Abstract
Measurement of the level of a specific protein can be an important parameter to discern as that can change and reflect disease status. A number of methods have been developed to quantitate the level of a protein, some amenable to high throughput screening. A method is described to measure the total level of the tumor suppressor p53 using scintillation proximity assay (SPA) beads and radiolabeled streptavidin. Three different cell extracts were used, with one used to develop the standard curve for the amount of p53. This method allows the specific detection of p53 in the range of 50 to 300 pg in 10 μl of an extract. While this detection is less than what can be detected by commercially available enzyme linked immunosorbant assay (ELISA) kits, the SPA compares favorably on time required and cost. This new assay also has the potential to be coupled with measurements for p53 DNA binding, a unique aspect of this approach.
Keywords: DNA binding, ELISA, p53, SPA, streptavidin, tumor suppressor
1. Introduction
A large number of transcription factors have been implicated in the control of the cell cycle and if one of these is mutated, it can result in uncontrolled proliferation, namely cancer. One of these transcription factors is the p53 protein. This phosphonuclear protein is found primarily at low levels in normal cells, but the amount of this protein can rapidly increase once the cells are treated with DNA damaging agents (Kruse and Gu 2009). The rapid change in the level of the p53 protein is in part due to a reduction in the ubiquitinylation and subsequent degradation of the protein mediated by the Mdm-2 protein (Marine and Lozano 2009). The p53 protein can bind to DNA and regulate a variety of genes including those controlling the cell cycle and implementing DNA repair. Mutation in the gene for the p53 protein is also linked to cancer. Some mutant p53 proteins show a reduced degradation of the protein due to changes in the binding to Mdm-2 (Marine and Lozano 2009). Accumulation of the p53 protein in tumor samples using immunohistochemistry has been used as one indicator of the transformation of cells (Hall and McCluggage 2006). Thus, the level of the total p53 protein is an important parameter to measure when analyzing tumor cells.
There are a variety of means for measuring the level of a specific protein such as enzyme linked immunosorbant assay (ELISA), western blots or immunohistochemistry. None of these three methods are ideal for screening large numbers of tumor samples quickly and getting quantitative numbers. Because of our experience with scintillation proximity assay (SPA) beads (Gal et al. 2006; Chandrachud and Gal, 2009), we were interested in setting up a method using these beads to measure the amount of p53 protein. SPA beads contain embedded scintillant that produces light when radioactive compounds are brought in proximity to the bead. Binding of these radioactively labeled compounds to the beads can occur through specific antibodies or other binding components (reviewed in Glickman et al. 2008). Since unbound material does not provide significant signal and need not be removed, this assay is easily automated since in many cases all of the components can be added together. This assay has been used to measure growth factors as well as other molecules and enzymes (Khawaja 2007; reviewed in Glickman et al. 2008). These beads have recently been used to develop an SPA that measures DNA binding of human p53 protein (Gal et al. 2006; Chandrachud and Gal, 2009) and are successfully used here to create an assay to measure the total level of this protein in human cell extracts.
2. Materials and methods
2.1 Preparation of nuclear extracts and ELISA for p53
Three human thyroid cancer cell lines were used in this study which each have a different p53 gene mutation. The ARO cells have an arginine to histidine mutation at codon 273, the WRO cells have mutation at codon 223 (proline to leucine) and the NPA cells have a glycine to glutamine mutation at codon 266 (Fagin et al. 1993). Thyroid cancer cell lines ARO, NPA and WRO (provided by Fran Carr now at the University of Vermont) were grown in RPMI-1640 (Lonza, Walkersville, MD) with 10% fetal calf serum. The method for preparing nuclear extracts was based on the one used by Jagelská and colleagues (2002). The total protein concentration of the cell extract was determined using bicinchoninic acid assay (Sigma St. Louis, MO). To determine the concentration of the p53, ELISA's (p53 ELISA Kit Pantropic, Calbiochem-EMD, La Jolla CA) were performed. The WRO cell extract had p53 that was not detectable by ELISA, but western blots done using the WRO cell extract showed the presence of this protein. Hence for this extract, the concentration of the p53 protein was determined by comparing the intensities of the p53 protein detected using western blots from the WRO cell extract to that in other cell extracts that had been quantitated using ELISA.
