Abstract
Cleft lip/palate is a defect of craniofacial development. In previous reports, chromosome 6q has been suggested as a candidate region for cleft lip/palate. A multipoint posterior probability of linkage analysis of multiplex families from the Philippines attributed an 88% probability of harboring a cleft-susceptibility gene to a narrower region on bands 6q14.2-14.3. We genotyped 2732 individuals from families and unrelated individuals with and without clefts to investigate the existence of possible cleft-susceptibility genes in this region. We found association of PRSS35 and SNAP91 genes with cleft lip/palate in the case-control cohort and in Caucasian families. Haplotype analyses support the individual associations with PRSS35. We found Prss35 expression in the head and palate of mouse embryos at critical stages for palatogenesis, whereas Snap91 was expressed in the adult brain. We provide further evidence of the involvement of chromosome 6q in cleft lip/palate and suggest PRSS35 as a novel candidate gene.
Keywords: cleft lip/palate, fine mapping, chromosome 6q, polymorphism
Introduction
Cleft lip with or without cleft palate (cleft lip/palate) results from defects in growth and patterning of the facial primordia. Over 300 syndromes, including some that are either chromosomal or Mendelian, might present a cleft of the lip and/or the palate as a feature, and comprise about 30% of all cleft cases. The remaining 70% are attributed to isolated, non-syndromic clefts, without any associated structural anomaly (Gorlin et al., 2001). In the United States, approximately 6800 children with cleft lip/palate are born each year, representing $101,000/each of lifetime costs (Centers for Disease Control and Prevention, 2006).
The etiology of non-syndromic cleft lip/palate is multifactorial, and from 3 to 14 genes may be involved in addition to environmental factors (Schliekelman and Slatkin, 2002). Several genes/loci have shown positive linkage and/or association results in cleft lip/palate, cleft palate only, or both. To date, the most remarkable finding was the association between IRF6 gene variants and non-syndromic cleft lip/palate (Zucchero et al., 2004), replicated in several other populations (reviewed by Vieira, 2008). Recent evidence attributed its effect to a point mutation in a TFAP2A binding site in an enhancer of the IRF6 gene promoter (Rahimov et al., 2008). Polymorphic variants or rare point mutations in other genes have also been found in individuals with clefts, including MSX1, TGFB3, FOXE1, PVR, PVRL1, and FGFs (reviewed by Vieira, 2008). Recently, a variant on the regulatory region of the PDGF-C gene has been shown to decrease transcriptional activity and to be associated with cleft lip/palate (Choi et al., 2008).
Genome-wide linkage scans of anonymous markers have provided important clues to narrowing down the number of candidate genes for cleft lip/palate. A genome scan of Chinese multiplex families revealed positive linkage results (LOD = 1.41) for cleft lip/palate near marker D6S1031 on chromosome 6q14.2 (Marazita et al., 2002). The imputed posterior probability of linkage (PPL) (Logue and Vieland, 2004) for multiplex families from the Philippines identified a peak at ~95cM on chromosome 6q14.2-14.3 to which a ~88% probability of harboring a cleft-susceptibility gene was attributed (Govil et al., unpublished observations). Interestingly, this linkage peak was located between D6S1031 and D6S1056, overlapping with the results of the Chinese genome scan (Marazita et al., 2002). Taken together, these point toward chromosome 6q as having a likely role in human clefting.
To investigate the existence of possible cleft-susceptibility genes in the 6q14.2-14.3 region, we used densely spaced markers to genotype 2732 individuals from families and unrelated persons with and without clefts. We also investigated embryonic expression of associated genes during mouse embryonic development.
Study Population & Methods
Study Population
The study population consisted of 2732 individuals (867 affected with cleft lip/palate) and unaffected family members and unrelated control individuals. Family-based samples came from multiplex families of the United States, Spain, Turkey, Guatemala, and China, and from family trios ascertained through the ECLAMC (Latin American Collaborative Study of Congenital Malformations) registry, a hospital-based birth defects registry with sites across South America (Castilla and Orioli, 2004). Case-control samples were all ethnically matched Caucasian individuals from Brazil (Table 1).
Table 1.
