Summary
Alternative splicing is a vast source of biological regulation and diversity that is misregulated in cancer and other diseases. To investigate global control of alternative splicing in human cells, we analyzed splicing of mRNAs encoding Bcl2-family apoptosis factors in a genome-wide siRNA screen. The screen identified many novel regulators of Bcl-x and Mcl1 splicing, notably an extensive network of cell cycle factors linked to aurora kinase A. Drugs or siRNAs that induce mitotic arrest promoted pro-apoptotic splicing of Bcl-x, Mcl1, and caspase-9, and altered splicing of other apoptotic transcripts. This response preceded mitotic arrest, indicating coordinated upregulation of pro-death splice variants that promotes apoptosis in arrested cells. These shifts corresponded to post-translational turnover of splicing regulator ASF/SF2, which directly binds and regulates these target mRNAs and globally regulates apoptosis. Broadly, our results reveal an alternative splicing network linking cell cycle control to apoptosis.
Introduction
Nearly all human pre-mRNAs undergo alternative splicing (AS), with tremendous variation and specificity across tissues, development and disease (Pan et al., 2008). This vast complexity is a formidable challenge to experimental and informatic analysis of AS and its physiological roles (Blencowe, 2006). One emergent concept is that functionally coherent transcript populations, termed RNA ‘regulons,’ are co-regulated by dedicated RNA-binding proteins (RBPs) to promote specific biological functions (Keene, 2007). These RBPs, notably SR proteins and hnRNPs, ‘decode’ transcript cis-elements and control stepwise assembly of spliceosomal snRNPs: U1 at the 5′-splice-site (ss), then U2 near the 3′-ss, and finally U5/4/6 (Wahl et al., 2009; Black, 2003). Splicing control is integrated with signal transduction pathways, promoting dynamic, context-driven regulation. Collectively, these features produce robust cell- and tissue-specific signatures of exon use that shape many aspects of cell fate (Moore and Silver, 2008). Exon signatures are radically transformed in tumors, but the causes and consequences are unknown.
In this study, we examine AS of Bcl2-family apoptosis regulators Bcl-x and Mcl1. Bcl2-like proteins contain up to four Bcl2-homology (BH) domains (BH1-4) (Hardwick and Youle, 2009). Factors possessing all four BH domains, including Bcl2, Bcl-xL, and Mcl1L, antagonize apoptosis by preventing mitochondrial outer membrane permeablization (MOMP), thus sequestering pro-apoptotic factors in mitochondria. Factors lacking one or more BH domain, including Bid, BAD, and BAX, are pro-apoptotic and promote MOMP. A finely tuned balance of pro- and anti-apoptotic Bcl2-like factors therefore controls mitochondrial integrity and hence downstream steps in apoptosis such as apoptosome formation and caspase activation (Wang and Youle, 2009). Remarkably, Bcl-x, Mcl1, and some other Bcl2-family mRNAs are alternatively spliced to yield both long (L) anti-apoptotic and short (S) pro-apoptotic forms. For Bcl-x, use of an alternative 5′-ss in exon 2 excludes the BH1 and BH2 domains (Akgul et al., 2004). For Mcl1, exon 2 skipping excludes the BH1 and BH2 domains and eliminates the downstream transmembrane domain via frame shift.
Many cis-regulatory elements and trans-acting factors exert combinatorial control of Bcl-x splicing. Most known regulators, including Sam68, ASF/SF2, hnRNP F/H, SRp30c, and RBM25, altered Bcl-x AS in vitro or when over-expressed in cell culture (Cloutier et al., 2008; Zhou et al., 2008; Paronetto et al, 2007; Garneau et al., 2005). In addition, in RNAi-based loss-of-function assays, depletion of Sam68 and hnRNPA1 favored Bcl-xL formation, while depletion of U2 snRNP component SF3B1/SAP155 favored Bcl-xS (Paronetto et al, 2007; Massiello et al., 2006). Comparably little is known of Mcl1 splicing regulation.
Beyond Bcl2-like factors, caspases, ‘death receptors,’ ligands and various adaptors are regulated by AS, suggesting broad roles in controlling apoptosis (Schwerk and Schulze-Osthoff, 2005). Many apoptosis regulators, including Bcl2-like proteins, are proto-oncogenes that contribute to apoptosis resistance in cancer (Letai, 2008; Fesik, 2005). Modulation of apoptotic factors by targeting the splicing machinery is thus an attractive strategy to facilitate tumor cell death. Furthermore, while the divergent functions of Bcl-x and Mcl1 isoforms in apoptosis are well-established, the physiological contexts and upstream regulation of their expression are poorly defined. These unanswered questions illustrate a pervasive challenge in defining physiological contexts of AS regulation, because strategies for systematic evaluation of upstream regulation are limited.
Genome-scale screening of RNA regulatory events is complicated by the difficulty of visualizing RNAs in vivo, and the technical infeasibility of high-throughput measurements by RT-PCR and other methods. Splicing-sensitive fluorescent reporters are an alternative strategy that produce a robust, visual output suitable for screening efforts (Stoilov et al., 2008; Warzecha, et al., 2006; Orengo et al., 2006). Here, we present high-throughput assays that recapitulate physiological regulation of Bcl-x and Mcl1 AS. In a whole-genome siRNA screen, we identified novel factors that regulate the balance of anti- and pro-apoptotic splice isoforms, with striking enrichment for cell cycle factors. These results define functional interactions between the cell cycle and splicing machineries in human cells that manifest in a coordinated program of AS controlling apoptosis.