2.2 Total p53 Scintillation Proximity Assay (SPA)
For each assay, 300 ng anti-human p53 antibody (pAb421, Calbiochem), 25ng biotinylated polyclonal anti-human p53 antibody (R&D Systems, Minneapolis, MN), 0.1 pmole 35S-labeled streptavidin (200–500Ci/mmole, GE Healthcare, Piscataway, NJ) and 0.5–10 μl of cell extract or buffer were mixed in a final volume of 30 μl in low salt buffer (20mM Hepes, pH 7.5, 1mM EDTA, 1mM DTT, 10mM (NH4)2SO4, 10mM KCl, 0.2% Tween-20). The mixed samples were added into a white 96-well plate (Matrix Technologies Corporation; Hudson, NH) and incubated at room temperature for 5–30 minutes, followed by the addition of 1 mg SPA PVT Protein A or anti-mouse beads (GE Healthcare) diluted in 100 μl of 1× high salt buffer (25mM Tris, pH7.5, 1mM EDTA, 1mM DTT, 10mM (NH4)2SO4, 130mM NaCl, 0.2% Tween) to each well. The plate was counted in a TopCount NXT TM (Packard Instrument Company, Meriden, CT) for 2–5 hours using the 35S-PVT-SPA program. Assays were duplicated for each cell extract. Statistical analysis of the data was done using one way ANOVA followed by a pairwise Student's t-test.
3. Results and Discussion
3.1 Description of the assay and initial tests
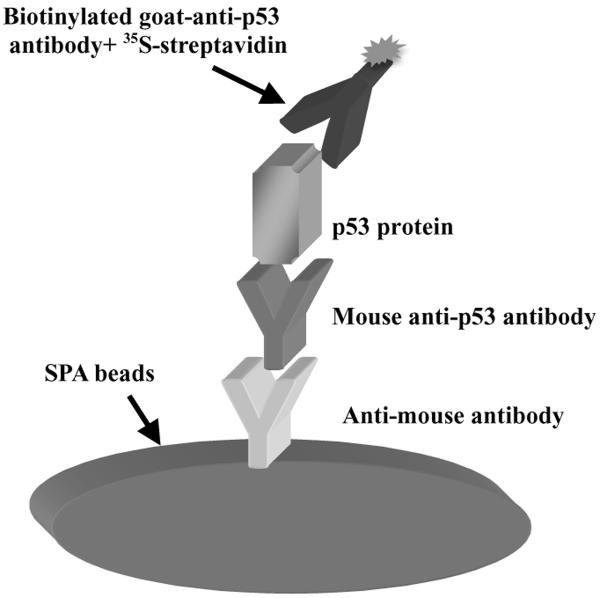
A DNA binding assay for analyzing the tumor suppressor protein p53 using 3H-labeled DNA and SPA beads has been developed (Gal et al. 2006). This assay is sensitive and fast and can work with recombinant p53 as well as p53 in nuclear extracts of human tissue culture cells (Chandrachud and Gal 2009). Because the level of the p53 protein can change dramatically under different stress conditions or in different cancers, a means to measure the total level of this protein in a manner that potentially could be compatible with the DNA binding assay is desirable. This assay was configured much like an ELISA with a monoclonal antibody on the SPA beads acting as capture antibody and a biotinylated anti-p53 polyclonal antibody acting as detector antibody (Figure 1A). The SPA beads are induced to produce light because of the presence of 35S-streptavidin binding to the biotinylated antibody. The DNA binding assay is arranged similarly with the mouse monoclonal antibody binding to the protein A on the SPA beads but with 3H-DNA added instead (Gal et al. 2006). The detector antibody chosen was polyclonal and derived from goat since that antibody should have minimal interaction with the protein A on the SPA beads (Thermoscientific technical information). However, there was evidence for direct interaction of this antibody with the protein A SPA beads (data not shown), as noted in another source (Eliasson et al. 1988), and so SPA beads with anti-mouse antibodies were used to reduce that background.
