglycan disaccharide alters the dimer dissociation constant of the chemokine MIP-1 beta. Biochemistry. 2004;43:10090–10101. doi: 10.1021/bi049751u. [DOI] [PubMed] [Google Scholar]
- 33.LiWang AC, Wang ZX, Sun Y, Peiper SC, LiWang PJ. The solution structure of the anti-HIV chemokine vMIP-II. Protein Science. 1999;8:2270–2279. doi: 10.1110/ps.8.11.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao W, Fernandez E, Wilken J, Thompson DA, Siani MA, West J, Lolis E, Schweitzer BI. Accessibility of selenomethionine proteins by total chemical synthesis: structural studies of human herpesvirus-8 MIP-II. FEBS Letters. 1998;441:77–82. doi: 10.1016/s0014-5793(98)01520-8. [DOI] [PubMed] [Google Scholar]
- 35.Shao W, Fernandez E, Sachpatzidis A, Wilken J, Thompson DA, Schweitzer BI, and Lolis E. CCR2 and CCR5 receptor-binding properties of herpesvirus-8 vMIP-II based on sequence analysis and its solution structure. European Journal of Biochemistry. 2001;268:2948–2959. doi: 10.1046/j.1432-1327.2001.02184.x. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez EJ, Wilken J, Thompson DA, Peiper SC, Lolis E. Comparison of the structure of vMIP-II with eotaxin-1, RANTES, and MCP-3 suggests a unique mechanism for CCR3 activation. Biochemistry. 2000;39:12837–12844. doi: 10.1021/bi001166f. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Liu D, Cao R, Kumar S, Dong C, An J, Wilson SR, Gao YG, Huang Z. Crystal structure of chemically synthesized vMIP-II. Proteins. 2007;67:243–246. doi: 10.1002/prot.21172. [DOI] [PubMed] [Google Scholar]
- 38.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C, 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 39.Delaglio F, Grzesiek S, Vuister GW, Hu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Garrett DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Garret DS, Seok Y, Peterkofsky A, Clore GM, Gronenborn AM. Identification by NMR of the Binding Surface for the Histidine-Containing Phosphocarrier Protein HPr on the N-Terminal Domain of Enzyme I of the Escherichia coli Phosphotransferase System. Biochemistry. 1997;36:4393–4398. doi: 10.1021/bi970221q. [DOI] [PubMed] [Google Scholar]
- 42.Mayer KL, Stone MJ. NMR solution structure and receptor peptide binding of the CC chemokine eotaxin-2. Biochemistry. 2000;39:8382–8395. doi: 10.1021/bi000523j. [DOI] [PubMed] [Google Scholar]
- 43.Kouchakdjian EM, Johnson F, Grollman AP, Patel DJ. Structural features of an exocyclic adduct positioned opposite an abasic site in a DNA duplex. Biochemistry. 1991;30:3262–3270. doi: 10.1021/bi00227a014. [DOI] [PubMed] [Google Scholar]
- 44.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophysical Journal. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Correia JJ, Chacko BM, Lam SS, Lin K. Sedimentation Studies Reveal a Direct Role of Phosphorylation in Smad3:Smad4 Homo- and Hetero-Trimerization. Biochemistry. 2001;40:1473–1482. doi: 10.1021/bi0019343. [DOI] [PubMed] [Google Scholar]
- 46.Nilges M, O'Donoghue SI. Ambiguous NOEs and automated NOE assignment. Progress in Nuclear Magnetic Resonance Spectroscopy. 1998;32:107–139. [Google Scholar]
- 47.Schüttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta crystallographica. Section D, Biological crystallography. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 48.Daura X, Gademann K, Jaun B, Seebach D, Van Gunsteren WF, Mark AE. Peptide Folding: When Simulation Meets Experiment. Angewandte Chemie International Edition. 1999;38:236–240. [Google Scholar]
- 49.Veldkamp CT, Peterson FC, Pelzek AJ, Volkman BF. The monomer-dimer equilibrium of stromal cell-derived factor-1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Science. 2005;14:1071–1081. doi: 10.1110/ps.041219505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laurence JS, Blanpain C, Parmentier M, Burgner JW, LiWang PJ. The CC chemokine MIP-1β can function as a monomer and depends on Phe13 for receptor binding. Biochemistry. 2000;39:3401–3409. doi: 10.1021/bi9923196. [DOI] [PubMed] [Google Scholar]