Results
Reporter Assays for Bcl-x and Mcl1 Alternative Splicing
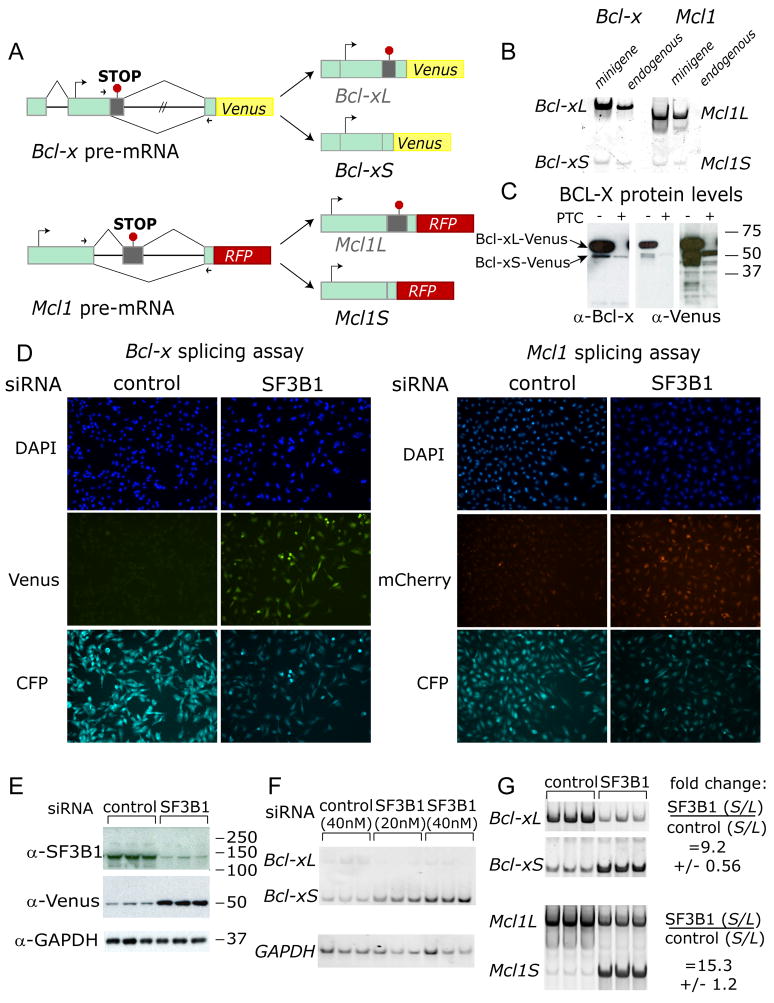
To develop splicing assays for high-throughput analysis, we designed splicing-sensitive reporters for the Bcl2-family apoptosis regulators Bcl-x and Mcl1. 5′-UTR, ORF, and intervening intron sequences for Bcl-x and Mcl1 were cloned in C-terminal fusions with Venus (yellow) and mCherry (red) cDNAs, respectively (Figure 1A). In HeLa cells, these constructs expressed long and short spliced mRNAs at ratios similar to endogenous mRNAs (Figure 1B). To render constructs splicing-sensitive, premature termination codons (PTCs) were introduced in alternative exon regions exclusive to long splice forms. As expected, PTCs eliminated expression of long protein variants, but short forms were retained (Figure 1C).
Figure 1. Bcl-x and Mcl1 alternative splicing reporters.
(A) Bcl-x and Mcl1 minigene splicing reporters are schematized. Green rectangles are exons, with dark gray as alternative regions. Black lines depict introns. Large arrows denote translation start sites, and arrowheads show primer sets used for qPCR. Inserted PTCs, exclusive to long (L) isoforms, are shown by red ‘stop signs.’
(B) Minigene constructs are spliced like endogenous mRNAs. Splice isoforms from minigene- or mock-transfected (endogenous) HeLa cells were analyzed by RT-PCR with primer sets from (A). Due to high transient minigene expression, cDNA from minigene samples was diluted 1:105 relative to mock (endogenous). At this dilution, endogenous mRNAs do not contribute to products in the ‘minigene’ lanes.
(C) A PTC introduced to the alternative exon region eliminated Bcl-xL-Venus but not Bcl-xS-Venus protein expression. HeLa cells were transfected with Bcl-x-Venus (PTC-) or Bcl-x-Venus-PTC (PTC+) constructs, and lysates were analyzed by Western with indicated antibodies. Migration of isoforms is indicated to the left of images (last two lanes overexposed to show Bcl-xS-Venus).
(D) SF3B1 knockdown activated pro-apoptotic splicing of Bcl-x and Mcl1 reporters. Stable reporter lines were transfected with SF3B1 or non-targeting siRNA pools and imaged in the indicated channels. SF3B1 knockdown increased minigene expression versus controls in both cases. SF3B1 depletion also decreased CFP modestly, possibly reflecting a role in splicing the EF1α intron. Subsequent experiments showed this effect on CFP expression was restricted to SF3B1 (see Figures S2B, S2F).
(E) Upregulation of Bcl-xS by SF3B1 depletion was confirmed by Western analysis. Lysates from cells treated as in (D) were analyzed with antibodies indicated to the left of images. SF3B1-knockdown depleted SF3B1 and increased Bcl-xS-Venus relative to controls.
(F) Results in (D) and (E) were verified at the RNA level by RT-PCR. Migration of Bcl-xS, Bcl-xL (PTC causes nonsense-mediated decay), and GAPDH (loading control) are indicated to left of images.
(G) Results in (D), (E), and (F) model the regulation of endogenous Bcl-x and Mcl1 mRNAs. Unmodified HeLa cells were transfected with SF3B1 or control siRNAs. RT-PCR analysis confirmed that SF3B1 knockdown favored short, pro-apoptotic forms. S/L ratios are quantified to the right of images; values are the mean of 3 independent measurements +/− SD, normalized to controls.
To produce screen assay cell-lines, splicing reporters were stably transfected into HeLa cells along with a constitutive mCerulean fluorescent protein (CFP) construct. All constructs used the human EF1α promoter, which contains a 5′-UTR intron, allowing dual measurements of minigene splicing and a constitutively spliced CFP reporter under identical control. To test the Bcl-x reporter line, we verified that siRNA-depletion of known regulator SF3B1 increased Bcl-xS-Venus expression relative to a non-targeting control (Figure 1D-left panels; Massiello et al., 2006). Immunoblotting confirmed efficient siRNA-knockdown of SF3B1, and upregulation of Bcl-xS-Venus reporter protein (Figure 1E). RT-PCR confirmed upregulation of the Bcl-xS-Venus mRNA (Figure 1F). Finally, RT-PCR analysis of endogenous Bcl-x transcript in HeLa cells verified that SF3B1-knockdown shifted splicing toward Bcl-xS, demonstrating congruous regulation of minigene and endogenous splicing (Figure 1G, upper panel).
We had no a priori knowledge of Mcl1 regulators, but SF3B1 knockdown also upregulated Mcl1S-mCherry in the splicing assay, establishing a positive assay control (Figure 1D-right panels). Analysis of endogenous Mcl1 verified this shift toward Mcl1S (Figure 1G, lower panel).
High-Throughput siRNA Screens for Alternative Splicing Regulators
To identify regulators of Bcl-x AS, >21,000 siRNA pools targeting known and predicted human genes were screened for upregulation of the Bcl-x reporter (Figure 2A). 369 positive hits were identified using a Support Vector Machine (SVM) model that determined reproducibility (i.e. ‘confidence’) across triplicates and signal ‘strength’ relative to positive and negative control siRNAs (Figures 2B, S1, Table S1). Hits had strong gene ontology (GO) enrichments for mRNA splicing/processing, protein kinase signaling, cytoskeleton association, and cell cycle functions (Figure 2C). Importantly, the screen blindly recovered positive control SF3B1 and several of its interactors.
Figure 2. Whole-genome siRNA screen for regulators of Bcl-x alternative splicing.
(A) Screening, analysis, and hit-selection are schematized. Of 369 primary hits, 274 re-validated. The pie chart indicates how many of 274 validated hits re-confirmed with 1, 2, 3, or 4 siRNAs. 160 ‘high-confidence’ hits validated with 2 or more siRNAs (see Supplementary Data for complete analysis methods).
