1. Tissue engineering and biomimetic strategies overview
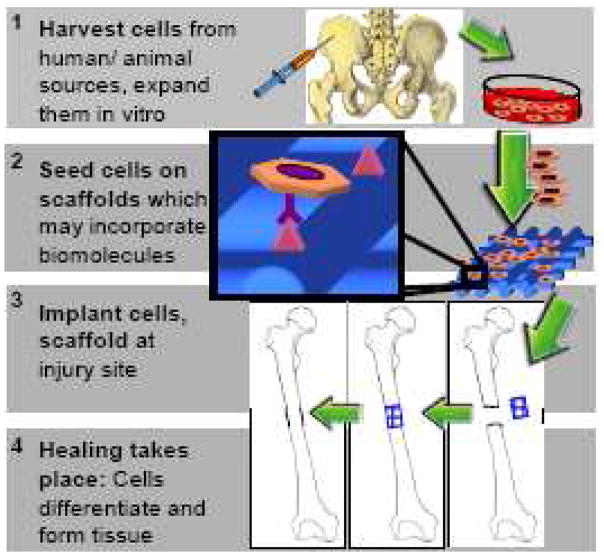
The main goal of tissue engineering and regenerative medicine strategies is to restore the function of damaged tissues by delivering a combination of cells, biological factors and a biomaterial scaffold on which these cells must adhere, organize and develop similarly to native tissue (Fig 1). In vivo, cell fates are determined by a complex interaction of nanoscale physical and chemical signals. Therefore, scaffolds for tissue engineering often incorporate biosignals to create a controlled, bioinspired extracellular environment to direct tissue-specific cell responses. The intention is that when presented with appropriate biological cues, cell receptors will bind to these signaling biomolecules and transmit the signals intracellularly by activating signaling cascades. These cascades will modulate gene expression and determine important cell fate processes such as differentiation to ultimately regenerate functioning tissue. As nanotechnology can recapitulate the submicron-scale spatial orientation of extracellular signaling molecules, it may be a powerful tool for enhancing cell-biomaterial communication and inducing desired cell behaviors.
Fig 1.
Tissue engineering paradigm.
2. Cellular interactions with extracellular environment
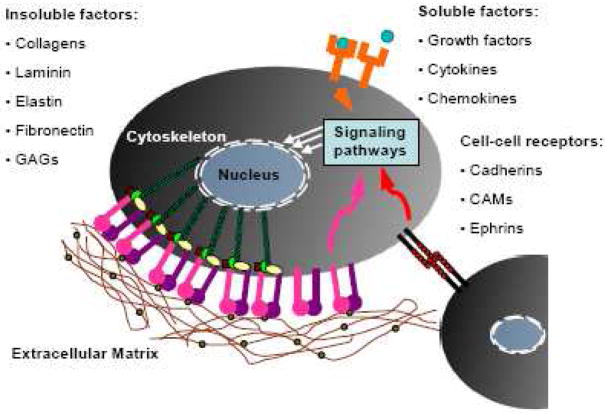
Signals from the extracellular microenvironment that may be incorporated into biomaterials (Fig 2) fall into three major categories: (1) insoluble extracellular matrix (ECM) macromolecules, (2) diffusible/soluble molecules, and (3) cell-cell receptors.
Fig 2.
Bioactive signals found within the extracellular environment.
2.1 ECM
2.1.1 ECM structure and function in vivo
There is a great diversity of insoluble ECM molecules including structural proteins such as collagens, elastin and laminin, glycoproteins such as fibronectin and vitronectin, as well as glycosaminoglycans such as chondroitin sulfate [1]. In vivo, these secreted ECM proteins form a meshwork of fibers or fibrils with ECM glycoproteins incorporated into them. The resulting matrix functions as both a structural and signaling scaffold to cells. ECM composition, immobilization and spatial arrangement varies for each tissue type. For example, bone ECM consists mostly of collagen I [2], mineral and non-collagenous proteins such as osteocalcin, fibronectin and vitronectin [3]. However, cartilage ECM is predominantly composed of collagen II and aggrecans [4]. This tissue-specific difference in ECM composition may be instructive to tissue engineering because different ECM macromolecules regulate cell growth and differentiation by selectively stimulating different signaling pathways through ECM interactions with various cell receptors [5].
2.1.2 Cell-ECM Interaction - Integrins
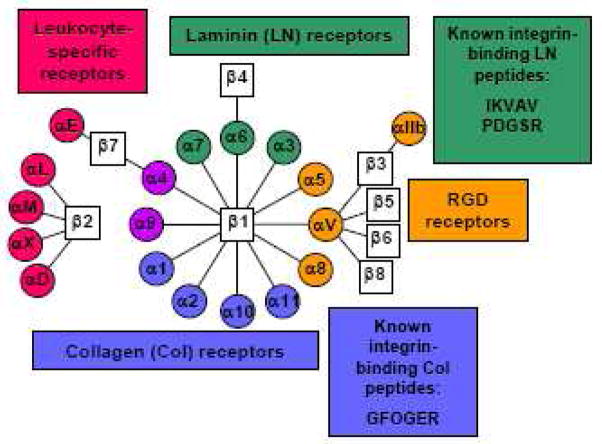
Transmission of chemical and mechanical signals from the ECM is primarily mediated by integrins. Integrins are a family of cell-surface transmembrane receptors, each of which consists of α and β subunits. So far, 8 β and 18 α integrin subunits have been found. These integrin subunits associate to form 24 distinct αβ combinations, and each of these integrins has unique binding characteristics [6] (Fig. 3). Most integrins bind to several types of ECM molecules and conversely, most ECM bind to more than one integrin. Integrins also can also undergo bidirectional signaling. That is, when ECM binds to the extracellular domain of integrins, it activates intracellular signaling (outside-in). Conversely, intracellular signaling can affect the conformation of an integrin, which modulates its affinity to its ligand (inside-out) [7].
Fig 3.
Integrin alpha and beta subunit combinations and binding specificity. Adapted from [6].
Both α and β integrin subunits pass through the cell membrane once and have large 700–1100 residue extracellular domains and small 30-50 residue cytoplasmic domains. The extracellular domains of integrins serve to recognize and bind ECM. Upon ECM binding, integrins cluster and their cytoplasmic domains associate with both cytoskeletal and intracellular signal transduction molecules. The association of integrins with the cellular signaling network initiates downstream signaling cascades such as the protein kinase C, Rac, Rho and MAPK pathways. The coordinated clustering of ECM ligands, integrins and cytoskeletal components forms macromolecular aggregates known as focal adhesions on the inside and outside of the cell membrane [8]. These integrin-ECM interactions govern cell survival, growth, migration and differentiation [7, 9, 10] and are therefore useful targets of biomimetic tissue engineering strategies. Furthermore, because focal adhesions occur on submicron to nanometer size scales [11] and integrins are approximately 10 nm in diameter [12] and have 20 nm long extracellular domains [13, 14], integrin-ECM based biomaterial strategies are especially relevant applications for nanofabrication and nanopatterning technologies.
2.1.3 Integrin-binding adhesive peptide sequences within ECM
Although ECM macromolecules such as collagens and fibronectin have long protein backbones consisting of thousands of amino acids, integrins recognize and bind to only a few short peptide sequences within the ECM molecules, triggering cell adhesion, signaling and spreading. In collagens I, II and III, cells bind to the GFOGER [15, 16] peptide sequence, while in fibronectin, the RGD [17], PHSRN [18], REDV[19], and LDV [20] sequences are responsible for cell binding. Recognition sequences within laminin include RGD, as well as IKVAV [21], YIGSR [22] and PDSGR [23] (Fig 3).
2.1.4 Biomaterial strategies utilizing ECM-derived adhesive peptides
Integrin interactions with ECM peptide ligands trigger complex signaling pathways which regulate crucial cell behaviors such as proliferation and differentiation as well as tissue-level responses such as morphogenesis, homeostasis and regeneration [6]. Therefore, coatings of ECM macromolecules such as collagen and laminin or their recognition peptides such as RGD or IKVAV have been used to biofunctionalize surfaces or implants and drive tissue-specific cell responses. Although naturally derived ECM molecules have proved fairly successful in some applications (for a review, see [24]), extracting and purifying matrix polymers in large scale is challenging, and animal-derived ECM may elicit an immune response. Furthermore, natural ECM biomaterials are difficult to modify, characterize and control. These limitations have driven the need for synthetic non-fouling materials functionalized with ECM-derived peptides [25] which are easily synthesized, immobilized, may be presented at unnaturally high densities, and may be tailored in composition for each tissue-specific application. Although there are many ECM-derived cell-binding motifs, most bioadhesive tissue engineering strategies have been restricted to modifying materials with RGD, GFOGER, IKVAV and YIGSR and PHSRN. Of these studies, the majority have focused on RGD due to its status as a universal and ‘promiscuous’ adhesion peptide which is found in numerous ECM molecules and binds to multiple integrins (αvβ3, α5β1, αvβ1, α8β1, αvβ8, αvβ6, αvβ5 and αIIbβ3) [6].
2.2 Diffusible/Soluble signals
Besides a broad host of ECM-mimetic studies, other bioinspired approaches have focused on incorporating soluble signals into tissue engineering scaffolds. Soluble signals include growth factors such as epidermal growth factor (EGF), vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF), as well as cytokines and chemokines. Growth factors are naturally occurring protein hormones which may act through autocrine or paracrine mechanisms and have potent effects on cell growth, proliferation, and differentiation. Growth factors are often stored and sequestered in the ECM and interact with cells through receptor tyrosine kinases (RTKs). Growth factor signaling pathways overlap to a large extent with integrin signaling pathways, and cell responses to many growth factors are dependent on integrin-mediated adhesion. Considerable efforts have been focused within the tissue engineering and regenerative medicine fields on delivering or immobilizing growth factors to biomaterials promote stem cell proliferation and differentiation (for reviews, see [26, 27]). Growth factor delivery strategies increasingly feature the use of nanoparticles and nanotechnology (reviewed here [28]). Although biomaterials incorporating growth factors will not be directly addressed in this article, growth factors can be used in combination with adhesive peptides to direct cell functions for tissue engineering.