Details of Family-based and Case-Control Cohorts Investigated in the Study
| Affected Individuals by |
Families |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Regional Group | Ancestral Origin | Number of Families | Number of Individuals | Affected Individuals | Unaffected Individuals | Cleft Lip | CL/Pb | CLO+CL,CLP (no CPO affecteds)c | CLO+CLP+CL, CLP (no CPO affecteds)d |
| Family-based Cohorts | |||||||||
| United States | Pittsburgh | 68 | 387 | 101 | 286 | 31 | 70 | 35 | 51 |
| Saint Louis | 21 | 104 | 33 | 71 | 6 | 27 | 10 | 19 | |
| Central America | Guatemala | 77 | 514 | 93 | 421 | 20 | 73 | 22 | 77 |
| South America | ECLAMCa | 171 | 513 | 171 | 342 | — | — | — | — |
| Europe | Spain | 36 | 136 | 43 | 93 | 10 | 33 | 9 | 33 |
| Turkey | 29 | 288 | 38 | 250 | 17 | 21 | 14 | 29 | |
| East Asia | China | 60 | 180 | 60 | 120 | 14 | 46 | 13 | 60 |
| Subtotal Families | 462 | 2122 | 539 | 1583 | 98 | 270 | 103 | 269 | |
| Case-Control Cohort | |||||||||
| South America | Brazil | 0 | 610 | 328 | 282 | 4 | 324 | — | — |
| TOTAL | 462 | 2732 | 867 | 1865 | 103 | 594 | 103 | 269 | |
ECLAMC, Estudio Colaborativo de Malformaciones Congenitas (Latin American Collaborative Study of Congenital Malformations).
CLO + CL,CLP (no CPO affecteds) = all families where all affecteds are CLO + all families where at least one affected is CLO, and one affected is CLP, excluding any family where an affected is CPO.
CLO + CLP + CL,CLP (no CPO affecteds) = all families where all affecteds are CLO + all families where all affecteds are CLP + all families where at least one affected is CLO, and one affected is CLP, excluding any family where an affected is CPO.
All cases had non-syndromic cleft lip with or without cleft palate. Non-syndromic status was determined according to patient records. Families were ascertained through probands, and additional relatives were recruited. Individuals presenting cleft palate only or unknown cleft types, and controls with family history of clefting were excluded. The study was approved by the local review boards and accredited by the University of Pittsburgh Institutional Review Board as an umbrella to the international collaboration efforts. Informed consent was obtained from participants and from parents/guardians of children under 15 yrs of age. Saliva samples were collected as the source of genomic DNA. DNA extraction followed established protocols.
Selection of Candidate Loci and Genetic Markers and Genotyping Procedures
Single-nucleotide polymorphisms spanning the chromosome 6q14.2-14.3 region were selected by means of the International HapMap Project database (http://www.hapmap.org). We generated a linkage disequilibrium plot of the candidate region where 5 genes and 528 polymorphisms were identified (Appendix). We used the function “Download tag SNP data” and selected 26 polymorphisms as representative of the polymorphisms in the region. We selected polymorphisms that maximally represent the linkage disequilibrium structure of a given region, to avoid redundant information (Carlson et al., 2004). Preference was given to polymorphisms with high heterozygosity levels and different minor allele frequencies, to avoid intermarker linkage disequilibrium.
Genotyping was performed with Taqman chemistry (Ranade et al., 2001) on an automatic sequence-detection instrument (ABI Prism 7900HT, Applied Biosystems, Foster City, CA, USA).
Details of the selected polymorphisms are available in the Appendix.
Statistical Analyses
Chi-square and Fisher exact tests were used for case-control comparisons and determination of fit to Hardy-Weinberg equilibrium. Genotype and allele frequencies of each polymorphism were compared between cases and controls. We used Bonferroni correction to adjust for multiple testing, considering a significance level of 0.05 divided by the number of independent tests (26) to give a corrected p-value (α = 0.002).
For family-based analyses, we tested for linkage disequilibrium between marker alleles and cleft lip/palate using the Family Based Association Test (FBAT) (Horvath et al., 2001). We examined the transmission of alleles from heterozygous parents to affected offspring. We tested each population individually and as a pooled Caucasian (US, Madrid, and Turkey) and total family data set (US, Guatemala, Spain, Turkey, and China). We further analyzed the families according to cleft subgroups: (1) families where all affecteds have cleft lip only plus families where at least one affected has cleft lip only, and one affected has cleft lip and palate; and (2) families where all affecteds have cleft lip only plus families where all affecteds have cleft lip and palate plus families where at least one affected has cleft lip only, and one affected has cleft lip and palate.