(B) SVM probability scores are plotted for the whole-genome siRNA screen: confidence on the x-axis, strength on the y-axis. The blue region shows the SVM cut-off for hit selection (see Figures S1–S3 and Tables S1, S2 for detailed analysis and validation).
(C) GO functional enrichments from primary, re-validated (1 or more siRNA), and high-confidence (2 or more siRNAs) hits are shown, plotted by relative statistical significance.
For validation, hits were re-tested in the screen assay with 4 individual siRNAs from deconvoluted SMARTpools. 274 of 369 factors validated with at least one siRNA, and 160 validated with 2 or more (Figure 2A, Table S2). In the primary and validation screens, Bcl-x-Venus expression correlated significantly with cell death (Figure S2A, S2C). Venus showed no systematic correlation to CFP, indicating that non-specific promoter effects were not a major source of positives (Figures S2B, S2F). In the validation screen, apoptosis was tracked by Annexin-V-Cy5 staining, and showed strong coupling to pro-apoptotic Bcl-x splicing and cell death (Figures S2D, S2E). Functional enrichments, notably splicing and cell cycle regulation, were similar between validated hits and the primary screen (Figure 2C). Subsequent analyses focused on ‘high-confidence’ factors validated with 2 or more siRNAs. We focused specifically on aurora kinase A (AURKA) and other mitotic regulators because of their strong functional enrichment and their novelty in the context of splicing regulation.
Physiological function for screen hits was confirmed by analyzing endogenous Bcl-x AS in HeLa cells. siRNAs against 19 factors were tested, and 16 (~85%) significantly shifted Bcl-x splicing toward Bcl-xS (Figure S3A). The magnitude of these shifts matched or exceeded fold-changes that promote apoptosis in various cell types, indicating physiological significance (Mercatante et al., 2002; Taylor et al., 1999). As further indication of this significance, AURKA knockdown strongly shifted endogenous Bcl-x protein toward Bcl-xS (Figure S3B). This shift was larger than the corresponding mRNA shift, suggesting an amplification effect during translation. Regulation of endogenous Bcl-x by AURKA and other hits also confirmed in MCF7 (breast adenocarcinoma) and PANC1 (pancreatic carcinoma) cells (Figure S3C). These lines differ markedly in steady state AURKA levels, with MCF7 (like HeLa) expressing high levels, and PANC1 levels closer to ‘normal’ tissue (Ross et al., 2000). Thus, splicing regulation by AURKA inhibition was not restricted to HeLa cells, and occurred irrespective of steady state AURKA levels.
Apoptosis Suppression Defines Direct Regulators of Bcl-x Alternative Splicing
In screening experiments, pro-apoptotic Bcl-x splicing correlated to apoptosis induction and increased cell death (Figure S2). However, screen hits are likely to include both direct regulators of Bcl-x splicing, and factors that influence splicing indirectly via upstream activation of pro-apoptotic pathways (Figure 3A). To define systematically the relationship of screen hits to apoptosis, all 274 validated regulators were re-tested in Bcl-x reporter cells engineered to over-express Bcl2. Bcl2 over-expression suppresses mitochondrial permeablization and hence canonical apoptosis progression (Kroemer, 1997). Importantly, Bcl2 directly antagonizes the function of Bcl-xS, so this strategy also suppresses potential downstream effects of Bcl-xS-Venus produced by the reporter construct. We predicted that Bcl2 overexpression would attenuate the effects of pro-apoptotic siRNAs that alter Bcl-x splicing as part of a general pro-apoptotic response, but not siRNAs that target more direct regulators of splicing. Supporting the latter prediction, depleting direct regulator SF3B1 increased reporter expression despite Bcl2 overexpression (Figure 3B).
Figure 3. Apoptosis inhibition by Bcl2 suppressed a subset of Bcl-x regulators.
(A) Screen hits (lightning bolts) may affect Bcl-x AS by many potential mechanisms. Regulation may be direct, such as (1) direct suppression of Bcl-xS formation or (2) facilitation of Bcl-xL. Indirect or downstream regulators may (3) promote cell survival pathways or (4) suppress pro-apoptotic pathways. We predicted that Bcl2 overexpression would suppress the splicing regulatory effect of siRNAs targeting indirect mechanisms, but not direct regulators.
(B) Bcl2 overexpression did not suppress the effect of SF3B1 depletion, consistent with direct regulation of Bcl-x splicing.
(C) Bcl2 suppressed the effect of siRNAs against mRNA export factors NXF1 and NUPL2, but not AURKA, on Bcl-x splicing (compare Venus images between ‘wild-type’ and ‘Bcl2 overexpression’). By contrast, Bcl2 suppressed cell death in all cases (compare DAPI images).
(D) 33% were suppressed by Bcl2 overexpression, while 67% showed partial to no suppression. GO functional enrichments within these subsets are shown with p-values (see Table S3).
33% of 160 high-confidence Bcl-x regulators were totally suppressed by Bcl2 over-expression (Table S3). For example, Bcl2 suppressed both cell death and changes in Bcl-x splicing in response to depleting mRNA export factors NXF1 or NUPL2 (Figure 3C). By contrast, 67% of factors, including AURKA, showed partial or no suppression of pro-apoptotic Bcl-x splicing, despite the fact that cell counts showed efficient suppression of cell death. Thus, AURKA, not previously implicated in splicing regulation, functioned independently of Bcl2, whereas mRNA export factors, which directly interact with the spliceosome, were suppressed. Overall, most mRNA splicing and cell cycle regulators showed no or partial suppression (Figure 3D). Suppressed factors were enriched for functions in transcription, while signaling factors spanned both categories.