Figure 1.A. Arrangement of Total SPA and Initial values.
A. Arrangement of total SPA to measure the amount of p53 protein. Scintillation proximity assay (SPA) beads having immobilized anti-mouse antibodies bind a mouse monoclonal antibody to p53 that can then bind the p53 protein in an extract. Subsequent binding of a biotinylated polyclonal anti-p53 antibody with 35S-labeled streptavidin to the immobilized p53 can bring the radiolabel in close proximity to the SPA beads. The beta-particle emission from the radiolabel then excites the SPA beads to produce light detected by a scintillation counter like the TopCount.
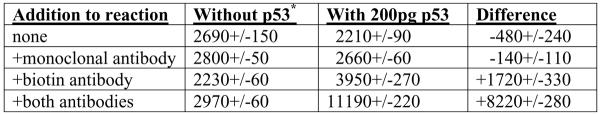
B. Values for initial controls of the SPA. All samples contained 35S-streptavidin, buffers and SPA beads, but to some reactions, the indicated antibodies and 200 pg p53 from a nuclear cell extract of ARO cells (quantitated using an ELISA) were added as described in Section 2. At 240 minutes after addition of the SPA beads, counts were taken of duplicate samples to give the presented values. A significant difference (p<0.005) between samples with both antibodies and samples with only one antibody in the presence of p53 was seen. The data presented are from a representative experiment of 4 similar ones performed.
This reaction was first performed using a nuclear extract from a thyroid cancer cell line, ARO. These extracts contain a significant amount of p53 detected using western blots (Chandrachud and Gal, 2009). First, several controls with this reaction were done to determine the background counts detected when certain components of the assay were absent (Figure 1B). When only 35S-streptavidin and anti-mouse SPA beads in the presence or absence of the p53 protein were added, low average background counts were obtained (between 2200 and 2700 cpm; see Figure 1B). The addition of the mouse monoclonal anti-p53 antibody did not increase the counts significantly. However, when the biotinylated goat polyclonal anti-p53-antibody was added, the counts increased in the presence of p53 protein even without the mouse monoclonal anti-p53 antibody (compare 3950 cpm with 2230 cpm; see Figure 1B). This result suggested that some of the biotinylated goat polyclonal anti-p53-antibodies may bind weakly to the anti-mouse antibodies on the SPA beads. If these immobilized antibodies then bound the p53 protein and other antibodies attach to a different epitope of p53, there may exist an exposed biotin allowing the 35S-streptavidin to come in proximity to the SPA beads. This apparent sandwich formation could not be disrupted by the presence of 10 μg BSA which showed the specificity of the assay for the presence of p53 (data not shown). In the presence of both antibody types, there was a more significant increase in counts that was dependent on the presence of the p53 protein (compare 11190 cpm with 2970 cpm, see Figure 1B). Standard curves with different amounts of the p53 protein in the presence of both antibodies and the anti-mouse SPA beads were next performed.