We performed haplotype analyses using the function ‘hbat’ of the FBAT software. We created haplotypes using 2-, 3-, and 4-sliding windows.
Gene Expression Analyses
We used real-time PCR to investigate the expression of Prss35 and Snap91 during embryonic development. We used total, head, and palate mRNA of mouse embryos (Zyagen Laboratories, San Diego, CA, USA) at different stages of pregnancy [embryonic days (ED) 10-18]. As a positive control for Prss35 expression, we used cDNA obtained from the ovaries of mice undergoing a stimulated estrous cycle (the period of peak Prss35 expression) (Miyakoshi et al., 2006). Brain cDNA (Clontech, Mountain View, CA, USA) served as a positive control for Snap91. β-actin (Actb) was used as endogenous control. [Primer sequences and reaction conditions are available in the Appendix.] Products were resolved in agarose gel electrophoresis stained with ethidium bromide. Images were captured by means of a GelDoc 2000 system and Quantity One software (BioRad Laboratories, Hercules, CA, USA). Products were purified and sequenced at the Oregon National Primate Research Center Molecular and Cell Biology Core Laboratory (Beaverton, OR, USA) for confirmation of the identity of the amplicons.
Results
Association Analyses
We tested 26 single-nucleotide polymorphisms spanning the 5 genes present on chromosome 6q14.2-14.3 (PRSS35, SNAP91, 401268, C6orf159, and CYB5R4) for association with cleft lip/palate in a large sample cohort. After adjusting for multiple testing, we found significant associations between an intronic marker in PRSS35 (rs7753918) with cleft lip/palate in the case-control cohort (p = 0.00001). In addition, 2 intergenic markers near SNAP 91 (rs6454338 and rs10943957) showed significant association in the US (p = 0.001) and pooled Caucasian (p = 0.002) families. Borderline associations were also seen with rs10943957 in the US (p = 0.007) families, with CYB5R4 (rs6940766) in the case-control (p = 0.005), and with SNAP91 (rs217325) in the ECLAMC cohort (p = 0.006) (Table 2).
Table 2.
Summary of Results for Association Tests with Markers in the Chromosome 6q14.2-14.3 Region and Cleft Lip/Palate in the Studied Populations
| Family-baseda |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case-Controla |
Individual Populations |
Groups |
|||||||||
| SNP | Gene | Brazil | Pittsburgh | St. Louis | Guatemala | Spain | Turkey | China | ECLAMC | Caucasian | Pooledb |
| rs10943944 | PRSS35 | 0.01 | 0.86 | 0.65 | 0.97 | 1.00 | 1.00 | 0.35 | 0.18 | 1.00 | 0.65 |
| rs7753918 | PRSS35 | 0.00001 | 0.73 | 0.32 | 0.35 | 0.25 | 0.74 | 0.56 | 0.18 | 0.69 | 0.98 |
| rs9449648 | PRSS35 | 0.98 | 0.22 | 0.18 | 0.67 | 0.81 | 0.08 | 0.19 | — | 0.17 | 0.06 |
| rs1171114 | PRSS35 | 0.06 | 0.75 | 0.93 | 0.51 | 0.68 | 0.06 | 0.07 | — | 0.58 | 0.26 |
| rs512140 | PRSS35 | 0.17 | 0.04 | 0.33 | 0.35 | 0.68 | 0.44 | 0.49 | — | 0.09 | 0.04 |
| rs1171105 | — | 0.10 | 0.36 | 0.39 | 0.80 | 0.85 | 0.69 | 0.32 | — | 0.34 | 0.30 |
| rs2023238 | SNAP91 | 0.55 | 0.22 | 0.93 | 0.55 | 0.48 | 0.41 | 0.11 | — | 0.41 | 0.57 |
| rs9294279 | SNAP91 | 0.12 | 0.18 | 0.69 | 0.23 | 0.85 | 0.35 | 0.16 | — | 0.25 | 0.35 |
| rs3798867 | SNAP91 | 0.23 | 0.55 | 0.74 | 0.74 | 0.14 | 0.03 | 0.02 | — | 0.22 | 0.03 |
| rs217325 | SNAP91 | 0.05 | 0.11 | 0.92 | 0.71 | 0.32 | 0.16 | 0.16 | 0.006 | 0.23 | 0.38 |
| rs217308 | SNAP91 | 0.60 | 0.55 | 0.74 | 0.54 | 0.18 | 0.06 | 0.02 | — | 0.42 | 0.04 |
| rs755101 | SNAP91 | 0.50 | 0.29 | 0.88 | 0.23 | 0.43 | 0.05 | 0.45 | — | 0.04 | 0.02 |