2.3 Cell-cell interations
Cell-cell interactions are primarily mediated by cadherins, ephrins and CAMs. Intercellular receptors have not been widely used in biomimetic tissue engineering with a few notable exceptions. For example, Beckstead et al. immobilized the Notch ligand, Jagged-1 to a biomaterial surface to direct stem cell differentiation [29] and Moon et al. functionalized hydrogels with ephrin A-1 to promote angiogenesis [30].
3. From micro to nano in biomaterials
Over the past few decades, techniques for creating nanoscale features, patterns and particles have emerged. Although these techniques were initially applied to electronics fabrication, they have more recently been used to pattern and immobilize proteins and peptides with nanoscale precision for applications such as tissue engineering, drug delivery and biosensing. Like nanotechnology, the interdisciplinary field of tissue engineering is also fledgling, and began approximately two decades ago with the idea that engineering and biology principles could be applied to the design of cell-based artificial constructs which would restore tissue and organ function [31]. The application of nanotechnology to tissue engineering thus far has mainly focused on recapitulating non-biochemical aspects of ECM. Examples include nanofiber scaffolds which recapitulate the architecture of structural proteins within ECM [32], substrates with nanoscale features which model native ECM nanotopography [33], and nanocomposites which recreate the mineral content and structure of bone ECM [34]. While these strategies represent promising avenues of research which may be combined with bioadhesive approaches, they fall beyond the scope of this article, which will focus solely on nanoscale biomaterial approaches using peptide or small protein ECM-derivatives for tissue regeneration.
4. Techniques to nanopattern peptides and proteins
Nanoscale control of ECM-derived peptides and proteins have primarily been used for non-regenerative medicine applications, including biosensing, drug delivery and for model systems to study cell functions such as adhesion and spreading. However, given that integrins and focal adhesions exist on submicron size scales, there is a compelling rationale for using nanotechnology in bioadhesive tissue engineering applications. The following strategies have been used to control and pattern proteins and peptides on a nanoscale for a range of applications and may be used for bioadhesive strategies as well. In the following section, the approaches may be equally applied to patterning either peptides or proteins unless stated otherwise. However for brevity, the term ‘peptides’ will be used to refer to ‘peptides and/or proteins’.
4.1 Self-assembly
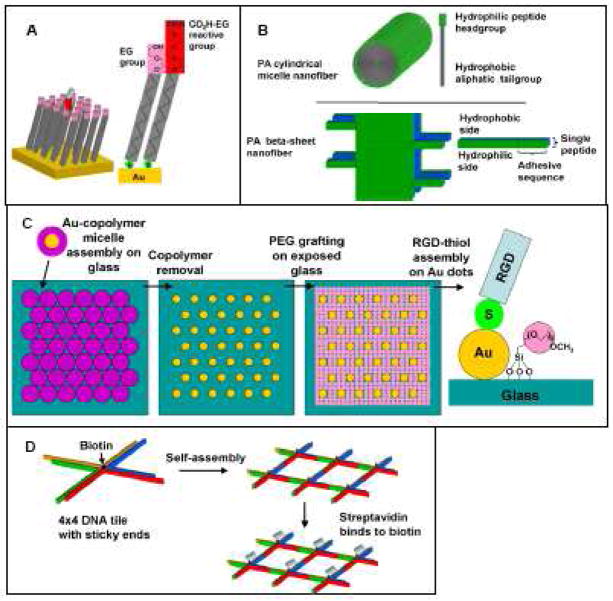
Self-assembly is the spontaneous formation of ordered structures and is an important nanotechnology tool which may be utilized for spatially orienting peptides with nanoscale precision. Self-assembly is a ‘bottom-up’ approach in which smaller building block molecules associate with each other in a coordinated fashion to form larger, more complex supramolecules. The organization of these building blocks into supramolecules is governed by molecular recognition due to non-covalent interactions such as hydrogen bonding, as well as electrostatic and hydrophobic interactions. Commonly used peptide self-assembly methods include self-assembled monolayers, amphiphatic peptide self-assembly, polymer assisted templating and DNA templating (Fig 4).
Fig 4.
Schematic of self-assembly-based peptide nanopatterning techniques. (A) Self-assembled monolayers (SAMs), (B) Peptide amphiphile (PA) self-assembly structures, (C) Polymer-assisted patterning of Au nanodots, (D) DNA templating using 4×4 DNA tiles.
4.1.1 Self-assembled monolayers (SAMs)
Self assembled monolayers are densely-packed two-dimensional arrays of long-chained molecules in which one end of the molecule, the head group, has a high affinity for a specific surface while the hydrophobic tail group may have a terminal functional group. The chemical properties of the surface can therefore be varied by using a mixture of SAM molecules with different terminal functional groups. The most commonly used SAMs for peptide conjugation are alkanethiols on gold, reviewed here [35]. Although self-assembled monolayers do not in themselves form nanopatterns, they are a platform technology often used in combination with other nanoscale techniques to generate peptide nanopatterns (see sections 2.1, 3.1 and 3.2) [36–38]. Peptide-functionalized SAMs are widely used in cell biology and tissue engineering because protein-resistant oligo(ethylene-glycol) (EG)-terminated SAMs can be used to create inert background surfaces patterned with peptides. EG SAMs prevent non-specific adsorption of proteins to the surface so that the surface is well defined and cells interact only with the functionalized peptide. A mixture of SAM molecules with non-fouling EG terminal groups and reactive terminal groups such as EG-carboxyl can be assembled on a surface, allowing peptides to adsorb onto or covalently bind to the reactive group (Fig 4A).
4.1.2 Amphiphilic peptides
Peptide amphiphile molecules consist of a peptide-containing hydrophilic region, as well as a hydrophobic region which is usually aliphatic. These peptide amphiphiles may self-assemble based on hydrophobic interactions into various structures including cylindrical micelle nanofibers, beta-sheet nanofibers, micelles [39, 40], vesicles [41], monolayers and bilayers (Fig 4B). Peptide amphiphiles cylindrical micelle nanofibers usually contain a single 10–22 carbon tail and have been studied as potential contrast agents [42], drug delivery vehicles [43, 44] and biomaterials for tissue engineering [45–48]. Peptide amphiphile beta sheets consist of alternating hydrophilic and hydrophobic peptide amino acids and have distinct polar and non-polar sides. These structures have been used as materials for protein release [49] and tissue engineering [50–52]. The peptide amphiphile self-assembly technique is compatible only with peptide sequences and not full proteins.
4.1.3 Polymer-assisted patterning
Numerous works have utilized polymers to control peptide presentation. Several groups have used varying molar ratios of peptide-modified and unmodified polymers together with computational modeling or estimates to predict the average peptide nanospacing within these mixtures [53–56]. However, peptide spacings are not precisely controlled in these peptide-polymer mixtures. In contrast, Spatz and coworkers developed a method to generate regular hexagonal nanopatterns of gold nanodots by self-assembly of diblock copolymer micelles on glass [57] (Fig 4C). Following micelle assembly, the polymer is completely removed by a gas plasma treatment, leaving behind small (<8 nm diameter) gold dots that are precisely positioned at spacings as small as 28, 58, 73 or 85 nm. The gold dots are functionalized with thiol-terminated peptides and the glass background is passivated with silane-PEG. These peptide-gold dot patterns have been applied extensively to study cell adhesion [58–60].
4.1.4 DNA templating
Yan et al. designed a protein nanopatterning technique in which a nanostructure was created using cross-shaped four-armed branched DNA with a square aspect ratio [61]. These 4 × 4 tiles of DNA with sticky ends self-assemble to form two-dimensional nanogrids. Periodic streptavidin protein arrays were fabricated by incorporating biotin into the center of the DNA tiles, and streptavidin density was controlled by having half or all the DNA tiles biotinylated [62] (Fig 4D). This DNA tile technique has also been combined with aptamers to link thrombin to the DNA lattice [63–65]. Using a different DNA templating method, Turberfield and coworkers have encapsulated proteins within DNA tetrahedra, with one cytochrome c molecule per DNA cage [66]. The tetrahedral DNA nanostructure can also be modified with multiple proteins per tetrahedron using click chemistry [67].
An advantage of self-assembly methods of nanopatterning is that extremely high resolution patterns may be obtained. However, the pattern itself may not be amenable to much modification, as it is predetermined by the self-assembling molecules used. It may also be difficult to pattern more than one peptide using self-assembly methods.
4.2 Stamping
In contrast to the ‘bottom-up’ self-assembly approach, stamping, like scanning probe and electron beam patterning methods to be described in the following sections, is a ‘top-down’ approach in which nanoscale patterns are created by starting with large, bulk materials. Stamping methods all involve a contact step between two surfaces to create a nanopattern.