Discovery of Mcl1 Alternative Splicing Regulators
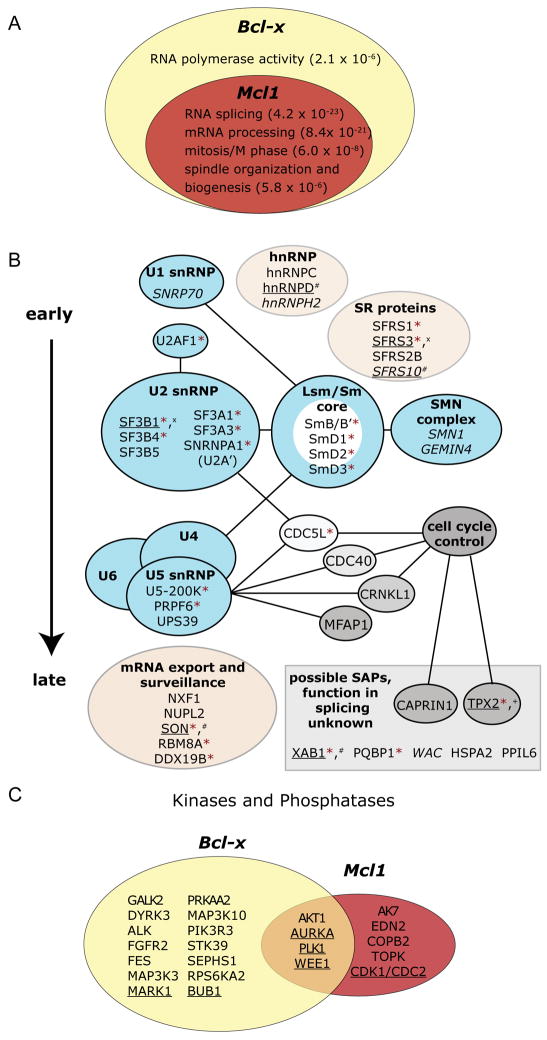
To explore functional coordination of apoptotic AS events, we determined all Bcl-x regulators that also control functionally analogous AS of Mcl1. 52 of 160 ‘high-confidence’ factors regulated both events, and we validated a subset on endogenous Mcl1 (Figures 4A and S4, Table S4). Cell cycle functions were enriched for factors that co-regulated Bcl-x and Mcl1, but not factors that regulated only Bcl-x (Figure 4A). Similarly, common regulators were disproportionately enriched for splicing functions versus those that only regulated Bcl-x. This result indicates extensive co-regulation of Bcl-x and Mcl1 by cell cycle and splicing factors. In contrast, transcription regulators were enriched only for Bcl-x. Figure 4B schematizes hits in the Bcl-x and Mcl1 screens with known or postulated functions in mRNA metabolism. Hits included ‘core’ spliceosome components in Sm ring domains and snRNPs, as well as hnRNP and SR proteins. Interestingly, factors acting late in spliceosome assembly, including U5 snRNP components and factors previously linked to cell cycle regulation (e.g. CDC40, CDC5L), scored positively for both Bcl-x and Mcl1.
Figure 4. Coordinated regulation of Bcl-x and Mcl1 alternative splicing.
(A) Bcl-x screen hits were tested for Mcl1 regulation to identify common regulators. High confidence hits are depicted as a Venn Diagram, with GO enrichments listed with p-values in the subsets where they were most significantly enriched (see Figure S4 and Table S4).
(B) Spliceosome-associated proteins that were Bcl-x screen hits are shown with established physical interactions and functional roles. Blue ellipses represent known RNP or protein complexes; pink ellipses contain functionally or structurally related factors. Low-confidence hits are italicized. Factors in gray rectangle have unknown roles in splicing, but were identified in proteomic analyses or are similar to known splicing factors (Chen et al., 2007). Factors are marked as follows: * = high-confidence hit for Mcl1; underlined = phosphorylated in mitosis; + = putative AURKA target; x = putative CDK1 target; # = putative PLK1 target (Dephoure et al., 2008).
(C) Hits from a kinase/phosphatase screen for Mcl1 splicing regulation are shown, with corresponding Bcl-x regulators. A cutoff of p<10−5 in three replicates, or p<10−7 in two replicates was applied (see Table S5). Cell cycle factors are underlined.
As an unbiased analysis of Mcl1 AS, we tested siRNA pools for >700 human kinases and phosphatases in the Mcl1 screening assay. Common regulators for Bcl-x and Mcl1 included cell cycle kinases AURKA, PLK1, and WEE1 (Figure 4C, Table S5). Further, BUB1 scored strongly for Bcl-x and weakly positive for Mcl1, though the latter did not surpass the confidence threshold. Conversely, CDK1 (CDC2) was strongly positive for Mcl1, and weakly so for Bcl-x. These data independently confirm Mcl1 AS regulation by cell cycle disruption.
Coupling of Cell Cycle Control and Alternative Splicing
The screen identified many cell cycle factors, including cancer therapy targets AURKA, PLK1, and survivin (BIRC5). Protein interaction network analysis of screen hits revealed AURKA-centered interactions spanning the cell-cycle, spliceosome, and tumor suppressors (Figure 5A). As independent validation of this regulation, nocodazole, an inhibitor of microtubule polymerization, and aurora kinase inhibitors ZM447439 and VX-680 strongly induced Bcl-xS formation in a dose-dependent manner (Figure 5B). By contrast, the broad-spectrum kinase inhibitor staurosporine induced apoptosis but not Bcl-xS formation, demonstrating specific regulation by cell cycle inhibitors. ZM447439 and VX-680 inhibit the 3 mammalian aurora kinases AURKA, AURKB, and germline-restricted AURKC. However, ZM447439 is ~20-fold more potent toward AURKB than AURKA in vitro, while VX-680 is ~30-fold more potent against AURKA (Ditchfield et al., 2003; Harrington et al., 2004). Since Bcl-x splicing regulation in screening assays was AURKA-specific, VX-680 was used subsequently due to its higher specificity for AURKA. Importantly, nocodazole and VX-680 treatment also shifted endogenous Bcl-x and Mcl1 splicing (Figure S5).
Figure 5. Alternative splicing regulation is coupled to cell cycle control.
(A) Screen hits span an AURKA-centered protein interaction network. Circles (‘nodes’) depict proteins, connected by lines (‘edges’) representing validated protein-protein interactions. A maximum of two non-hit bridges was permitted to connect hits to AURKA, as described in Supplemental Data.
(B) Drugs disrupting the cell-cycle promote pro-apoptotic Bcl-x AS. Bcl-x reporter cells were treated with nocodazole, aurora inhibitors VX-680 or ZM447439, or staurosporine for 18 hours. 3-fold dilution series were tested with the following maximum concentrations: 200 nM nocodazole, 10 μM VX-680, 30 μM ZM447439, 33nM staurosporine. %-Venus-positive values for drug treatments were normalized to DMSO-treatments; plotted values reflect means of 8–12 measurements +/− SD. Nocodazole, VX-680, and ZM447439, but not staurosporine, caused significant pro-apoptotic shifts in Bcl-x splicing (p<0.001). LD50 ranges derived from cell counts are shown below the x-axis. Wells with >33nM staurosporine had >95% death, so they were not quantified (see Figure S5).
(C) Aurora inhibitors induced mitotic arrest coupled with pro-apoptotic Bcl-x splicing. Bcl-x reporter cells were treated with 3 μM VX-680, 10 μM ZM447439 or DMSO, stained with propidium iodide, and analyzed for DNA content and Venus fluorescence by flow cytometry.
(D) Asynchronous, actively cycling cells from (C) show little to no Bcl-x splicing fluctuations relative to arrested cells.
(E) S-phase arrest by double thymidine block did not alter Bcl-x splicing, indicating specificity for mitotic arrest. Cells were analyzed as in (C).
(F) Bcl-x splicing regulation precedes mitotic arrest. Asynchronous or thymidine-synchronized cells from (E) were treated with the indicated inhibitors or DMSO. Cells were analyzed by automated microscopy 16 hours after thymidine release, precluding an intervening round of mitosis for synchronized cells. The response in pre-synchronized cells confirms a splicing shift upstream of mitotic arrest.