| rs217290 | SNAP91 | 0.65 | 0.55 | 0.74 | 0.64 | 0.26 | 0.03 | 0.02 | — | 0.28 | 0.03 |
| rs217289 | SNAP91 | 0.27 | 0.63 | 0.74 | 0.80 | 0.32 | 0.03 | 0.04 | — | 0.28 | 0.05 |
| rs624076 | — | 0.18 | 0.02 | 0.32 | 0.16 | 0.51 | 0.82 | 0.45 | — | 0.12 | 0.03 |
| rs614565 | — | 0.13 | 0.09 | 0.56 | 0.74 | 0.37 | 0.81 | 0.50 | 0.80 | 0.09 | 0.07 |
| rs6454338 | — | 0.22 | 0.001 | 1.00 | 0.91 | 0.24 | 0.43 | 0.82 | — | 0.01 | 0.11 |
| rs10943957 | — | 0.25 | 0.007 | 1.00 | 0.69 | 0.16 | 0.41 | 0.81 | — | 0.002 | 0.09 |
| rs1325474 | — | 0.09 | 0.37 | 1.00 | 0.11 | 0.32 | 0.16 | 0.30 | 0.34 | 0.38 | 0.04 |
| rs9350989 | C6orf159 | 0.02 | 0.56 | — | 0.17 | 0.32 | 0.56 | 0.20 | — | 0.59 | 0.15 |
| rs9353149 | CYB5R4 | 0.24 | 0.32 | 0.56 | 0.08 | 0.53 | — | 0.90 | — | 0.32 | 0.14 |
| rs2324482 | CYB5R4 | 0.17 | 0.26 | — | 0.17 | 0.32 | 0.56 | 0.53 | — | 0.37 | 0.11 |
| rs6940766 | CYB5R4 | 0.005 | 0.28 | 0.41 | 0.30 | 0.16 | 0.02 | 0.32 | 0.74 | 0.45 | 0.47 |
| rs1998742 | CYB5R4 | 0.04 | 0.75 | 0.56 | 0.13 | 0.86 | 0.74 | 0.07 | — | 0.69 | 0.46 |
| rs1325469 | CYB5R4 | 0.12 | 0.28 | 0.32 | 0.16 | 0.26 | 0.08 | 0.39 | — | 0.48 | 0.08 |
| rs7770749 | CYB5R4 | 0.42 | 0.59 | — | 0.78 | 0.71 | — | — | — | 0.83 | 1.00 |
Bold indicates statistically significant difference (α = 0.002). Italic indicates borderline association.
Comprises analysis of Pittsburgh, St. Louis, Guatemala, Spain, Turkey, and Beijing families pooled together. Cells with no numerical values represent untyped or uninformative markers in the respective population.
Under a nominal value of 0.05, markers in PRSS35 (rs1171114 and rs512140) and markers in or flanking SNAP91 (rs9294279, rs624076, rs10943957) were associated with families where all affecteds have cleft lip only plus families where at least one affected has cleft lip only, and one affected has cleft lip and palate in Chinese and Caucasian families. For families where all affecteds have cleft lip only plus families where all affecteds have cleft lip and palate plus families where at least one affected has cleft lip only, and one affected has cleft lip and palate, marker rs10943957 showed association in Caucasian families. [Detailed results are available in the Appendix.]
Haplotype analyses support the individual associations found for PRSS35 and cleft lip/palate in Caucasian families (p = 0.008 and p = 0.003, for 3- and 4-window haplotypes, respectively; Table 3).
Table 3.
Results of Haplotype Analyses for PRSS35 Markers in Caucasian Families (US, Madrid, and Turkey)
|
PRSS35 Markers |
||||||
|---|---|---|---|---|---|---|
| Haplotype Window | rs10943944 | rs7753918 | rs9449648 | rs1171114 | rs512140 | rs1171105 |
| 0.62 TA | ||||||
| Window=2 | 0.09 AA | |||||
| 0.03 AT | ||||||
| 0.07 TA | ||||||
| 0.48 AT | ||||||
| 0.63 TAA | ||||||
| Window=3 | 0.30 AAT | |||||
| 0.008 ATA | ||||||
| 0.12 TAC | ||||||
| 0.15 CGAC | ||||||
| Window=4 | 0.37 AGTA | |||||
| 0.003 GCGC | ||||||
Expression Analyses
We used RT-PCR to determine whether Prss35 and Snap91 are expressed during the periods of mammalian craniofacial development. Expression of Prss35 was evident at ED10, decreased at ED11, then increased and peaked at ED12 and ED13. Lower levels of expression were noted each day from ED14 through ED18 (Fig., A). In contrast, Snap91 gene expression was undetectable in the embryonic material analyzed, but was significant in the adult brain (Fig., B). Limited levels of Snap91expression were noted only in embryo-derived cDNA samples after an additional 5 to 10 PCR cycles (data not shown).