4.2.1 Nanocontact printing
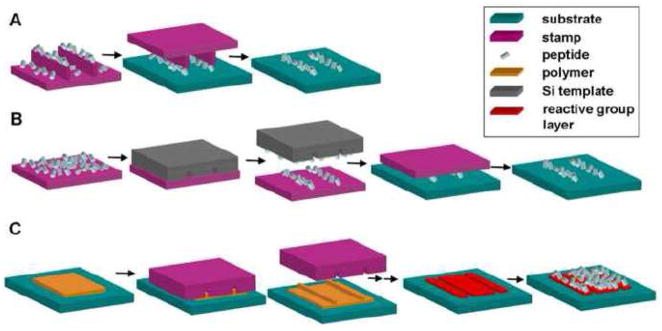
Nanocontact printing adapts the technique of microcontact printing, which was first used by Whitesides and coworkers, to create nanoscale patterns. In nanocontact printing, an elastomeric stamp is typically fabricated from poly(dimethylsiloxane) (PDMS) with raised nanoscale features corresponding to the desired pattern. The stamp is dipped in ‘ink’, which is the peptide solution and then brought into contact with the substrate to be patterned. The stamping process transfers the protein on the raised features of the stamp onto the contacted substrate [68–70] (Fig 5A). However, a limitation of nanocontact printing is that the nanoscale raised features of the soft stamps may buckle during contact, resulting in distortion of the protein pattern.
Fig 5.
Schematic of stamping nanopatterning techniques. (A) Nanocontact printing, (B) Subtractive printing, (C) Nanoimprint lithography.
4.2.2 Subtractive printing
Subtractive printing addresses the problem of stamp feature deformation by employing a flat stamp. In this method, a flat PDMS stamp is inked with protein followed by subtraction of protein from the stamp in a brief contact step with a silicon nanotemplate. The remaining protein pattern on the flat stamp is then transferred to the substrate (Fig 5B). Coyer et al. used subtractive printing to create lines of TRITC labeled antibodies on glass with sub-100 nm widths and 280 nm squares [71]. In the same study, it was also demonstrated that the subtractive printing method can be used to create overlapping and non-overlapping patterns of two different antibodies.
4.2.3 Nanoimprint lithography
Like nanocontact printing and subtractive printing, nanoimprint lithography also employs a stamp which, in this case, is typically a silicon (Si) nanotemplate. The raised features of the Si stamp create an imprint in the substrate by contacting a thin layer of heated polymer. The residual polymer on the imprinted surface is etched to create an inverse pattern on the underlying surface, and a reactive group layer is then added to generate a protein pattern [72–75] (Fig 5C).
Stamping methods in general allow rapid patterning of large areas or multiple surfaces compared to serial methods such as scanning probe and electron beam patterning and are low-cost as they do not require specialized equipment.
4.3 Atomic Force Microscope (AFM)-based methods
Atomic Force Microscopes (AFMs) may be modified to allow for patterning of surfaces. Peptide nanopatterning techniques such as nanografting, dip-pen nanolithography and nanopen utilize AFM tips.
4.3.1 Nanografting
Nanografting involves creating patterns by scratching molecules of a protein-resistant EG self assembled monolayer (SAM) off the substrate using an atomic force microscope (AFM) tip (Fig 6A). When the SAM molecules are removed, reactive SAM molecules [76, 77], DNA [78, 79] or peptides [37, 80, 81] in solution replace the grafted SAMs and assemble in the patterned area. In the case of reactive SAM or DNA replacement strategies, peptides then adsorb to or react chemically with the molecules in the grafted area. This process thereby generates a pattern of adsorbed protein localized to the regions initially scraped by the AFM tip.
Fig 6.
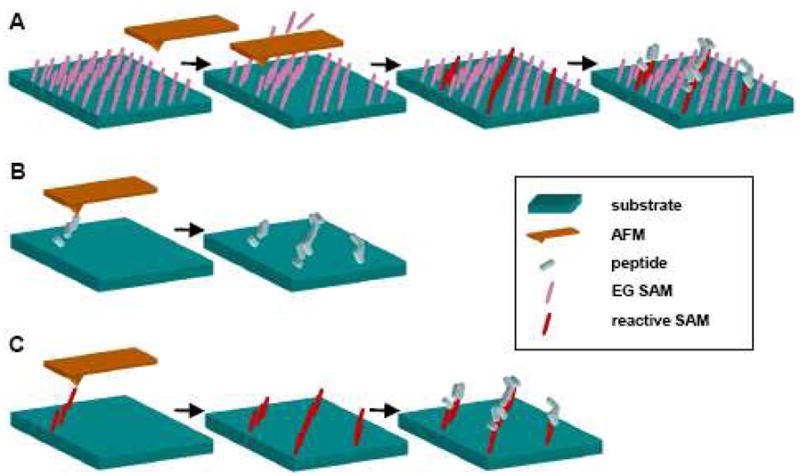
Schematic of scanning microscopy nanopatterning techniques. (A) Nanografting (B) Direct dip-pen nanolithography (C) Indirect dip-pen nanolithography. Adapted from [133].
4.3.2 Dip-pen nanolithography
The dip-pen nanolithography technique involves dipping an AFM tip into an ‘ink’ and then transferring the molecules in that ‘ink’ to a surface in a manner analogous to using a quill to write. Protein patterning using this method may be direct or indirect. In the direct method, the AFM tip is dipped in a protein solution and the protein is directly deposited on the substrate in a desired pattern (Fig 6B). In contrast, the indirect method involves dipping the AFM tip in a solution of protein adherent SAM molecules and delivering the molecules to a surface. Typically the SAM molecule is an alkanethiol which is deposited on a gold surface. The surface is then incubated with a protein, which adsorbs with higher affinity to the areas on which the protein adherent molecules were patterned [82–86] (Fig 6C).
4.3.3 Nanopen
The nanopen method of patterning biomolecules is analogous to writing with a fountain pen as a protein solution flows through a hollow ‘nanopipette’ which is spatially manipulated over a surface with nanoscale control [87]. The advantage of this method over dip-pen nanolithography is that it allows for complex patterns involving multiple ‘inks’ to be created as different proteins may be delivered without re-dipping the tip. However, because the ‘ink’ diffuses out of the pipet tip, the minimal feature size that can be generated using the nanopen will be limited by the pipette size, which is approximately 100 nm.
The scanning probe patterning methods generally allow smaller feature sizes and more complex patterns to be created than with stamping methods, but also take longer to pattern and require expensive scanning probe equipment.
4.4 Electron beam lithography
Electron beam lithography is a maskless process which uses an electron beam to scan across a substrate. For protein patterning, the electron beam is typically used to ablate a pattern into a self-assembled monolayer [88], to crosslink peptide-reactive molecules to a surface [89, 90], or to pattern a SiO2 surface using a PMMA resist [91, 92]. Electron beam lithography can be used to create extremely high resolution patterns. However, like scanning probe methods, electron beam lithography is also a low throughput process.
4.5 Three-dimensional (3D) peptide patterning
The peptide nanopatterning techniques described earlier in sections 4.1– 4.4 are used for patterning on a two-dimensional (2D) surface. However, because cells in most tissues exist in a 3D microenvironment, there is growing interest in creating 3D peptide patterns within artificial matrices for tissue engineering applications. Although little work has been done on 3D peptide nanopatterning, techniques similar to those mentioned in sections 4.1 – 4.4 have been applied to the 3D micropatterning of peptides in matrices such as hydrogels. Examples of these 3D patterning methods include additive multilayered photolithography using UV-opaque masks [93, 94], laser scanning photolithography [95, 96], microfluidic lithography [97, 98], 3D printing [99], micromolding [100] and electrochemical deposition [101].
5. Nanoscale engineering of proteins and peptides for bioadhesion
How can nanoscale engineering of ECM-derived adhesive peptides be used to enhance cellular response for tissue regeneration? Although this field is admittedly in its infancy, some significant works have already been completed which demonstrate the promise of this approach. Nanoscale bioadhesive tissue engineering strategies which have been employed can be classified under three major categories:
Nanoscale control of adhesive and modulatory peptide domains to retain full bioactivity of parental protein
Nanoscale patterning of adhesive peptides for high density presentation
Nanoscale engineering of multivalency of adhesive peptide/protein
5.1 Nanoscale control of co-presentation of adhesive and modulatory peptide domains to maintain bioactivity of parental protein
Our lab has previously engineered FNIII7-10, a 39 kD recombinant fragment of fibronectin (FN) which incorporates both the FN-derived RGD adhesion site and the PHSRN synergy site while maintaining the correct spacing and relative angle between them with nanoscale precision. Because FNIII7-10 presents RGD and PHSRN in their native structural orientation, like full-length FN, FNIII7-10 demonstrates preferential binding to the α5β1 integrin with high affinity as well as higher cell adhesion than on surfaces with RGD-only or RGD-PHSRN peptides which fail to recapitulate the native conformation of both binding sites [102]. It should be noted that because FNIII7-10 is a 39 kD fragment of FN, it can be presented on a surface with greater control than full length FN and is easier to produce, while still retaining the full biofunctionality of the parental molecule. Furthermore, we have shown that FNIII7-10-modified surfaces enhance osteoblast adhesion strength and differentiation in vitro, as well as titanium implant osseointegration in vivo when compared to RGD peptide surfaces [103]. In addition, this fragment exhibits enhanced activities compared to FN because other domains that may have antagonistic effects are not included in its structure [104]. These studies demonstrate that ECM-mimetic peptide constructs which reproduce the in vivo structure of their adhesive and modulatory domains on a nanoscale can greatly enhance the outcome for tissue engineering applications.