3.2 Detection limits of the assay
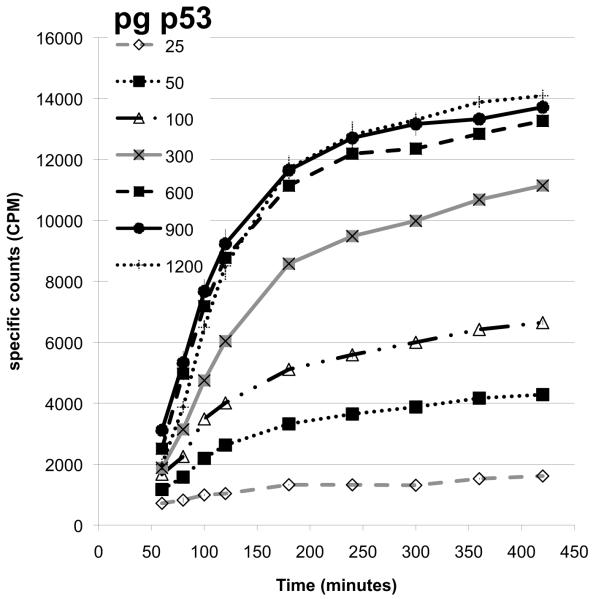
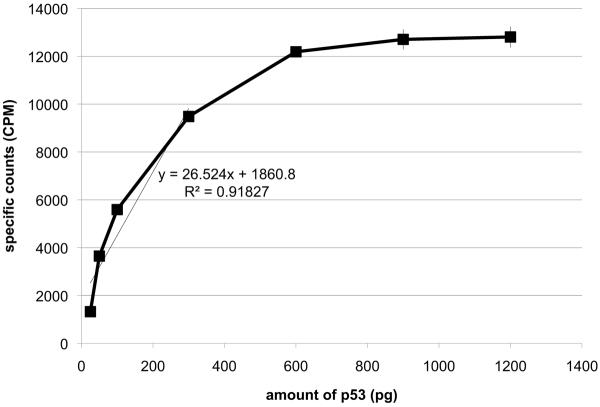
The amount of the p53 protein in the ARO nuclear cell extracts was first measured using ELISA and western blots, then, this protein was added in a range of 25 to 1200 pg to the total p53 SPA (Figure 2A). The minimal amount of p53 protein detected given the reagents was 50 pg (2× counts over background), while the counts maximized for 300 pg of p53 (Figure 2B). This remained consistent over several different experiments. It was interesting to note that the time to equilibrium binding appeared to indirectly vary with the amount of p53 added (Figure 2A). This makes sense if the process is a concentration driven reaction so that at lower p53 protein levels, binding to the 2 different antibodies takes longer. In any case, the assay allowed detection of the p53 protein in the extract in a concentration dependent manner to produce a linear standard curve in this extract between 50 and 300 pg of the p53 protein (Figure 2B). Increased concentrations of the monoclonal antibody and increased amounts of SPA beads were tested, but this only modestly changed the detection limits and was deemed an unnecessary use of material. Therefore, with this amount of antibodies and SPA beads, the range of detection of p53 from the ARO cell nuclear extract appeared to be between 50 and 300 pg.
Figure 2. Variation of SPA counts with different amounts of ARO cell extract.
A. Counts at various times with different amounts of nuclear cell extract. To a reaction, as configured in Section 2, different amounts (25–1200 pg of p53) of an ARO nuclear cell extract were added and the counts determined at regular intervals after the addition of the SPA PVT anti-mouse beads.
B. Standard curve of the data in panel A at 240 minutes. This curve indicates saturation of the assay above 300pg of p53. A trendline for the first 4 points was calculated and is presented on the graph with the equation. The data presented are from a representative experiment of 3 similar ones performed.
When comparing the total p53 SPA described here with commercially available ELISA kits, the former assay is less sensitive for detecting the p53 protein. The commercial ELISAs indicate that they can detect as little as 10 pg of p53 in a volume of 100 μl (Calbiochem literature). In the present configuration of the SPA, the p53 protein sample is in a small volume (10 μl) that limits significantly the amount of material that can be detected. Minimally 50 pg of p53 in that volume can be detected using the SPA. Thus, this assay is about 50-fold less sensitive than some commercial ELISA kits in measuring accurately the amount of this tumor suppressor protein. The linear range for detection using the SPA is similar to that for the ELISA, namely 50–300 pg for the SPA versus 10–150 pg for the ELISA. Despite being less sensitive, the SPA compares favorably on price, user time and time to information when examined relative to a commercial ELISA kit. To quantitate a total of 42 samples in duplicate with 4 controls and blanks in duplicate, the costs of the SPA are about $300 while the single ELISA kit costs about $500. Because no washing steps are involved in the SPA, it takes much less user time and effort to conduct the assay when compared to a standard ELISA (5 minutes time post-plate set-up versus 1.5 hours for the ELISA). The SPA provides the information on the level of p53 within 4 hours of addition of all of the reagents while the standard ELISA requires an overnight incubation and other subsequent steps. These differences make the currently described SPA to measure the total p53 levels in extracts an attractive alternative to ELISA.