Figure.
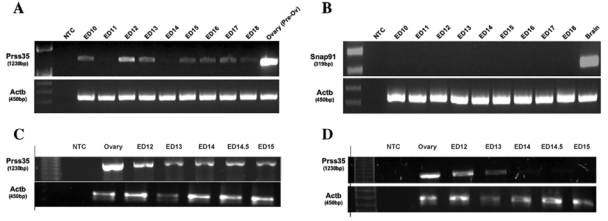
Analyses of Prss35 and Snap91 gene expression during craniofacial development in mice. Prss35 (A) and Snap91 (B) gene expression was performed with cDNA generated from whole embryos (ED 10 through ED 18). Prss35 expression in the head (C) and palate (D) of mouse embryos at critical stages for palate development (ED12-E15). β-Actin (Actb) was used as a normalization control. NTC, no template control; ED, embryonic day.
We then used cDNA from mouse head and palate to investigate Prss35 expression at periods critical for palate development (ED12-15). Prss35 was expressed at all stages in the developing mouse head (Fig., C). In the palate, Prss35 expression peaked at ED12 and ED13, then declined dramatically at ED14 and ED14.5. No expression was detected at ED15 (Fig., D, and Appendix).
Discussion
Chromosome 6 has been long considered a candidate for the etiology of oral-facial clefts. Studies have reported the presence of cleft lip/palate in individuals with deletions and translocations affecting chromosome 6 (Hopkin et al., 1997; Yu et al., 2005). More specifically, chromosome 6q was identified in a previous genome scan as a cleft-susceptibility region in Chinese individuals (Marazita et al., 2002), and a later meta-analysis supported these findings (Marazita et al., 2004). Imputed multipoint posterior probabilities of linkage for Filipino multiplex families revealed a region at 6q14.2-14.3, between markers D6S1031 and D6S1056, presenting an ~88% chance of harboring a cleft gene (Govil et al., unpublished observations). The posterior probability of linkage, a class of likelihood-based, model-free statistics, is designed for accumulation of evidence for or against linkage across multiple, heterogeneous sets of data (Vieland, 1998, 2006), and allows for measurement of the probability to have found true linkage.
To investigate the existence of possible cleft-susceptibility genes in the 6q14.2-14.3 region, we performed association tests with densely spaced single-nucleotide polymorphisms and 2732 individuals from eight different populations. We found association of a novel gene, PRSS35, with cleft lip/palate in a case-control cohort from Brazil. Studies with unrelated cases and controls always raise the question as to whether the individuals are appropriately matched, particularly when admixture is a feature of the population. To overcome possible confounding results of undetectable population stratification, we limited our case and control groups to include individuals of only Caucasian descent. We also found association of PRSS35 with cleft subgroups in our family cohorts, more specifically in Guatemalan, Chinese, and Caucasian populations. Analyses of PRSS35 marker haplotypes in the Caucasian families further support the associations found in the case-control cohort. Although our target region does not completely overlap with the region previously described in Chinese families (Marazita et al., 2002), the fact that both studies found positive results in different populations reinforces the possibility that cleft-susceptibility genes may be located on chromosome 6q.
We observed clearly detectable levels of Prss35 expression in the head and palate of mouse embryos during the periods of craniofacial development, and these increased particularly at embryonic days 12 and 13. We speculate that Prss35 may be involved in the early stages of palatogenesis, and that, if disturbed, it may impose a risk to proper elevation of the palatal shelves rather than palatal fusion. PRSS35 codes for a serine protease, belonging to a group of structurally and functionally diverse proteins critical for essential biological processes. For instance, proteases are determinants of cellular proliferation and migration during embryonic development (including palate development) for their ability to remodel the extracellular matrix. Similarly, other proteases, such as matrix metalloproteinases, have been postulated to play critical roles during palate formation (Iamaroon et al., 1996; Morris-Wiman et al., 1999, 2000; Blavier et al., 2001; Brown et al., 2002). Moreover, variations in one MMP gene, MMP3, have been reported in association with cleft lip/palate (Letra et al., 2007). Taken together, these observations warrant additional research to determine the role of a protease such as PRSS35 in palate development.