The full-length fibronectin (FN) molecule contains both an RGD adhesion site and a PHSRN synergy site spaced approximately 3.2 nm apart from each other (Fig 7 and 8). The presence of the PHSRN synergy site on the FNIII9 module enhances the affinity of the α5β1 integrin to the RGD loop on FNIII10 over forty-fold in FN [18]. In contrast, multiple integrins, including αvβ3, also bind to the RGD site on FN, but are not influenced by the presence of the PHSRN site [105]. Furthermore, cell adhesion strength on bioadhesive surfaces presenting RGD alone is reduced compared to full FN [106–108]. The synergy site therefore functions to allow α5β1 integrin to bind preferentially to FN over other integrins. The co-presentation of RGD and PHSRN may therefore be used to create α5β1 integrin-specific bioadhesive surfaces. These integrin-specific surfaces may be especially applicable to regenerative medicine of connective tissues, in particular bone, for several reasons. Interactions between fibronectin, which binds α5β1, and osteoblasts are essential for osteoblast differentiation [109, 110]. Furthermore, α5β1 expression is positively correlated with viability and osteogenic differentiation of human mesenchymal stem cells and primary osteoblasts [111–114]. Lastly, RGD alone binds non-specifically to multiple integrins, but mostly αvβ3, and αvβ3 has been shown to negatively modulate bone mineralization and osteoblast differentiation [115, 116].
Fig 7.
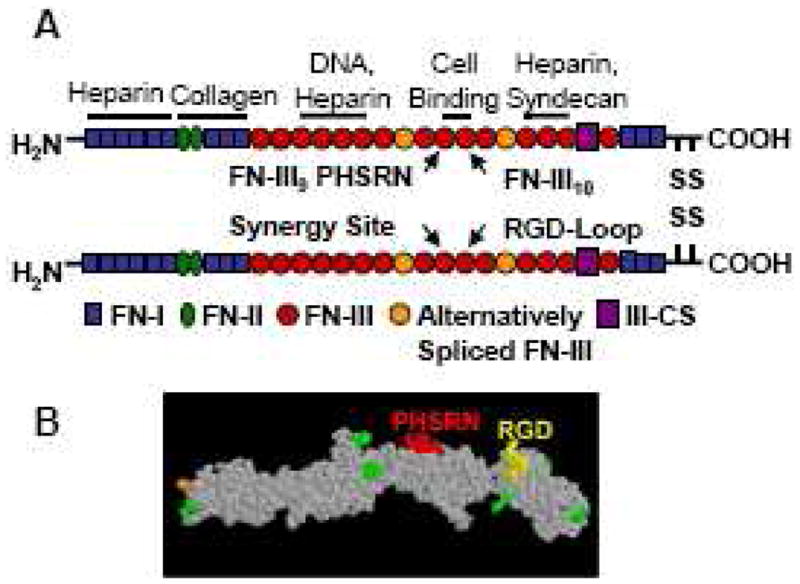
(A)Structure of plasma fibronectin and location of major binding sites. Adapted from [134]. (B) Space-filling model of FNIII7-10 recombinant fragment of fibronectin.
Fig 8.
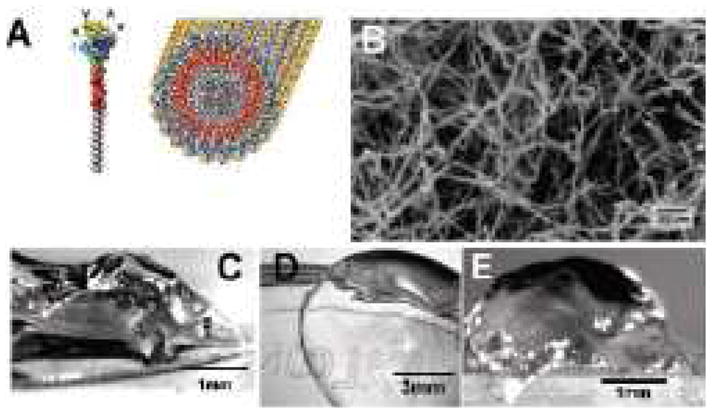
(A) Illustration of IKVAV-containing peptide amphiphile (PA) molecule and self-assembled PA nanofiber. (B) SEM image of IKVAV nanofiber network. (C and D) Micrographs of gel formed by adding to IKVAV PA solutions (C) cell culture media and (D) cerebral spinal fluid. (E) Micrographs of an IKVAV nanofiber gel extracted from an enucleated rat eye after intraocular injection of the peptide amphiphile solution. Reproduced with permission from [127].
Other attempts to mimic FN’s bioactivity have featured peptide designs in which RGD and PHSRN were co-presented by either immobilizing a combination of RGD and PHSRN peptides to a substrate [117], or by incorporating the RGD and PHSRN sequences on the same molecule separated by polyglycine linkers [118–120], a PEG spacer [121] or a 3.7 nm spacer [122, 123] . However, many of these studies have failed to show cell adhesion and integrin specificity characteristics that are equivalent to FN. This may be because even minor alterations in the spacing, relative angle and sequences separating the 9th and 10th FNIII repeats will result in losses of bioactivity compared to FN [124–126]. Mimicking the nanoscale spacings and orientations of cell binding sequences within ECM macromolecules may therefore be essential to recapitulating the function of native ECM.
5.2 Nanoscale patterning of adhesive peptides for high density presentation
Nanopatterning or nanoscale self-assembly of adhesive peptides may be used to present adhesive peptides at a much higher density than would naturally occur in the extracellular matrix to elicit a stronger cell response.
Silva et al. found that culturing neural progenitor cells in a hydrogel of cylindrical nanofibers formed by the self-assembly of peptide amphiphiles incorporating the laminin-derived IKVAV peptide promoted their neural differentiation, while suppressing astroglial differentiation [127]. The self-assembled nanofibers present the IKVAV peptide at near van der Waal’s packing density (Fig 8). Neural progenitor cells encapsulated in the IKVAV peptide amphiphile nanofibers showed higher expression of neural marker beta-tubulin and lower expression of astrocyte marker glial fibrillary acidic protein than cells cultured on polylysine or laminin coated surfaces. However, progenitor cells cultured in nanofibers incorporating a biologically irrelevant EQS peptide instead of IKVAV did not differentiate into neural cells, indicating that the cell response was induced by the presentation of the bioactive IKVAV sequence. Tysseling-Mattiace et al. used the same IKVAV peptide amphiphile nanofibers in a clip compression mouse model of spinal cord injury and showed that the treatment with IKVAV nanofibers 24 hours after injury reduced astrogliosis and cell death at the injury site and promoted the regeneration of motor and sensory axons [46]. Furthermore, mice treated with IKVAV nanofibers showed improved functional recovery over sham, glucose and non-bioactive EQS peptide amphiphile nanofiber injections as assessed by the BBB locomotor scale.
Several other works have utilized self-assembled molecules incorporating the RGD sequence and nanopatterned RGD to enhance osteogenic differentiation of cells in vitro. These approaches include RGD-modified Ac(RADA)4 peptides forming beta-sheet nanofibers seeded with pre-osteoblast MC3T3-E1 cells [52], RGD-peptide amphiphile nanofibers seeded with rat mesenchymal stem cells [45] and RGD-coupled alginate coils seeded with pre-osteoblasts MC3T3-E1 cells [55].
5.3 Nanoscale engineering of multivalent adhesion peptides
Another important application of nanoscale engineering to adhesive biomaterials may be in the multivalent presentation of adhesion peptides. Multivalent presentation of adhesion peptides may enhance cell adhesion and cell signaling for two reasons. First, polyvalent molecules collectively bind to their receptors with a much higher affinity than the same monovalent molecule. For example, an engineered trivalent system for vancomycin binding to D-Ala-D-Ala has a dissociation constant of 4 × 10−17 M, in contrast to 1 × 10−6 M for the monovalent interaction [128]. Second, both occupancy and clustering of integrins is necessary for the full range of integrin-mediated cell signaling. Therefore, polyvalent ligands can be used to promote integrin clustering and thereby control downstream cell responses. Nanoscale engineering of adhesion site position on a multivalent construct is necessary to promote avidity and force integrin clustering since integrins are approximately 10 nm in diameter [12].
Examples of enhancements in cell behavior induced by polyvalent adhesion peptides include work by Maheswari et al. using star polymers with 1, 5 or 9 immobilized RGD peptides per star to present RGD clustered on an approximately 50 nm scale [53]. This study showed that cell adhesion and motility was increased with clustered RGD over non-clustered monovalent presentation of RGD, even when the overall average RGD surface density was approximately equal. Similarly, nanoscale RGD clustering enhanced cell adhesion to substrates in Koo et al.’s work with polymer combs [54]. However, it should be noted that in both the previous studies, the RGD clusters were not patterned, and therefore, the within-cluster RGD spacing, inter-cluster spacing and overall RGD density were not precisely controlled on a nanoscale. Instead, the average values were estimated based on the molar ratios of RGD-functionalized to unfunctionalized star polymers or comb polymers.
Multimeric adhesion peptides have also been used in drug delivery applications to target specific cell types which over-express the corresponding integrin. Sancey et al. and Carlson et al. used multivalent RGD molecules to target tumor cells which over-express the αvβ3 integrin [129, 130]. While this approach has not been used for tissue engineering applications, the success of these targeting studies indicate that multivalent ECM-derived peptides could potentially be used to promote adhesion of a specific cell type based on its unique integrin expression profile.
Although polyvalency has not yet been shown to enhance peptide effects on cellular behavior by orders of magnitude, polyvalency has demonstrated this effect in the field of viral inhibitors. The dramatic results seen with polyvalent viral inhibitors may be instructive to and demonstrate the potential of similar efforts with ECM-mimetic peptides for tissue engineering applications. Mourez et al. attached multiple copies of the P1 anthrax inhibitor peptide to a flexible polyacrylamide, with one P1 peptide functionalized on average per 40 acrylamide monomers. The P1 peptide inhibits assembly the anthrax toxin by preventing two toxin components, PA63 and LF protease, from binding each other by competitively binding to PA63. The polyvalent P1 molecule inhibited binding of PA63 to LF with a 7,500 fold increase in efficacy on a per peptide basis when compared to free P1 peptide inhibitor [131] and also inhibited toxicity in CHO cells and in a rat model. This study highlights the power of multivalency to enhance the strength of peptide-ligand interactions and thereby improve the therapeutic efficacy of the approach by many orders of magnitude. However, it should be noted that in this application, viral inhibition requires only high-affinity binding of the P1 inhibitor molecule to PA63, while forced clustering of the peptide-ligand is not necessary. It is probably for this reason that large increases in efficacy were observed despite the use of a peptide-functionalized flexible polymer system with no control of nanoscale peptide spacing. In regenerative medicine applications however, controlled peptide spacing will likely be crucial to enhancing cell responses by forcing integrin clustering.