3.3 Using the assay with other extracts
The detection of the p53 was next tested using nuclear extracts from 2 other human thyroid cancer cell lines NPA and WRO that have different mutations in the p53 protein (Fagin et al. 1993). The DNA binding in these extracts using the SPA as well as a streptavidin magnetic bead assay with biotinylated DNA has been measured (Chandrachud and Gal, 2009). As above, the amount of the p53 protein in nuclear extracts of these cells was first measured using an ELISA or western blots, then those protein extracts were tested in the total p53 SPA using the p53 from the ARO cells as a standard. In this way, comparison of the level of the p53 in the SPA with the values from the ELISA and western blots could be done. Overall the assays do agree with some extract specific differences (Table I). Two different nuclear extracts from the NPA cells were tested at two different amounts in the SPA, and it was found that the SPA indicated a higher total amount of the p53 protein than the ELISA or western blots. The amount detected in the latter assays was within the margin of error for the SPA for one of the extracts (Table I). For the WRO extract, the ELISA did not detect the protein potentially due to the nature of the mutation and the recognition specificity of the antibodies, but the detection of the p53 by the western blot and the SPA were very similar for two different amounts of the extract. For this data, one monoclonal antibody (pAb421) was used for capturing the p53 onto the anti-mouse SPA beads. Two other monoclonal antibodies, DO-1 and DO-7 (from Calbiochem) were tested, and the former could detect the p53 from both ARO and NPA extracts but neither worked for the protein from WRO cells. All three antibodies could detect the p53 protein from these cells on western blots (data not shown). It is possible the difference in recognition of the SPA is due to the differences in the antigens detected by the antibodies and the conformation of the mutant p53 proteins.
Table I.
Determination of p53 protein amounts from different nuclear cell extracts.
| Different methods for determining p53 amounts | |||
|---|---|---|---|
| Cell source | ELISA | western | SPA |
| NPA 1 | 50+/−2 | 55+/−5 | 80+/−20 |
| 100+/−5 | 110+/−10 | 166+/−8 | |
| NPA 2 | 50+/−2 | 55+/−5 | 70+/−20 |
| 100+/−5 | 110+/−10 | 120+/−20 | |
| WRO | ND | 42+/−8 | 50+/−10 |
| ND | 80+/−10 | 73+/−7 | |
The amount of p53 (in pg) in the nuclear cell extracts was determined using 3 different methods- ELISA, western blots and SPA as described in Section 2. For the SPA, the values were determined from a standard curve with one ARO nuclear cell extract and then the counts from the other 3 extracts were compared to that standard curve. The equation for the SPA determined standard curve 240 minutes after adding the SPA beads was y=29.9x+1151 with an R2 value of 0.9926. These data are from one representative experiment out of a total of 6 similar experiments performed. ND- the p53 in the WRO cell extract was not detectable by ELISA.
This new method for quantitating the level of a specific protein takes advantage of the available antibodies for p53 and scintillation proximity assay (SPA) beads to allow detection of radioactive substances. Improvements to the SPA as described can certainly be envisaged for the future. The use of the specific antibodies chosen allowed detection of 3 different mutant proteins and the wild-type protein (not shown) in extracts. Many other anti-p53 antibodies exist and could be tested in this SPA for ones that are mutant p53 specific or can detect a broader range of p53 mutant proteins. To reduce the background from direct interaction of the detector polyclonal antibodies with the SPA beads, one could prepare Fab fragments and label them with biotin.