In flow cytometry experiments, aurora inhibitors induced mitotic arrest concomitant with upregulation of the Bcl-x reporter (Figure 5C). Bcl-xS-Venus levels in actively cycling G2/M cells were slightly higher than in G1, but the difference was not significant compared to arrested cells (Figure 5D). In addition, S-phase-arrest by double-thymidine block did not affect Bcl-xS-Venus levels, so the splicing response was specific to mitotic arrest (Figure 5E). Since transcription and splicing are primarily silenced in M phase (Shin and Manley, 2002), we considered two possibilities: 1.) splicing changes preceded arrest or 2.) a subset of cells underwent an abnormal division prior to full arrest, leading to a pro-apoptotic stress response. To test the latter scenario, cells were synchronized by double-thymidine block, released, and treated with VX-680. Venus fluorescence was measured 16 hours after thymidine release, thus ruling out an intervening mitotic cycle. Figure 5F shows the splicing response to VX-680 in synchronized cells was comparable to asynchronous cells. Apoptosis induction by staurosporine caused no splicing response, regardless of synchronization. These results show that stimuli that activate G2/M and spindle checkpoints trigger pro-apoptotic Bcl-x splicing, but the response precedes actual arrest.
Downstream Mechanisms of Splicing Regulation
To identify direct splicing regulators downstream of cell cycle inhibition, we examined expression of splicing factors upon AURKA knockdown, focusing on strong screen hits or factors previously linked to Bcl-x splicing (Figure S6A). Among these factors, only ASF/SF2 (SFRS1)—an SR protein previously linked to cell cycle regulation and a strong hit in the primary screen—showed specific downregulation upon AURKA knockdown (Figure 6A; Li et al., 2005). Inhibition of AURKA with VX-680 also resulted in dose-responsive downregulation of ASF/SF2 (Figure 6B).
Figure 6. ASF/SF2 regulates splicing downstream of AURKA.
(A) siRNA depletion of AURKA downregulated ASF/SF2. Cells were transfected with two different siRNAs for AURKA or AURKB, or control siRNAs. Lysates were analyzed by Western analysis for indicated factors. No other splicing factors measured in these samples showed significant changes (Figure S6A), indicating specificity for ASF/SF2.
(B) VX-680 treatment downregulated ASF/SF2 in a dose dependent manner.
(C) ASF/SF2 is downregulated post-translationally upon AURKA inhibition (see also Figure S6B). HeLa cells were transfected with a GFP-ASF/SF2 construct, then treated with VX-680 or DMSO. Western analysis showed that VX-680 downregulated exogenous ASF/SF2, indicating post-translational turnover. ASF/SF2 downregulation also occurred in apoptosis-resistant cells over-expressing Bcl2 (see Figures S6C).
(D) Visualization of ASF/SF2-RNA complexes by CLIP is shown. RNP complexes were UV-cross-linked in live HeLa cells, ASF/SF2 was immunopurified, and RNA was end-labeled with γ-32P-ATP. Complexes were run on SDS-PAGE and visualized by autoradiography. Non-cross-linked ASF/SF2 (UV- lane) was also labeled, as observed in Sanford et al., 2009.
(E) ASF/SF2 directly binds Bcl-x and Mcl1 mRNAs. RT-PCR analysis of ASF/SF2-bound RNAs showed significant enrichment of Bcl-x, Mcl1, and known target TPX2 versus an irrelevant IgG control.
(F) Exogenous expression of ASF/SF2 attenuated the effect of AURKA inhibition on Bcl-x splicing. Bcl-x reporter cells were transfected with mCherry-tagged ASF/SF2, a mutant lacking the second RRM domain (ΔRRM2), or mCherry alone. 18 hours post-transfection, cells were treated with 3 μm VX-680 or DMSO for 24 hours. The fraction of Venus-positive cells among mCherry-positive cells was determined by automated microscopy. VX-680-treated samples were background-corrected by subtracting values from DMSO treatment. Values are means of 3 separate measurements +/− SD, expressed relative to the mCherry-transfected control. ASF/SF2-WT attenuated the Bcl-x response to VX-680 by ~50% versus ΔRRM2. CFP expression was not affected, indicating specific attenuation of Bcl-x splicing.
(G) ASF/SF2-ΔRRM2 shows deficient association with Bcl-x and Mcl1 mRNAs. CLIP analysis of exogenous, GFP-tagged ASF/SF2-WT or - ΔRRM2 was performed with α-GFP antisera.
ASF/SF2 mRNA was not significantly reduced upon AURKA inhibition, ruling out transcriptional downregulation or effects on RNA stability (Figure S6B). However, VX-680 treatment reduced exogenous GFP-tagged ASF/SF2 in a manner similar to endogenous ASF/SF2, indicating post-translational turnover as the main source of ASF/SF2 depletion (Figure 6C). AURKA inhibition also induced ASF/SF2 downregulation in apoptosis-resistant cells that over-express Bcl2 (Figure S6C). Therefore, this regulatory pathway functions in cells where apoptosis is suppressed, consistent with our finding that AURKA knockdown shifts Bcl-x splicing when apoptosis is suppressed (Figure 3C).
To determine if Bcl-x is a direct target of ASF/SF2, we used the CLIP method (UV cross linking and immunoprecipitation) to analyze RNAs bound directly by ASF/SF2 in vivo. RNP complexes were covalently cross-linked in living cells by UV exposure, and ASF/SF2-RNA complexes were immunopurified (Figure 6D). RT-PCR analysis showed significant enrichment of Bcl-x mRNA in ASF/SF2-RNA IPs versus an irrelevant IgG control, along with Mcl1 and previously known target TPX2 (Figure 6E; Sanford et al., 2009). APAF1 and TNFSF13 mRNAs were detectable in input but not ASF/SF2 IPs, demonstrating that cross-linking was specific.
Our data show direct regulation of Bcl-x and Mcl1 splicing by ASF/SF2, and downregulation of ASF/SF2 by AURKA inhibition. To test if ASF/SF2 functions downstream of AURKA inhibition, we examined whether exogenous ASF/SF2 expression could ‘rescue’ the splicing response to AURKA inhibition. Bcl-x reporter cells were transfected with mCherry-tagged constructs expressing wild-type (WT) ASF/SF2 or a variant lacking the second RNA recognition motif (ΔRRM2) domain. After VX-680 treatment, the Bcl-x splicing response in cells expressing ASF/SF2-WT was reduced ~50% relative to cells expressing ΔRRM2 or mCherry alone (Figure 6F). CLIP of exogenous GFP-tagged ASF/SF2 constructs confirmed deficient recognition of endogenous Bcl-x mRNA by the ΔRRM2 mutant (Figure 6G; Cáceres et al., 1997). Therefore, enforced ASF/SF2 expression rescued the effect of AURKA inhibition on Bcl-x splicing, and rescue was dependent on intact splicing regulatory function.