We also found significant association of intergenic markers near the SNAP91 gene with cleft lip/palate in the US and pooled Caucasian families. Although unlikely to be of functional significance with respect to a phenotype, intergenic polymorphisms may localize to regulatory regions such as gene promoters and enhancers and affect gene function through transcriptional or translational regulation and ultimately be associated with complex diseases (Mottagui-Tabar et al., 2005). Borderline association was also found for a marker in SNAP91 and the ECLAMC data set. SNAP91 (synaptosomal-associated protein) encodes a synapse-associated protein with highest expression detected in the brain (Ishikawa et al., 1998). We did not detect Snap91 expression in the embryonic periods analyzed in our study. Notwithstanding, the association with SNAP91, a central-nervous-system-associated gene, and cleft lip/palate raises intriguing questions. Brain abnormalities have been reported in patients with oral clefts as an additional phenotype. Individuals with clefts often present an increased incidence of brain structural anomalies that were further correlated to cognitive function and lower intelligence quotient (Nopoulos et al., 2000, 2002, 2007). Problems with visual perceptual skills and higher incidence of reading disability among children with clefts have also been reported (Richman et al., 1988). Although there may be a genetic link among SNAP91, brain abnormalities, and oral clefts, this remains to be solved.
In summary, the accumulated evidence for linkage of chromosomal region 6q14.2-14.3 with cleft lip/palate has driven us to pursue additional research within that particular region. Our results corroborate these findings. To our knowledge, this is the first report of an association of PRSS35 and SNAP91 with cleft lip/palate. The expression of Prss35 mRNA at the time of palate formation further supports its role as a more suitable candidate gene for oral clefts at this time. Nonetheless, additional studies are necessary to explain the functional role of both genes in the susceptibility for human clefting.
Supplementary Material
Acknowledgments
We thank the individuals and families for their valuable collaboration.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This work was supported by grants from the National Institutes of Health: K99-DE018954 (to A.L.); K99-DE018413 (to R.M.); K99-DE018085 (to M.G.); R01-DE016148, P50-DE0-16215, R21-DE016930, R01-DE09886, and R01-DE12472 (to M.L.M.); R21-DE16718 (to A.R.V.); R01-HD42000, RR00163, and U54-HD55744 (to J.D.H.); by FAPERJ 26/152.831/2006, CNPq 308885/2006-0 and 401467/2004-0 (to I.M.O.); and by CAPES, Brazil (to R.F.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research or the National Institutes of Health. This article is partly based on a thesis submitted to the graduate faculty, Federal University of Rio de Janeiro, in partial fulfillment of the requirements for the PhD degree (for RFF).
References
- Blavier L, Lazaryev A, Groffen J, Heisterkamp N, Declerck YA, Kaartinen V. (2001). TGF-beta3-induced palatogenesis requires matrix metalloproteinases. Mol Biol Cell 12:1457-1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Yarram SJ, Mansell JP, Sandy JR. (2002). Matrix metalloproteinases have a role in palatogenesis. J Dent Res 81:826-830 [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. (2004). Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla EE, Orioli IM. (2004). ECLAMC: The Latin American Collaborative Study of Congenital Malformations. Community Genet 7:76-94 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2006). Cleft lip and palate. Atlanta, GA: The Center [Google Scholar]
- Choi SJ, Marazita ML, Hart PS, Sulima PP, Field LL, McHenry T, et al. (2008). The PDGF-C regulatory region SNP rs2899109 decreases promoter transcriptional activity and is associated with CL/P. Eur J Hum Genet 17:774-784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM, Hennekam RCM, editors (2001). Syndromes of the head and neck. New York: Oxford University Press [Google Scholar]
- Hopkin RJ, Schorry E, Bofinger M, Milatovich A, Stern HJ, Jayne C, et al. (1997). New insights into the phenotypes of 6q deletions. Am J Med Genet 70:377-386 [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. (2001). The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet 9:301-306 [DOI] [PubMed] [Google Scholar]