- 51.Hoover DMBC, Yang D, Oppenheim JJ, Tucker K, Lu W, Lubkowski J. The structure of human macrophage inflammatory protein-3alpha /CCL20. Linking antimicrobial and CC chemokine receptor-6-binding activities with human beta-defensins. J Biol Chem. 2002;277:37647–37654. doi: 10.1074/jbc.M203907200. [DOI] [PubMed] [Google Scholar]
- 52.Lortat-Jacob H GA, Imberty A. Structural diversity of heparan sulfate binding domains in chemokines. Proc Natl Acad Sci U S A. 2002;99:1229–1234. doi: 10.1073/pnas.032497699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crump MP, Gong J-H, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier J-L, Baggiolini M, Sykes B, Clark-Lewis I. Solution structure and the basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shao W, Jerva LF, West J, Lolis E, Schweitzer BI. Solution structure of murine macrophage inflammatory protein-2. Biochemistry. 1998;37:8303–8313. doi: 10.1021/bi980112r. [DOI] [PubMed] [Google Scholar]
- 55.Handel TM, Johnson Z, Crown SE, Lau EK, Proudfoot AE. Regulation of protein function by glycosaminoglycans--as exemplified by chemokines. Annual Review of Biochemistry. 2005;74:385–410. doi: 10.1146/annurev.biochem.72.121801.161747. [DOI] [PubMed] [Google Scholar]
- 56.Kuschert GS, Coulin F, Power CA, Proudfoot AE, Hubbard RE, Hoogewerf AJ, Wells TN. Glycosaminoglycans interact selectively with chemokines and modulate receptor binding and cellular responses. Biochemistry. 1999;38:12959–12968. doi: 10.1021/bi990711d. [DOI] [PubMed] [Google Scholar]
- 57.Proudfoot AEI, Fritchley S, Borlat F, Shaw JP, Vilbois F, Zwahlen C, Trkola A, Marchant D, Clapham PR, Wells TNC. THe BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J. Biol. Chem. 2001;276:10620–10626. doi: 10.1074/jbc.M010867200. [DOI] [PubMed] [Google Scholar]
- 58.Paavola CD, Hemmerich S, Grunberger D, Polsky I, Bloom A, Freedman R, Mulkins M, Bhakta S, McCarley D, Wiesent L, Wong B, Jarnagin K, Handel TM. Monomeric monocyte chemoattractant protein-1 (MCP-1) binds and activates the MCP-1 receptor CCR2B. J. Biol. Chem. 1998;273:33157–33165. doi: 10.1074/jbc.273.50.33157. [DOI] [PubMed] [Google Scholar]
- 59.Dominiguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. Journal of American Chemistry Society. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 60.Schieborr U, Vogtherr M, Elshorst B, Betz M, Grimme S, Pescatore B, Langer T, Saxena K, Schwalbe H. How much NMR data is required to determine a protein-ligand complex structure? Chembiochem. 2005;6:1891–1898. doi: 10.1002/cbic.200500092. [DOI] [PubMed] [Google Scholar]
- 61.Crump MP, Rajarathnam K, Kim K-S, Clark-Lewis I, Sykes BD. Solution structure of eotaxin, a chemokine that selectively recruits eosinophils in allergic inflammation. J. Biol. Chem. 1998;273:22471–22479. doi: 10.1074/jbc.273.35.22471. [DOI] [PubMed] [Google Scholar]
- 62.Lowman HB, Fairbrother WJ, Slagle PH, Kabakoff R, Liu J, Shire S, Hebert CA. Monomeric variants of IL-8: Effects of side chain substitutions and solution conditions upon dimer formation. Protein Sci. 1997;6:598–608. doi: 10.1002/pro.5560060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim S, Jao S-c, Laurence JS, LiWang PJ. Structural comparison of monomeric variants of the chemokine MIP-1b having differing ability to bind the receptor CCR5. Biochemistry. 2001;40:10782–10791. doi: 10.1021/bi011065x. [DOI] [PubMed] [Google Scholar]
- 64.Kim K-S, Rajarathnam K, Clark-Lewis I, Sykes BD. Structural characterization of a monomeric chemokine:Monocyte chemoattractant protein-3. FEBS Lett. 1996;395:277–282. doi: 10.1016/0014-5793(96)01024-1. [DOI] [PubMed] [Google Scholar]
- 65.Lodi PJ, Garrett DS, Kuszewski J, Tsang ML-S, Weatherbee JA, Leonard WJ, Gronenborn AM, Clore GM. High-resolution solution structure of the β chemokine hMIP-1β by multidimensional NMR. Science. 1994;263:1762–1767. doi: 10.1126/science.8134838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.