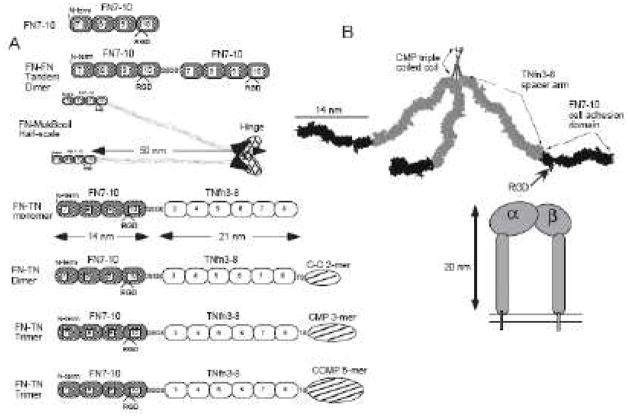
Precise nanoscale control of multivalent ECM-derived adhesive protein fragments has been achieved using self-assembly techniques. Coussen et al. designed a monomer, dimer, trimer and pentamer of FN7-10, an adhesive protein fragment derived from fibronectin [132] (Fig 9). The multimeric constructs consisted of 14 nm long FN7-10 fragments attached at the N-terminus, followed by 21 nm long TNfn3-8 spacer arms, followed by C-terminus dimer, trimer or pentamer-forming self-assembly sequences: C-C-2, CMP and COMP respectively. These multimeric FN7-10 constructs were attached to small 40nm gold beads to address the question of how many integrins are required to form a cluster that will attach to the cytoskeleton. Beads functionalized with FN7-10 trimers and pentamers, but not monomers or dimers localized to the actin cytoskeleton and displayed sustained cell binding and rearward linear movement. Coussen et al. also found that a five-fold excess of monomeric FN7-10 could compete with binding of trimeric FN7-10, demonstrating the avidity effects of multivalent FN7-10. While this study used nanoscale engineered multivalent adhesive ECM fragments to investigate cell motility, these multimers could also be functionalized to biomaterials to direct cell responses for tissue regeneration.
Fig 9.
(A) Constructs for multimeric FN7-10. (B) A scale model of the FN7-10 trimer. Reproduced with permission from [132].
6. Conclusion and future outlook
The fast-evolving fields of tissue engineering and nanotechnology have begun to converge, giving rise to new methods of directing cell fates by controlling the presentation of ECM-derived peptides with nanoscale precision. In recent decades, a range of exciting new strategies for peptide nanopatterning have been developed with important benefits and capabilities such as patterning more complex, higher resolution, multi-peptide patterns with greater spatial control, as well as rapid and low-cost pattern fabrication. Concurrently, tissue engineering and cell biology studies have demonstrated the potential of nanoscale control of adhesive peptide-modified materials to control cell behaviors in vitro and in select cases, to promote tissue healing in rigorous vivo models as well. While the application of nanotechnology to regenerative medicine is still in its infancy, there has been significant interest and progress in both protein nanopatterning technology and nanobioadhesive tissue engineering. Although these synergistic developments provide opportunities to harness nanotechnology to enhance tissue repair, numerous questions and challenges remain. We have yet to fully understand the mechanisms by which variations in nanoscale spacing, orientation and co-presentation of ECM ligands modulate cell responses. A better understanding of these topics would elucidate principles for the rational design of adhesive biomaterials. Major challenges for the future include using nanoscale engineering of bioadhesive surfaces and implants to control specific ligand-cell receptor interactions, targeting multiple receptors using multivalent peptide presentation, as well as to incorporating greater spatiotemporal, mechanical and 3D architectural control of cell-material interactions .
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Molecular biology of the cell. 4. Garland Science; New York: 2002. [Google Scholar]
- 2.Rossert J, de Crombrugghe B, John PB, Lawrence GR, Gideon AR. Type I Collagen: Structure, Synthesis, and Regulation, Principles of Bone Biology. 2. Academic Press; San Diego: 2002. pp. 189–210. [Google Scholar]
- 3.Robey PG, John PB, Lawrence GR, Gideon AR. Bone Matrix Proteoglycans and Glycoproteins, Principles of Bone Biology. 2. Academic Press; San Diego: 2002. pp. 225–237. [Google Scholar]
- 4.Zhang H, Marshall KW, Tang H, Hwang DM, Lee M, Liew CC. Profiling genes expressed in human fetal cartilage using 13,155 expressed sequence tags. Osteoarthritis Cartilage. 2003;11:309–319. doi: 10.1016/s1063-4584(03)00032-3. [DOI] [PubMed] [Google Scholar]
- 5.Aplin AE, Hogan BP, Tomeu J, Juliano RL. Cell adhesion differentially regulates the nucleocytoplasmic distribution of active MAP kinases. J Cell Sci. 2002;115:2781–2790. doi: 10.1242/jcs.115.13.2781. [DOI] [PubMed] [Google Scholar]
- 6.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 7.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 8.Petit V, Thiery JP. Focal adhesions: structure and dynamics. Biol Cell. 2000;92:477–494. doi: 10.1016/s0248-4900(00)01101-1. [DOI] [PubMed] [Google Scholar]
- 9.Bourdoulous S, Orend G, MacKenna DA, Pasqualini R, Ruoslahti E. Fibronectin matrix regulates activation of RHO and CDC42 GTPases and cell cycle progression. J Cell Biol. 1998;143:267–276. doi: 10.1083/jcb.143.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 11.Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- 12.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 13.Carrell NA, Fitzgerald LA, Steiner B, Erickson HP, Phillips DR. Structure of human platelet membrane glycoproteins IIb and IIIa as determined by electron microscopy. J Biol Chem. 1985;260:1743–1749. [PubMed] [Google Scholar]
- 14.Nermut MV, Green NM, Eason P, Yamada SS, Yamada KM. Electron microscopy and structural model of human fibronectin receptor. EMBO J. 1988;7:4093–4099. doi: 10.1002/j.1460-2075.1988.tb03303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emsley J, Knight CG, Farndale RW, Barnes MJ. Structure of the integrin alpha2beta1-binding collagen peptide. J Mol Biol. 2004;335:1019–1028. doi: 10.1016/j.jmb.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 16.Knight CG, Morton LF, Onley DJ, Peachey AR, Messent AJ, Smethurst PA, Tuckwell DS, Farndale RW, Barnes MJ. Identification in collagen type I of an integrin alpha2 beta1-binding site containing an essential GER sequence. J Biol Chem. 1998;273:33287–33294. doi: 10.1074/jbc.273.50.33287. [DOI] [PubMed] [Google Scholar]
- 17.Leahy DJ, Aukhil I, Erickson HP. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 18.Aota S, Nomizu M, Yamada KM. The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. J Biol Chem. 1994;269:24756–24761. [PubMed] [Google Scholar]
- 19.Humphries MJ, Akiyama SK, Komoriya A, Olden K, Yamada KM. Identification of an alternatively spliced site in human plasma fibronectin that mediates cell type-specific adhesion. J Cell Biol. 1986;103:2637–2647. doi: 10.1083/jcb.103.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komoriya A, Green LJ, Mervic M, Yamada SS, Yamada KM, Humphries MJ. The minimal essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J Biol Chem. 1991;266:15075–15079. [PubMed] [Google Scholar]
- 21.Tashiro K, Sephel GC, Weeks B, Sasaki M, Martin GR, Kleinman HK, Yamada Y. A synthetic peptide containing the IKVAV sequence from the A chain of laminin mediates cell attachment, migration, and neurite outgrowth. J Biol Chem. 1989;264:16174–16182. [PubMed] [Google Scholar]
- 22.Graf J, Ogle RC, Robey FA, Sasaki M, Martin GR, Yamada Y, Kleinman HK. A pentapeptide from the laminin B1 chain mediates cell adhesion and binds the 67,000 laminin receptor. Biochemistry. 1987;26:6896–6900. doi: 10.1021/bi00396a004. [DOI] [PubMed] [Google Scholar]
- 23.Kleinman HK, Graf J, Iwamoto Y, Sasaki M, Schasteen CS, Yamada Y, Martin GR, Robey FA. Identification of a second active site in laminin for promotion of cell adhesion and migration and inhibition of in vivo melanoma lung colonization. Arch Biochem Biophys. 1989;272:39–45. doi: 10.1016/0003-9861(89)90192-6. [DOI] [PubMed] [Google Scholar]
- 24.Badylak SF, Freytes DO, Gilbert TW. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 25.Shakesheff K, Cannizzaro S, Langer R. Creating biomimetic micro-environments with synthetic polymer-peptide hybrid molecules. J Biomater Sci Polym Ed. 1998;9:507–518. doi: 10.1163/156856298x00596. [DOI] [PubMed] [Google Scholar]
- 26.Chen RR, Mooney DJ. Polymeric growth factor delivery strategies for tissue engineering. Pharm Res. 2003;20:1103–1112. doi: 10.1023/a:1025034925152. [DOI] [PubMed] [Google Scholar]
- 27.Anitua E, Sanchez M, Orive G, Andia I. Delivering growth factors for therapeutics. Trends Pharmacol Sci. 2008;29:37–41. doi: 10.1016/j.tips.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, Uludag H. Nanoparticulate systems for growth factor delivery. Pharm Res. 2009;26:1561–1580. doi: 10.1007/s11095-009-9897-z. [DOI] [PubMed] [Google Scholar]