ASF/SF2 regulates apoptosis globally in response to cell cycle inhibition. Consistent with previous reports, siRNA depletion of ASF/SF2 activates markers of apoptosis (e.g. Annexin-V staining, DNA fragmentation, and caspase cleavage) (data not shown; Li et al., 2005; Karni et al., 2007). To analyze apoptosis regulation by ASF/SF2 in the context of cell cycle disruption, ASF/SF2 was depleted by RNAi and apoptosis markers were analyzed upon AURKA inhibition with VX-680. ASF/SF2 depletion enhanced caspase activation and Annexin-V staining relative to a non-targeting siRNA control (Figures 7A, 7B). In addition, we compared the effects of ASF/SF2 depletion to the depletion of two splicing factors (SRPK1 and SRNP70) that had no significant effect on endogenous Bcl-x splicing, and a third factor (Sam68) that has antagonistic effects to ASF/SF2 on Bcl-x splicing (Figure S3A, Table S1; Paronetto et al., 2007). ASF/SF2 depletion sensitized cells to VX-680-induced apoptosis relative to SRPK1, SNRP70, or Sam68 depletion. In contrast, ASF/SF2 depletion did not enhance staurosporine-induced apoptosis relative to SRPK1, SNPR70, or Sam68 depletion (Figure S6D). We could not reliably analyze the effects of enforced ASF/SF2 expression on apoptotic markers, because over-expression was toxic in the time-frame required for this analysis. In sum, ASF/SF2 loss-of-function triggers apoptosis, and specifically sensitizes cells to apoptosis induced by AURKA inhibition.
Figure 7. An apoptotic alternative splicing program linked to cell cycle control.
(A) ASF/SF2 downregulation triggers apoptosis, and sensitizes cells to apoptosis induction by AURKA inhibition. HeLa cells were transfected with indicated siRNAs, incubated to allow factor depletion, then treated with varying concentrations of VX-680. Apoptosis was measured via cleavage of a fluorescent caspase-3 substrate. Depletion of ASF/SF2, but not SRPK1, SNRP70, or Sam68, sensitized cells to VX-680-induced apoptosis. This selective sensitization by ASF/SF2 depletion was not observed for staurosporine-induced apoptosis (see Figure S6D). Data reflect means of 3 independent transfections +/− SDs.
(B) Analysis in (A) was repeated using Annexin-V staining as the apoptosis marker.
(C) CASP9 and CASP2 AS were analyzed in HeLa cells by qPCR following transfection with the indicated siRNAs. Relative levels of pro-apoptotic isoform (L) normalized to negative controls are shown. Values are means of 3 independent measurements +/− SD.
(D) Results of AS profiling by real-time qPCR are shown in a heat-map of z-scores averaged across four biological replicates, along with unsupervised hierarchical clustering (see Figure S7 for an expanded view and validation of this data).
(E) A model of cell cycle arrest coupled to apoptosis via AS is shown. We propose anticipatory, pro-apoptotic splicing decisions in interphase that later promote apoptosis in arrested cells. Lightning bolts represent siRNAs, drugs, or other stimuli that disrupt the cell cycle and promote pro-apoptotic splicing of Bcl-x, Mcl1, CASP9, and other targets.
To examine broader functions of AS in this apoptotic response, we analyzed caspase-9 (CASP9) and caspase-2 (CASP2) mRNAs, which produce short (S), anti-apoptotic or long (L), pro-apoptotic isoforms (Schwerk and Schulze-Osthoff, 2005). Depletion of ASF/SF2 and AURKA favored pro-apoptotic CASP9 splicing (Figure 7C, Massiello and Chalfant, 2006). In addition, ASF/SF2 depletion favored pro-apoptotic CASP2 splicing, although AURKA knockdown had no significant effect. We extended our analysis to additional factors with diverse apoptotic functions and AS patterns, including exon skipping, alternative 5′-ss use, and intron retention. Splice isoforms were measured with an isoform-sensitive qPCR strategy upon knockdown of various screen hits, revealing widespread changes in splicing upon silencing of cell cycle regulators (Figures 7D). This approach is only semi-quantitative, so we focused on identifying biologically robust events by z-scores analysis of 4 independent experiments (Figure S7A). Hierarchical clustering of knockdown profiles revealed overlapping regulation by cell cycle kinases AURKA, CDC2, and BUB1. In addition, splicing factor profiles grouped by validated spliceosomal interactions; the U2 snRNP factors SF3B1 and SF3A1 overlapped significantly, as did U5 factors PRPF6 and U5-200K. Interestingly, ASF/SF2 targets clustered with AURKA and kinetochore component NDC80, suggesting overlapping regulation in this specific set of mRNAs. As confirmation of direct regulation, binding of ASF/SF2 to selected targets CASP9, CASP2, ATG4, and BMF was established by CLIP (Figure S7D).
Discussion
Genome-wide Identification of Alternative Splicing Regulators in Human Cells
We developed a strategy for systematic, high-throughput analysis of AS regulation in human cells. Our siRNA screens identified many spliceosomal regulators of Bcl-x and Mcl1 splicing, as well as functional connections to cell cycle control, cytoskeleton dynamics, and signal transduction (Figure 2C). Strong functional enrichment for known splicing factors was an encouraging indication of specificity. In addition, a high validation rate of screen hits on endogenous AS indicated faithful modeling of physiological regulation in our reporter assays (Figure S3). Broadly, our efforts expand the catalog of factors linking mRNA metabolism and apoptosis, and provide a platform for future high-throughput analyses of mRNA processing in living cells.
Screen hits spanned essential processes, such as transcription, cell division, and pro-survival signaling, with extensive mechanistic links to apoptosis, implicating splicing regulation in diverse pro-apoptotic pathways. The strong correlation of Bcl-xS formation with apoptosis induction and cell death further support a physiological function for splicing regulation in apoptosis (Figure S2). To investigate the functional connection between splicing regulation and apoptosis, factors were retested in cells rendered resistant to apoptosis by exogenous Bcl2 expression (Figure 3). Surprisingly, cell cycle factors, like most spliceosomal factors, were generally not affected by Bcl2 expression. Therefore, functional coupling of cell cycle disruption to pro-apoptotic splicing remained intact in cells where the canonical apoptosis pathway was suppressed. This finding suggests that coordinated, pro-apoptotic splicing could be exploited to target apoptosis-resistant cell populations, such as tumors.
Our screening experiments also revealed functional coordination of pro-apoptotic splicing events. 52 of 160 ‘high-confidence’ Bcl-x regulators affected functionally analogous splicing of Mcl1, and common regulators were disproportionately enriched for splicing and cell cycle functions (Figure 4A). Overlapping spliceosomal regulators of Bcl-x and Mcl1 included ‘alternative splicing factors’ (e.g. hnRNPs and SR proteins), and ‘core’ spliceosome components spanning most steps in spliceosome assembly (Figure 4B). Interestingly, U5 snRNP and associated factors that function late in spliceosome assembly affected splice-site choice for Bcl-x, Mcl1, and other mRNAs (Figures 4B, 7C, and 7D). Current mechanistic models support splice-site pairing early in spliceosome assembly upon U2 snRNP recruitment (Dassah et al., 2009). Our findings suggest that perturbations of late steps in spliceosome function may promote remodeling of splice-site choice downstream of the rate-limiting steps that normally determine splicing decisions.