Using this SPA arrangement for detecting the amount of p53, it may also be possible to measure other functional aspects of the protein. Preliminary data indicate this arrangement is amenable to measuring both DNA binding and the amount of p53 protein (Gal, unpublished data). Thus, this new assay has some distinct advantages over classical ELISAs and could be used to measure the changes in p53 protein during cell transformation. The detection of DNA binding using the SPA beads ranges from 50–200 pg of p53 (Chandrachud and Gal, 2009; Gal et al. 2006), so the lower detection limit of the total p53 SPA described here is not a problem.
4. Conclusions
This article describes the development of a new method to measure the total level of the p53 protein using SPA beads. The general arrangement of this SPA is expected to allow the measurement of DNA binding of this protein at the same time. This configuration may be useful for other systems as well where a binding reaction showing activity and antibody binding to show total level of a protein could be measured simultaneously. There are a number of systems where this approach could be successfully applied in the future.
Acknowledgements
The work was supported by a National Institutes of Health grant (R15 CA101783-01A1) and by the US Department of Energy (Grant DE-FG02-06ER64281) as a subcontract from SUNY-Utica both to SG. The work on developing the SPA for DNA binding was originally funded by Amersham Pharmacia Biotech, the original commercial source of the SPA beads used in this work. Since that time, the authors have had no financial support from the company or it's new entity, GE Healthcare.
Abbreviations
- cpm
counts per minute
- ELISA
enzyme linked immunosorbant assay
- ND
not detected
- pg
picograms-
- SPA
scintillation proximity assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note added in preparation During the later stage of this research, the company from which the 35S- labeled streptavidin was obtained discontinued this product. Another source of this material has not yet been found. It is however likely that the polyclonal antibodies or the streptavidin could be synthesized with a radiolabel like 14C or 35S, but that has not yet been fully explored by this group. During the preparation of the manuscript, GE Healthcare sold the rights to the SPA beads to Perkin Elmer who now supply this product.
References
- Chandrachud U, Gal S. Three assays show differences in binding of wild-type and mutant p53 to unique gene sequences. Technology in Cancer Research and Treatment. 2009;8:445. doi: 10.1177/153303460900800606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson M, Olsson A, Palmcrantz E, Wiberg K, Inganäs M, Guss B, Lindberg M, Uhlèn M. Chimeric IgG-binding receptors engineered from staphylococcal protein A and streptococcal protein G. Journal of Biological Chemistry. 1988;263:4323. [PubMed] [Google Scholar]
- Fagin JA, Matsuo K, Karmakar A, Chen DL, Tang SH, Koeffler HP. High prevalence of mutations of the p53 gene in poorly differentiated human thyroid carcinomas. Journal of Clinical Investigations. 1993;91:179. doi: 10.1172/JCI116168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal S, Cook JR, Howells L. Scintillation proximity assay for DNA binding by human p53. Biotechniques. 2006;41:303. doi: 10.2144/000112222. [DOI] [PubMed] [Google Scholar]
- Glickman JF, Schmid A, Ferrand S. Scintillation proximity assays in High-Throughput screening. Assay and Drug Development Technologies. 2008;6:433. doi: 10.1089/adt.2008.135. [DOI] [PubMed] [Google Scholar]
- Hall PA, McCluggage WG. Assessing p53 in clinical contexts: unlearned lessons and new perspectives. Journal of Pathology. 2006;208:1. doi: 10.1002/path.1913. [DOI] [PubMed] [Google Scholar]
- Jagelská E, Brázda V, Pospisilová S, Vojtesek B, Palecek E. New ELISA technique for analysis of p53 protein/DNA binding properties. Journal of Immunological Methods. 2002;267:227. doi: 10.1016/s0022-1759(02)00182-5. [DOI] [PubMed] [Google Scholar]
- Khawaja XZ. Development of a scintillation proximity assay for human insulin-like growth factor-binding protein 4 compatible with inhibitor high-throughput screening. Analytical Biochemistry. 2007;366:80. doi: 10.1016/j.ab.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Lozano G. Mdm2-mediated ubiquitinylation: p53 and beyond. Cell Death and Differentiation. 2010;17:93. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]