- 29.Beckstead BL, Santosa DM, Giachelli CM. Mimicking cell-cell interactions at the biomaterial-cell interface for control of stem cell differentiation. J Biomed Mater Res A. 2006;79:94–103. doi: 10.1002/jbm.a.30760. [DOI] [PubMed] [Google Scholar]
- 30.Moon JJ, Lee SH, West JL. Synthetic biomimetic hydrogels incorporated with ephrin-A1 for therapeutic angiogenesis. Biomacromolecules. 2007;8:42–49. doi: 10.1021/bm060452p. [DOI] [PubMed] [Google Scholar]
- 31.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 32.Kumbar SG, James R, Nukavarapu SP, Laurencin CT. Electrospun nanofiber scaffolds: engineering soft tissues. Biomed Mater. 2008;3:034002. doi: 10.1088/1748-6041/3/3/034002. [DOI] [PubMed] [Google Scholar]
- 33.Bettinger CJ, Langer R, Borenstein JT. Engineering substrate topography at the micro- and nanoscale to control cell function. Angew Chem Int Ed Engl. 2009;48:5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christenson EM, Anseth KS, van den Beucken JJ, Chan CK, Ercan B, Jansen JA, Laurencin CT, Li WJ, Murugan R, Nair LS, Ramakrishna S, Tuan RS, Webster TJ, Mikos AG. Nanobiomaterial applications in orthopedics. J Orthop Res. 2007;25:11–22. doi: 10.1002/jor.20305. [DOI] [PubMed] [Google Scholar]
- 35.Mrksich M. Using self-assembled monolayers to model the extracellular matrix. Acta Biomater. 2009;5:832–841. doi: 10.1016/j.actbio.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jang SG, Choi DG, Kim S, Jeong JH, Lee ES, Yang SM. Nanoscopic Pd line arrays using nanocontact printed dendrimers. Langmuir. 2006;22:3326–3331. doi: 10.1021/la0525170. [DOI] [PubMed] [Google Scholar]
- 37.Nuraje N, Banerjee IA, MacCuspie RI, Yu L, Matsui H. Biological bottom-up assembly of antibody nanotubes on patterned antigen arrays. J Am Chem Soc. 2004;126:8088–8089. doi: 10.1021/ja048617u. [DOI] [PubMed] [Google Scholar]
- 38.Salazar RB, Shovsky A, Schonherr H, Vancso GJ. Dip-pen nanolithography on (bio)reactive monolayer and block-copolymer platforms: deposition of lines of single macromolecules. Small. 2006;2:1274–1282. doi: 10.1002/smll.200600235. [DOI] [PubMed] [Google Scholar]
- 39.Wiradharma N, Tong YW, Yang YY. Self-assembled oligopeptide nanostructures for co-delivery of drug and gene with synergistic therapeutic effect. Biomaterials. 2009;30:3100–3109. doi: 10.1016/j.biomaterials.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Accardo A, Tesauro D, Del Pozzo L, Mangiapia G, Paduano L, Morelli G. Micelles by self-assembling peptide-conjugate amphiphile: synthesis and structural characterization. J Pept Sci. 2008;14:903–910. doi: 10.1002/psc.1024. [DOI] [PubMed] [Google Scholar]
- 41.Marques BF, Schneider JW. Sequence-specific binding of DNA to liposomes containing di-alkyl peptide nucleic acid (PNA) amphiphiles. Langmuir. 2005;21:2488–2494. doi: 10.1021/la047962u. [DOI] [PubMed] [Google Scholar]
- 42.Bull SR, Guler MO, Bras RE, Meade TJ, Stupp SI. Self-assembled peptide amphiphile nanofibers conjugated to MRI contrast agents. Nano Lett. 2005;5:1–4. doi: 10.1021/nl0484898. [DOI] [PubMed] [Google Scholar]
- 43.Kim JK, Anderson J, Jun HW, Repka MA, Jo S. Self-assembling peptide amphiphile-based nanofiber gel for bioresponsive cisplatin delivery. Mol Pharm. 2009;6:978–985. doi: 10.1021/mp900009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kushwaha M, Anderson JM, Bosworth CA, Andukuri A, Minor WP, Lancaster JR, Jr, Anderson PG, Brott BC, Jun HW. A nitric oxide releasing, self assembled peptide amphiphile matrix that mimics native endothelium for coating implantable cardiovascular devices. Biomaterials. 2009 doi: 10.1016/j.biomaterials.2009.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosseinkhani H, Hosseinkhani M, Kobayashi H. Proliferation and differentiation of mesenchymal stem cells using self-assembled peptide amphiphile nanofibers. Biomed Mater. 2006;1:8–15. doi: 10.1088/1748-6041/1/1/002. [DOI] [PubMed] [Google Scholar]
- 46.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA. Self-assembling nanofibers inhibit glial scar formation and promote axon elongation after spinal cord injury. J Neurosci. 2008;28:3814–3823. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sargeant TD, Guler MO, Oppenheimer SM, Mata A, Satcher RL, Dunand DC, Stupp SI. Hybrid bone implants: self-assembly of peptide amphiphile nanofibers within porous titanium. Biomaterials. 2008;29:161–171. doi: 10.1016/j.biomaterials.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 48.Anderson JM, Kushwaha M, Tambralli A, Bellis SL, Camata RP, Jun HW. Osteogenic differentiation of human mesenchymal stem cells directed by extracellular matrix-mimicking ligands in a biomimetic self-assembled peptide amphiphile nanomatrix. Biomacromolecules. 2009;10:2935–2944. doi: 10.1021/bm9007452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koutsopoulos S, Unsworth LD, Nagai Y, Zhang S. Controlled release of functional proteins through designer self-assembling peptide nanofiber hydrogel scaffold. Proc Natl Acad Sci U S A. 2009;106:4623–4628. doi: 10.1073/pnas.0807506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kisiday J, Jin M, Kurz B, Hung H, Semino C, Zhang S, Grodzinsky AJ. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proc Natl Acad Sci U S A. 2002;99:9996–10001. doi: 10.1073/pnas.142309999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis ME, Hsieh PC, Takahashi T, Song Q, Zhang S, Kamm RD, Grodzinsky AJ, Anversa P, Lee RT. Local myocardial insulin-like growth factor 1 (IGF-1) delivery with biotinylated peptide nanofibers improves cell therapy for myocardial infarction. Proc Natl Acad Sci U S A. 2006;103:8155–8160. doi: 10.1073/pnas.0602877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horii A, Wang X, Gelain F, Zhang S. Biological designer self-assembling Peptide nanofiber scaffolds significantly enhance osteoblast proliferation, differentiation and 3-D migration. PLoS One. 2007;2:e190. doi: 10.1371/journal.pone.0000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Cell adhesion and motility depend on nanoscale RGD clustering. J Cell Sci. 2000;113(Pt 10):1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 54.Koo LY, Irvine DJ, Mayes AM, Lauffenburger DA, Griffith LG. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J Cell Sci. 2002;115:1423–1433. doi: 10.1242/jcs.115.7.1423. [DOI] [PubMed] [Google Scholar]
- 55.Comisar WA, Kazmers NH, Mooney DJ, Linderman JJ. Engineering RGD nanopatterned hydrogels to control preosteoblast behavior: a combined computational and experimental approach. Biomaterials. 2007;28:4409–4417. doi: 10.1016/j.biomaterials.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 56.Comisar WA, Hsiong SX, Kong HJ, Mooney DJ, Linderman JJ. Multi-scale modeling to predict ligand presentation within RGD nanopatterned hydrogels. Biomaterials. 2006;27:2322–2329. doi: 10.1016/j.biomaterials.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 57.Glass R, Moller M, Spatz JP. Block copolymer micelle nanolithography. Nanotechnology. 2003;14:1153–1160. [Google Scholar]
- 58.Huang J, Grater SV, Corbellini F, Rinck S, Bock E, Kemkemer R, Kessler H, Ding J, Spatz JP. Impact of order and disorder in RGD nanopatterns on cell adhesion. Nano Lett. 2009;9:1111–1116. doi: 10.1021/nl803548b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graeter SV, Huang J, Perschmann N, Lopez-Garcia M, Kessler H, Ding J, Spatz JP. Mimicking cellular environments by nanostructured soft interfaces. Nano Lett. 2007;7:1413–1418. doi: 10.1021/nl070098g. [DOI] [PubMed] [Google Scholar]
- 60.Cavalcanti-Adam EA, Volberg T, Micoulet A, Kessler H, Geiger B, Spatz JP. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys J. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 62.Park SH, Yin P, Liu Y, Reif JH, LaBean TH, Yan H. Programmable DNA self-assemblies for nanoscale organization of ligands and proteins. Nano Lett. 2005;5:729–733. doi: 10.1021/nl050175c. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y, Lin C, Li H, Yan H. Aptamer directed self assembly of proteins on a DNA nanostructure. Angew Chem, Int Ed. 2005;44:4333–4338. doi: 10.1002/anie.200501089. [DOI] [PubMed] [Google Scholar]