Cell Cycle Disruption Coupled to Pro-apoptotic Alternative Splicing
Our screen for AS regulators identified factors involved in mitotic spindle assembly, kinetochore and centromere dynamics, microtubule transport, centrosome duplication and dynamics, and cell division (Figure 5A). Well-characterized pharmacological inhibitors, notably for AURKA, confirmed functional coupling of cell cycle and splicing regulation (Figures 5B, S5). In flow cytometry experiments, pro-apoptotic Bcl-x splicing was coupled to mitotic arrest, but not S-phase arrest or G2/M in actively cycling cells (Figure 5C, 5D). Since prevailing evidence supports suppression of transcription and splicing in mitosis, it is unlikely that the observed response initiates in mitotically arrested cells (Shin and Manley, 2002). Moreover, when cells were pre-synchronized to preclude an intervening cell division, VX-680 still induced a robust splicing response (Figure 5E). Therefore, this splicing regulation precedes mitotic arrest, indicating accumulation of pro-death factors in anticipation of G2/M and/or mitotic spindle checkpoints. Since cell cycle disruption also promoted a pro-apoptotic switch in CASP9 splicing and altered splicing of other apoptotic regulators, this regulation likely extends to many co-regulated mRNAs (Figure 7C, 7D).
Many previous studies have linked cell cycle and splicing regulation, reaching back to the seminal isolation of cell-division cycle (CDC) loci in S. cerevisiae that encode spliceosome components, including factors (CDC5L and CDC40) identified here. Recent genetic and proteomic analyses of mitotic spindle assembly have re-asserted the notion that the cell cycle and splicing machineries are intertwined, but functional consequences have not been established (Bjorklund et al., 2006, Rines et al., 2006). We observe coordinated pro-apoptotic splicing upon inhibition of key cell cycle factors, but preceding mitotic arrest, indicating anticipatory upregulation of pro-death factors that may subsequently propagate apoptosis in arrested cells. Since arrested cells cannot mount de novo transcriptional and splicing responses, this finding suggests a novel genetic mechanism for cell cycle checkpoint response.
ASF/SF2 regulates an apoptotic splicing network
We present evidence that ASF/SF2 down-regulation mediates a pro-apoptotic splicing response to cell cycle disruption. ASF/SF2 was a hit in the Bcl-x and Mcl1 screens, and its overexpression was previously shown to favor Bcl-xL formation (Paronetto et al., 2007). AURKA inhibition triggered post-translational turnover of ASF/SF2, but not other SR proteins or splicing factors identified in the screen (Figures 6A–C, S6A). In addition, ASF/SF2 directly binds Bcl-x and Mcl1 mRNAs, and its enforced expression attenuated the effect of AURKA inhibition on Bcl-x splicing (Figures 6D–G).
ASF/SF2 regulation extends beyond Bcl-x to global regulation of apoptosis (Li et al., 2005). ASF/SF2 depletion, but not depletion of several splicing factors that were not validated screen hits, sensitized cells to apoptosis induction by AURKA inhibition (Figures 7A, 7B). In contrast, ASF/SF2 depletion did not specifically enhance apoptosis in response to staurosporine (Figure S6D). ASF/SF2-mediated splicing regulation extended to other functional targets, including CASP9, which underwent a pro-apoptotic splicing shift upon depletion of AURKA and ASF/SF2 (Figure 7C). Our analysis of some 25 additional AS targets revealed overlapping effects of AURKA and ASF/SF2 inhibition (Figure 7D). The fact that some mRNAs (e.g. CASP2) did not respond identically to AURKA and ASF/SF2 knockdown may reflect differential effects of targeting upstream versus direct regulators of splicing, or the activity of additional, unidentified splicing regulators. Nonetheless, these experiments strongly indicate that ASF/SF2 downregulation globally shifts alternative splicing patterns to promote apoptosis upon disruption of the cell cycle.
Alternative Splicing in a Global Context
Our data link coordinated pro-apoptotic splicing to activated cell-cycle checkpoints (Figure 7E). These findings are relevant to basic mechanisms of apoptosis in arrested cells, which are poorly defined, and the exploitation of these pathways in cancer therapy. Aberrant expression of AURKA and other pro-division factors promotes spindle checkpoint abrogation, chromosomal instability, and aneuploidy in tumors (Lapenna and Giordano, 2009). Our findings indicate that AURKA activation may also propagate oncogenesis by promoting anti-apoptotic splicing of crucial cell-death regulators. Conversely, AURKA inhibition elicits parallel upregulation of pro-death variants, which may contribute to its demonstrated efficacy as a cancer drug target. Importantly, this regulation remains intact in apoptosis-resistant cells, suggesting functional and potential therapeutic relevance in tumors.
ASF/SF2 downregulation triggers pro-apoptotic splicing and promotes apoptosis. Importantly, ASF/SF2 is a proto-oncogene in its own right, and its depletion is linked to genome instability and G2/M arrest (Karni et al., 2007; Li and Manley, 2005). Our data highlight RNA targets that may contribute to its transforming properties. Taken together, these findings suggest ASF/SF2 has multiple, synergistic functions in controlling cell arrest and apoptosis, underscoring its role as a central regulator of mRNA metabolism in cancer. Finally, it bears reiterating that our systematic screening approach was the original source of these biological insights, as well as many novel regulators and functional connections not expounded here. We believe future screening efforts will be instrumental in defining global roles for AS in physiological and disease contexts.
Experimental Procedures
siRNA Screening
Cells were transfected in triplicate in 384-well plates with >21,000 siRNA SMARTpools (Thermo-Fisher) for known and predicted human genes. 72 hours later, cells were fixed, DAPI-stained, and imaged on the Image Xpress Micro microscope (Molecular Devices). Cell counts (DAPI), % Venus-positive cells, and average per-cell CFP signal were determined with MetaXpress software. Wells were then assigned probabilistic ‘strength’ and ‘confidence’ scores in the Venus channel using a Support Vector Machine (SVM) model trained on positive and negative control data (Figure S1, Table S1, Supplemental Experimental Procedures). For independent verification, plate-wide, median-normalized z-scores were also tabulated. Wells with SVM strength scores with false discovery rate (FDR) < 10, or z > 2.6 were inspected visually.
369 positives were re-tested in a validation screen with 4 siRNAs from deconvoluted SMARTpools. siRNAs >2 SDs above negative controls in the Venus channel were deemed positive (Table S2). Bcl2-overexpression and Mcl1 screens were performed and analyzed as for the validation screen (Tables S3, S4).