- Iamaroon A, Wallon UM, Overall CM, Diewert VM. (1996). Expression of 72-kDa gelatinase (matrix metalloproteinase-2) in the developing mouse craniofacial complex. Arch Oral Biol 41:1109-1119 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Nagase T, Suyama M, Miyajima N, Tanaka A, Kotani H, et al. (1998). Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res 5:169-176 [DOI] [PubMed] [Google Scholar]
- Letra A, Silva RA, Menezes R, Astolfi CM, Shinohara A, de Souza AP, et al. (2007). MMP gene polymorphisms as contributors for cleft lip/palate: association with MMP3 but not MMP1. Arch Oral Biol 52:954-960 [DOI] [PubMed] [Google Scholar]
- Logue MW, Vieland VJ. (2004). A new method for computing the multipoint posterior probability of linkage. Hum Hered 57:90-99 [DOI] [PubMed] [Google Scholar]
- Marazita ML, Field LL, Cooper ME, Tobias R, Maher BS, Peanchitlertkajorn S, et al. (2002). Genome scan for loci involved in cleft lip with or without cleft palate, in Chinese multiplex families. Am J Hum Genet 71:349-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein T, et al. (2004). Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32-35. Am J Hum Genet 75:161-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakoshi K, Murphy MJ, Yeoman RR, Mitra S, Dubay CJ, Hennebold JD. (2006). The identification of novel ovarian proteases through the use of genomic and bioinformatic methodologies. Biol Reprod 75:823-835 [DOI] [PubMed] [Google Scholar]
- Morris-Wiman J, Du Y, Brinkley L. (1999). Occurrence and temporal variation in matrix metalloproteinases and their inhibitors during murine secondary palatal morphogenesis. J Craniofac Genet Dev Biol 19:201-212 [PubMed] [Google Scholar]
- Morris-Wiman J, Burch H, Basco E. (2000). Temporospatial distribution of matrix metalloproteinase and tissue inhibitors of matrix metalloproteinases during murine secondary palate morphogenesis. Anat Embryol (Berl) 202:129-141 [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Faghihi MA, Mizuno Y, Engström PG, Lenhard B, Wasserman WW, et al. (2005). Identification of functional SNPs in the 5-prime flanking sequences of human genes. BMC Genomics 6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopoulos P, Berg S, Canady J, Richman L, Van Demark D, Andreasen NC. (2000). Abnormal brain morphology in patients with isolated cleft lip, cleft palate, or both: a preliminary analysis. Cleft Palate Craniofac J 37:441-446 [DOI] [PubMed] [Google Scholar]
- Nopoulos P, Berg S, Van Demark D, Richman L, Canady J, Andreasen NC. (2002). Cognitive dysfunction in adult males with non-syndromic clefts of the lip and/or palate. Neuropsychologia 40:2178-2184 [DOI] [PubMed] [Google Scholar]
- Nopoulos P, Langbehn DR, Canady J, Magnotta V, Richman L. (2007). Abnormal brain structure in children with isolated clefts of the lip or palate. Arch Pediatr Adolesc Med 161:753-75817679656 [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, et al. (2008). Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet 40:1341-1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade K, Chang MS, Ting CT, Pei D, Hsiao CF, Olivier M, et al. (2001). High-throughput genotyping with single nucleotide polymorphisms. Genome Res 11:1262-1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman LC, Eliason MJ, Lindgren SD. (1988). Reading disability in children with cleft lip and/or palate. Cleft Palate J 25:21-25 [PubMed] [Google Scholar]
- Schliekelman P, Slatkin M. (2002). Multiplex relative risk and estimation of the number of loci underlying an inherited disease. Am J Hum Genet 71:1369-1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira AR. (2008). Unraveling human cleft lip and palate research (review). J Dent Res 87:119-125 [DOI] [PubMed] [Google Scholar]
- Vieland VJ. (1998). Bayesian linkage analysis, or: How I learned to stop worrying and love the posterior probability of linkage. Am J Hum Genet 63:947-954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieland VJ. (2006). Thermometers: something for statistical geneticists to think about. Hum Hered 61:144-156 [DOI] [PubMed] [Google Scholar]
- Yu M, Obringer AC, Fowler MH, Hummel M, Wenger SL. (2005). Prenatal detection of deletion 6q13q15 in a complex karyotype. Prenat Diagn 25:1084-1087 [DOI] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, et al. (2004). Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. New Engl J Med 351:769-780 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