- 64.Chhabra R, Sharma J, Ke Y, Liu Y, Rinker S, Lindsay S, Yan H. Spatially addressable multiprotein nanoarrays template by aptamer-tagged DNA nanoarchitectures. J Am Chem Soc. 2007;129:10304–10305. doi: 10.1021/ja072410u. [DOI] [PubMed] [Google Scholar]
- 65.Rinker S, Ke Y, Liu Y, Chhabra R, Yan H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat Nanotechnol. 2008;3:418–422. doi: 10.1038/nnano.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew Chem Int Ed Engl. 2006;45:7414–7417. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
- 67.Duckworth BP, Chen Y, Wollack JW, Sham Y, Mueller JD, Taton TA, Distefano MD. A universal method for the preparation of covalent protein-DNA conjugates for use in creating protein nanostructures. Angew Chem Int Ed Engl. 2007;46:8819–8822. doi: 10.1002/anie.200701942. [DOI] [PubMed] [Google Scholar]
- 68.von Philipsborn AC, Lang S, Bernard A, Loeschinger J, David C, Lehnert D, Bastmeyer M, Bonhoeffer F. Microcontact printing of axon guidance molecules for generation of graded patterns. Nat Protoc. 2006;1:1322–1328. doi: 10.1038/nprot.2006.251. [DOI] [PubMed] [Google Scholar]
- 69.Csucs G, Michel R, Lussi JW, Textor M, Danuser G. Microcontact printing of novel co-polymers in combination with proteins for cell-biological applications. Biomaterials. 2003;24:1713–1720. doi: 10.1016/s0142-9612(02)00568-9. [DOI] [PubMed] [Google Scholar]
- 70.Renault JP, Bernard A, Bietsch A, Michel B, Bosshard HR, Delamarche E, Kreiter M, Hecht B, Wild UP. Fabricating Arrays of Single Protein Molecules on Glass Using Microcontact Printing. J Phys Chem B. 2003;107:703–711. [Google Scholar]
- 71.Coyer SR, Garcia AJ, Delamarche E. Facile preparation of complex protein architectures with sub-100-nm resolution on surfaces. Angew Chem Int Ed Engl. 2007;46:6837–6840. doi: 10.1002/anie.200700989. [DOI] [PubMed] [Google Scholar]
- 72.Falconnet D, Pasqui D, Park SR, Eckert R, Schift H, Gobrecht J, Barbucci R, Textor M. A Novel Approach to Produce Protein Nanopatterns by Combining Nanoimprint Lithography and Molecular Self-Assembly. Nano Lett. 2004;4:1909–1914. [Google Scholar]
- 73.Hoff JD, Cheng LJ, Meyhofer E, Guo LJ, Hunt AJ. Nanoscale protein patterning by imprint lithography. 2004;4:853–857. [Google Scholar]
- 74.Maury P, Escalante M, Peter M, Reinhoudt DN, Subramaniam V, Huskens J. Creating nanopatterns of His-tagged proteins on surfaces by nanoimprint lithography using specific NiNTA-histidine interactions. Small. 2007;3:1584–1592. doi: 10.1002/smll.200700046. [DOI] [PubMed] [Google Scholar]
- 75.Schvartzman M, Nguyen K, Palma M, Abramson J, Sable J, Hone J, Sheetz MP, Wind SJ. Fabrication of Nanoscale Bioarrays for the Study of Cytoskeletal Protein Binding Interactions Using Nanoimprint Lithography. J Vac Sci Technol B Microelectron Nanometer Struct Process Meas Phenom. 2009;27:61–65. doi: 10.1116/1.3043472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wadu-Mesthrige K, Amro NA, Liu GY. Immobilization of proteins on self-assembled monolayers. Scanning. 2000;22:380–388. doi: 10.1002/sca.4950220607. [DOI] [PubMed] [Google Scholar]
- 77.Liu GY, Amro NA. Positioning protein molecules on surfaces: a nanoengineering approach to supramolecular chemistry. Proc Natl Acad Sci U S A. 2002;99:5165–5170. doi: 10.1073/pnas.072695699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bano F, Fruk L, Sanavio B, Glettenberg M, Casalis L, Niemeyer CM, Scoles G. Toward multiprotein nanoarrays using nanografting and DNA directed immobilization of proteins. Nano Lett. 2009;9:2614–2618. doi: 10.1021/nl9008869. [DOI] [PubMed] [Google Scholar]
- 79.Liu M, Liu GY. Hybridization with nanostructures of single-stranded DNA. Langmuir. 2005;21:1972–1978. doi: 10.1021/la047586d. [DOI] [PubMed] [Google Scholar]
- 80.Hu Y, Das A, Hecht MH, Scoles G. Nanografting de novo proteins onto gold surfaces. Langmuir. 2005;21:9103–9109. doi: 10.1021/la046857h. [DOI] [PubMed] [Google Scholar]
- 81.Case MA, McLendon GL, Hu Y, Vanderlick TK, Scoles G. Using Nanografting to Achieve Directed Assembly of de novo Designed Metalloproteins on Gold. Nano Lett. 2003;3:425–429. [Google Scholar]
- 82.Lee KB, Lim JH, Mirkin CA. Protein nanostructures formed via direct-write dip-pen nanolithography. J Am Chem Soc. 2003;125:5588–5589. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- 83.Lim JH, Ginger DS, Lee KB, Heo J, Nam JM, Mirkin CA. Direct-Write Dip-Pen Nanolithography of Proteins on Modified Silicon Oxide Surfaces. Angew Chem, Int Ed. 2003;42:2411–2414. doi: 10.1002/anie.200351256. [DOI] [PubMed] [Google Scholar]
- 84.Valiokas R, Vaitekonis S, Klenkar G, Trinkunas G, Liedberg B. Selective recruitment of membrane protein complexes onto gold substrates patterned by dip-pen nanolithography. Langmuir. 2006;22:3456–3460. doi: 10.1021/la053367w. [DOI] [PubMed] [Google Scholar]
- 85.Lee M, Kang DK, Yang HK, Park KH, Choe SY, Kang C, Chang SI, Han MH, Kang IC. Protein nanoarray on Prolinker surface constructed by atomic force microscopy dip-pen nanolithography for analysis of protein interaction. Proteomics. 2006;6:1094–1103. doi: 10.1002/pmic.200500392. [DOI] [PubMed] [Google Scholar]
- 86.Salaita K, Wang Y, Mirkin CA. Applications of dip-pen nanolithography. Nat Nanotechnol. 2007;2:145–155. doi: 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- 87.Taha H, Marks RS, Gheber LA, Rousso I, Newman J, Sukenik C, Lewis A. Protein printing with an atomic force sensing nanofountainpen. Appl Phys Lett. 2003;83:1041–. [Google Scholar]
- 88.Rundqvist J, Hoh JH, Haviland DB. Directed immobilization of protein-coated nanospheres to nanometer-scale patterns fabricated by electron beam lithography of poly(ethylene glycol) self-assembled monolayers. Langmuir. 2006;22:5100–5107. doi: 10.1021/la052306v. [DOI] [PubMed] [Google Scholar]
- 89.Hong Y, Krsko P, Libera M. Protein surface patterning using nanoscale PEG hydrogels. Langmuir. 2004;20:11123–11126. doi: 10.1021/la048651m. [DOI] [PubMed] [Google Scholar]
- 90.Christman KL, Vazquez-Dorbatt V, Schopf E, Kolodziej CM, Li RC, Broyer RM, Chen Y, Maynard HD. Nanoscale Growth Factor Patterns by Immobilization on a Heparin-Mimicking Polymer. J Am Chem Soc. 2008 doi: 10.1021/ja803676r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Senaratne W, Sengupta P, Jakubek V, Holowka D, Ober CK, Baird B. Functionalized surface arrays for spatial targeting of immune cell signaling. J Am Chem Soc. 2006;128:5594–5595. doi: 10.1021/ja058701p. [DOI] [PubMed] [Google Scholar]
- 92.Powell T, Yoon JY. Fluorescent biorecognition of gold nanoparticle-IgG conjugates self-assembled on E-beam patterns. Biotechnol Prog. 2006;22:106–110. doi: 10.1021/bp0501726. [DOI] [PubMed] [Google Scholar]
- 93.Mercey E, Obeid P, Glaise D, Calvo-Munoz ML, Guguen-Guillouzo C, Fouque B. The application of 3D micropatterning of agarose substrate for cell culture and in situ comet assays. Biomaterials. 31:3156–3165. doi: 10.1016/j.biomaterials.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 94.Liu Tsang V, Chen AA, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN. Fabrication of 3D hepatic tissues by additive photopatterning of cellular hydrogels. FASEB J. 2007;21:790–801. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- 95.Lee SH, Moon JJ, West JL. Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3D cell migration. Biomaterials. 2008;29:2962–2968. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hahn MS, Miller JS, West JL. Three-Dimensional Biochemical and Biomechanical Patterning of Hydrogels for Guiding Cell Behavior. Advanced Materials. 2006;18:2679–2684. [Google Scholar]
- 97.Panda P, Ali S, Lo E, Chung BG, Hatton TA, Khademhosseini A, Doyle PS. Stop-flow lithography to generate cell-laden microgel particles. Lab Chip. 2008;8:1056–1061. doi: 10.1039/b804234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee SA, Chung SE, Park W, Lee SH, Kwon S. Three-dimensional fabrication of heterogeneous microstructures using soft membrane deformation and optofluidic maskless lithography. Lab Chip. 2009;9:1670–1675. doi: 10.1039/b819999j. [DOI] [PubMed] [Google Scholar]
- 99.Mironov V, Kasyanov V, Drake C, Markwald RR. Organ printing: promises and challenges. Regen Med. 2008;3:93–103. doi: 10.2217/17460751.3.1.93. [DOI] [PubMed] [Google Scholar]