RNA Extraction and RT-qPCR
Cellular RNA was extracted with Trizol (Invitrogen) reagent, and 2.5–5 μg were reverse-transcribed with Superscript III (Invitrogen) and oligo-dT. Bcl-x, Mcl1, CASP9, and CASP2 isoforms were analyzed by qPCR with primers flanking alternative exon sequences. For each product, a linear range of cDNA input was determined empirically by running a two-fold dilution series in 26–28 cycle reactions. Products were run on 5% TBE-PAGE gels, stained with Sybr-Gold (Invitrogen), and quantified with QuantityOne software (Biorad).
Real-time qPCR primer sets were designed with Primer3 webware and tested by electrophoresis to verify correctly sized products (Table S6). Reactions were prepared with Sybr-Green PCR mix (ABI), run on the ABI 7900HT system, and quantified by a ΔΔC(t) method. Amplicons from alternative regions were first normalized to a constitutive amplicon from the same transcript. To correct for differential primer efficiencies, data were then normalized to negative controls. Z-scores were tabulated across four independent experiments and clustered with Eisen Cluster 2.0.
Splicing Assays with Drug Treatments
Sources for inhibitors are in Supplementary Data. Cells were treated in 96-well plates for 18–24 hours and analyzed by automated microscopy. For flow cytometry, cells were fixed in 3% PFA/1X PBS then 70% ethanol, permeablized in 1X PBS/0.1% Triton X-100, and stained with 20 μg/ml propidium iodide. DNA content and Venus fluorescence were analyzed on the LSRII system (BD Biosciences).
Western Blotting
Proteins were extracted in RIPA buffer (IX PBS, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1 % SDS, protease inhibitors) and quantified by Protein Dye-Binding Assay (Biorad). 10–50 μg total protein were run on 4–12% Bis-Tris polyacrylamide gels (Invitrogen), transferred to PVDF membranes, and probed with antibodies listed in Supplementary Data.
CLIP
UV cross-linking and RNP purification/visualization were performed as described (Sanford et al., 2009). For quantification, CLIP and input samples were treated with Proteinase K (Ambion), and RNA was extracted, reverse-transcribed, and analyzed by real-time qPCR.
Apoptosis Assays
Annexin-V-Cy5 (Abcam) staining was quantified by automated microscopy. Caspase activation was measured with the DEVD-Nucview 488 reagent (Biotum), which stains nuclei specifically after it is cleaved by caspase-3. Cells were incubated for 1 hour with 2.5 μM substrate, fixed, and analyzed by automated microscopy.
Supplementary Material
Acknowledgments
We thank Junying Yuan and Robin Reed for reagents, Jodene Moore for help with flow cytometry, and Jessica Hurt, Natalie Gilks-Farny, Daniel Ducat, William Senapedis, Faisal Aldaye, Deborah Flusberg, and Ian Swinburne for thoughtful critiques and discussions. RNAi screening capability was provided by the ICCB-Longwood screening facility at Harvard Medical School. We are indebted to ICCB-L staff Caroline Shamu, Sean Johnston, Stewart Rudnicki, Zac Cooper, and David Wrobel. M.J.M. was supported by funding from the National Science Foundation. Work was supported by grant NIHGM057476 to P.A.S. from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akgul C, Moulding DA, Edwards SW. Alternative splicing of Bcl-2-related genes: functional consequences and potential therapeutic applications. Cell Mol Life Sci. 2004;17:2189–2199. doi: 10.1007/s00018-004-4001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenperä J, Taipale J. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;14:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YI, Moore RE, Ge HY, Young MK, Lee TD, Stevens SW. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 35:3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P, Toutant J, Shkreta L, Goekjian S, Revil T, Chabot B. Antagonistic effects of the SRp30c protein and cryptic 5′ splice sites on the alternative splicing of the apoptotic regulator Bcl-x. J Biol Chem. 2008;283:21315–21324. doi: 10.1074/jbc.M800353200. [DOI] [PubMed] [Google Scholar]
- Dassah M, Patzek S, Hunt VM, Medina PE, Zahler AM. A genetic screen for suppressors of a mutated 5′ splice site identifies factors associated with later steps of spliceosome assembly. Genetics. 2009;182:725–734. doi: 10.1534/genetics.109.103473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beaosoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C, Keen N, Taylor SS. The Ipl1/Aurora kinase family: methods of inhibition and functional analysis in mammalian cells. Methods Mol Biol. 2005;296:371–81. doi: 10.1385/1-59259-857-9:371. [DOI] [PubMed] [Google Scholar]
- Fesik SW. Promoting apoptosis as a strategy for cancer drug discovery. Nat Rev Cancer. 2005;5:876–885. doi: 10.1038/nrc1736. [DOI] [PubMed] [Google Scholar]
- Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- Hardwick JM, Youle RJ. SnapShot: BCL-2 Proteins. Cell. 2009;138:404. doi: 10.1016/j.cell.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genetics. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Lapenna S, Giordano A. Cell cycle kinases as therapeutic targets for cancer. Nat Rev Drug Discov. 2009;8:547–566. doi: 10.1038/nrd2907. [DOI] [PubMed] [Google Scholar]
- Letai AG. Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat Rev Cancer. 2008;8:121–132. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Lührmann R. A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J. 23:2381–91. doi: 10.1038/sj.emboj.7600241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massiello A, Roesser JR, Chalfant CE. SAP155 binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 2006;20:1680–1682. doi: 10.1096/fj.05-5021fje. [DOI] [PubMed] [Google Scholar]
- Massiello A, Chalfant CE. SRp30a (ASF/SF2) regulates the alternative splicing of caspase-9 pre-mRNA and is required for ceramide-responsiveness. J Lipid Res. 2006;47:892–897. doi: 10.1194/jlr.C600003-JLR200. [DOI] [PubMed] [Google Scholar]
- Mercatante DR, Mohler JL, Kole R. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J Biol Chem. 2002;277:49374–82. doi: 10.1074/jbc.M209236200. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Silver PA. Global analysis of mRNA splicing. RNA. 2008;14:197–203. doi: 10.1261/rna.868008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo JP, Bundman D, Cooper TA. A bichromatic fluorescent reporter for cell-based screen of alternative splicing. Nucleic Acids Res. 2006;34:e148. doi: 10.1093/nar/gkl967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rines DR, Gomez-Ferreria MA, Zhou Y, DeJesus P, Grob S, Batalov S, Labow M, Huesken D, Mickanin C, Hall J, et al. Whole genome functional analysis identifies novel components required for mitotic spindle integrity in human cells. Genome Biol. 2008;9:R44. doi: 10.1186/gb-2008-9-2-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;3:227–35. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–94. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. doi: 10.1016/j.molcel.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci USA. 2008;105:11218–11223. doi: 10.1073/pnas.0801661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JK, Zhang QQ, Wyatt JR, Dean NM. Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat Biotechnol. 1999;11:1097–1100. doi: 10.1038/15079. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Ou AC, Cho A, Benz EJ, Jr, Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol Cell Biol. 2008;28:5924–36. doi: 10.1128/MCB.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.