- 100.Yeh J, Ling Y, Karp JM, Gantz J, Chandawarkar A, Eng G, Blumling J, 3rd, Langer R, Khademhosseini A. Micromolding of shape-controlled, harvestable cell-laden hydrogels. Biomaterials. 2006;27:5391–5398. doi: 10.1016/j.biomaterials.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 101.Fernandes R, Wu LQ, Chen T, Yi H, Rubloff GW, Ghodssi R, Bentley WE, Payne GF. Electrochemically Induced Deposition of a Polysaccharide Hydrogel onto a Patterned Surface. Langmuir. 2003;19:4058–4062. [Google Scholar]
- 102.Cutler SM, Garcia AJ. Engineering cell adhesive surfaces that direct integrin alpha5beta1 binding using a recombinant fragment of fibronectin. Biomaterials. 2003;24:1759–1770. doi: 10.1016/s0142-9612(02)00570-7. [DOI] [PubMed] [Google Scholar]
- 103.Petrie TA, Capadona JR, Reyes CD, Garcia AJ. Integrin specificity and enhanced cellular activities associated with surfaces presenting a recombinant fibronectin fragment compared to RGD supports. Biomaterials. 2006;27:5459–5470. doi: 10.1016/j.biomaterials.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 104.Petrie TA, Reyes CD, Burns KL, Garcia AJ. Simple application of fibronectin-mimetic coating enhances osseointegration of titanium implants. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2008.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 106.Garcia AJ, Schwarzbauer JE, Boettiger D. Distinct activation states of alpha5beta1 integrin show differential binding to RGD and synergy domains of fibronectin. Biochemistry. 2002;41:9063–9069. doi: 10.1021/bi025752f. [DOI] [PubMed] [Google Scholar]
- 107.Akiyama SK, Aota S, Yamada KM. Function and receptor specificity of a minimal 20 kilodalton cell adhesive fragment of fibronectin. Cell Adhes Commun. 1995;3:13–25. doi: 10.3109/15419069509081275. [DOI] [PubMed] [Google Scholar]
- 108.Garcia AJ, Reyes CD. Bio-adhesive surfaces to promote osteoblast differentiation and bone formation. J Dent Res. 2005;84:407–413. doi: 10.1177/154405910508400502. [DOI] [PubMed] [Google Scholar]
- 109.Moursi AM, Damsky CH, Lull J, Zimmerman D, Doty SB, Aota S, Globus RK. Fibronectin regulates calvarial osteoblast differentiation. J Cell Sci. 1996;109(Pt 6):1369–1380. doi: 10.1242/jcs.109.6.1369. [DOI] [PubMed] [Google Scholar]
- 110.Moursi AM, Globus RK, Damsky CH. Interactions between integrin receptors and fibronectin are required for calvarial osteoblast differentiation in vitro. J Cell Sci. 1997;110(Pt 18):2187–2196. doi: 10.1242/jcs.110.18.2187. [DOI] [PubMed] [Google Scholar]
- 111.Benoit DS, Durney AR, Anseth KS. The effect of heparin-functionalized PEG hydrogels on three-dimensional human mesenchymal stem cell osteogenic differentiation. Biomaterials. 2007;28:66–77. doi: 10.1016/j.biomaterials.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 112.Kundu AK, Khatiwala CB, Putnam AJ. Extracellular matrix remodeling, integrin expression, and downstream signaling pathways influence the osteogenic differentiation of mesenchymal stem cells on poly(lactide-co-glycolide) substrates. Tissue Eng Part A. 2009;15:273–283. doi: 10.1089/ten.tea.2008.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.El-Amin SF, Attawia M, Lu HH, Shah AK, Chang R, Hickok NJ, Tuan RS, Laurencin CT. Integrin expression by human osteoblasts cultured on degradable polymeric materials applicable for tissue engineered bone. J Orthop Res. 2002;20:20–28. doi: 10.1016/S0736-0266(01)00062-6. [DOI] [PubMed] [Google Scholar]
- 114.Schneider GB, Zaharias R, Stanford C. Osteoblast integrin adhesion and signaling regulate mineralization. J Dent Res. 2001;80:1540–1544. doi: 10.1177/00220345010800061201. [DOI] [PubMed] [Google Scholar]
- 115.Cheng SL, Lai CF, Blystone SD, Avioli LV. Bone mineralization and osteoblast differentiation are negatively modulated by integrin alpha(v)beta3. J Bone Miner Res. 2001;16:277–288. doi: 10.1359/jbmr.2001.16.2.277. [DOI] [PubMed] [Google Scholar]
- 116.Keselowsky BG, Collard DM, Garcia AJ. Integrin binding specificity regulates biomaterial surface chemistry effects on cell differentiation. Proc Natl Acad Sci U S A. 2005;102:5953–5957. doi: 10.1073/pnas.0407356102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aucoin L, Griffith CM, Pleizier G, Deslandes Y, Sheardown H. Interactions of corneal epithelial cells and surfaces modified with cell adhesion peptide combinations. J Biomater Sci Polym Ed. 2002;13:447–462. doi: 10.1163/156856202320253956. [DOI] [PubMed] [Google Scholar]
- 118.Benoit DS, Anseth KS. The effect on osteoblast function of colocalized RGD and PHSRN epitopes on PEG surfaces. Biomaterials. 2005;26:5209–5220. doi: 10.1016/j.biomaterials.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 119.Kao WJ. Evaluation of protein-modulated macrophage behavior on biomaterials: designing biomimetic materials for cellular engineering. Biomaterials. 1999;20:2213–2221. doi: 10.1016/s0142-9612(99)00152-0. [DOI] [PubMed] [Google Scholar]
- 120.Kim TI, Jang JH, Lee YM, Ryu IC, Chung CP, Han SB, Choi SM, Ku Y. Design and biological activity of synthetic oligopeptides with Pro-His-Ser-Arg-Asn (PHSRN) and Arg-Gly-Asp (RGD) motifs for human osteoblast-like cell (MG-63) adhesion. Biotechnology Letters. 2002;24:2029–2033. [Google Scholar]
- 121.Susuki Y, Hojo K, Okazaki I, Kamata H, Sasaki M, Maeda M, Nomizu M, Yamamoto Y, Nakagawa S, Mayumi T, Kawasaki K. Preparation and biological activities of a bivalent poly(ethylene glycol) hybrid containing an active site and its synergistic site of fibronectin. Chem Pharm Bull (Tokyo) 2002;50:1229–1232. doi: 10.1248/cpb.50.1229. [DOI] [PubMed] [Google Scholar]
- 122.Mardilovich A, Craig JA, McCammon MQ, Garg A, Kokkoli E. Design of a novel fibronectin-mimetic peptide-amphiphile for functionalized biomaterials. Langmuir. 2006;22:3259–3264. doi: 10.1021/la052756n. [DOI] [PubMed] [Google Scholar]
- 123.Craig JA, Rexeisen EL, Mardilovich A, Shroff K, Kokkoli E. Effect of linker and spacer on the design of a fibronectin-mimetic peptide evaluated via cell studies and AFM adhesion forces. Langmuir. 2008;24:10282–10292. doi: 10.1021/la702434p. [DOI] [PubMed] [Google Scholar]
- 124.Grant RP, Spitzfaden C, Altroff H, Campbell ID, Mardon HJ. Structural requirements for biological activity of the ninth and tenth FIII domains of human fibronectin. J Biol Chem. 1997;272:6159–6166. doi: 10.1074/jbc.272.10.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Altroff H, Schlinkert R, van der Walle CF, Bernini A, Campbell ID, Werner JM, Mardon HJ. Interdomain tilt angle determines integrin-dependent function of the ninth and tenth FIII domains of human fibronectin. J Biol Chem. 2004;279:55995–56003. doi: 10.1074/jbc.M406976200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 127.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 128.Rao J, Lahiri J, Isaacs L, Weis RM, Whitesides GM. A trivalent system from vancomycin.D-ala-D-Ala with higher affinity than avidin.biotin. Science. 1998;280:708–711. doi: 10.1126/science.280.5364.708. [DOI] [PubMed] [Google Scholar]
- 129.Sancey L, Garanger E, Foillard S, Schoehn G, Hurbin A, Albiges-Rizo C, Boturyn D, Souchier C, Grichine A, Dumy P, Coll JL. Clustering and internalization of integrin alphavbeta3 with a tetrameric RGD-synthetic peptide. Mol Ther. 2009;17:837–843. doi: 10.1038/mt.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Carlson CB, Mowery P, Owen RM, Dykhuizen EC, Kiessling LL. Selective tumor cell targeting using low-affinity, multivalent interactions. ACS Chem Biol. 2007;2:119–127. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]
- 131.Mourez M, Kane RS, Mogridge J, Metallo S, Deschatelets P, Sellman BR, Whitesides GM, Collier RJ. Designing a polyvalent inhibitor of anthrax toxin. Nat Biotechnol. 2001;19:958–961. doi: 10.1038/nbt1001-958. [DOI] [PubMed] [Google Scholar]
- 132.Coussen F, Choquet D, Sheetz MP, Erickson HP. Trimers of the fibronectin cell adhesion domain localize to actin filament bundles and undergo rearward translocation. J Cell Sci. 2002;115:2581–2590. doi: 10.1242/jcs.115.12.2581. [DOI] [PubMed] [Google Scholar]
- 133.Christman KL, Enriquez-Rios VD, Maynard HD. Nanopatterning proteins and peptides. Soft Matter. 2006;2:928–939. doi: 10.1039/b611000b. [DOI] [PubMed] [Google Scholar]
- 134.Krammer A, Craig D, Thomas WE, Schulten K, Vogel V. A structural model for force regulated integrin binding to fibronectin’s RGD-synergy site. Matrix Biol. 2002;21:139–147. doi: 10.1016/s0945-053x(01)00197-4. [DOI] [PubMed] [Google Scholar]