Abstract
A mesophilic bacterium, strain An4, was isolated from an underground gas storage reservoir with methanol as substrate and perchlorate as electron acceptor. Cells were Gram-negative, spore-forming, straight to curved rods, 0.5–0.8 μm in diameter, and 2–8 μm in length, growing as single cells or in pairs. The cells grew optimally at 37°C, and the pH optimum was around 7. Strain An4 converted various alcohols, organic acids, fructose, acetoin, and H2/CO2 to acetate, usually as the only product. Succinate was decarboxylated to propionate. The isolate was able to respire with (per)chlorate, nitrate, and CO2. The G+C content of the DNA was 42.6 mol%. Based on the 16S rRNA gene sequence analysis, strain An4 was most closely related to Sporomusa ovata (98% similarity). The bacterium reduced perchlorate and chlorate completely to chloride. Key enzymes, perchlorate reductase and chlorite dismutase, were detected in cell-free extracts.
Keywords: Sporomusa sp., Perchlorate, Underground gas storage
Introduction
Perchlorate and chlorate are used in a wide range of applications. Chlorate is used as an herbicide or defoliant. Perchlorate salts have been manufactured in large quantities and used as ingredients in solid rocket fuels, highway safety flares, air bag inflators, fireworks, and matches (Renner 1998; Logan 2001; Motzer 2001). Perchlorate is chemically very stable and has low reactivity even in highly reducing environments (Logan 1998). Inorganic perchlorate salts are generally soluble. Mishandling of these compounds has led to harmful concentrations in soil, groundwater, and drinking water supplies (Renner 1998; Urbansky and Schock 1999; Hogue 2003).
Perchlorate is reported to interrupt human thyroid hormone production (Urbansky 1998). Therefore, its occurrence in dairy and breast milk are of major concern (Kirk et al. 2005; Dyke et al. 2007; Dasgupta et al. 2008). Although perchlorate is chemically stable, many studies have proven that microorganisms can convert perchlorate to chloride under anaerobic conditions (i.e., Attaway and Smith 1993; Rikken et al. 1996; Bruce et al. 1999; Coates et al. 1999; Herman and Frankenberger 1999; Giblin et al. 2000; Logan et al. 2001).
Many mesophilic perchlorate-reducing mixed and pure cultures have been described, and so far, all of the isolates obtained are members of the Proteobacteria, with the majority of the isolates belonging to the Betaproteobacteria subclass (Wallace et al. 1996; Bruce et al. 1999; Herman and Frankenberger 1999; Logan 2001; Wu et al. 2001; Wolterink et al. 2002; 2005; Weelink et al. 2008; Thrash et al. 2010). Only recently, a (per)chlorate-reducing member, Moorella perchloratireducens, belonging to the Firmicutes, was isolated (Balk et al. 2008). This isolate is an acetogenic, thermophilic, and Gram-positive species. However, so far, perchlorate utilization has not been reported for acetogenic bacteria in this phylum that grow under mesophilic conditions. This report represents the description of a mesophilic acetogenic (per)chlorate-respiring bacterium. The isolation and characterization of strain An4 from underground gas storage in Russia are described.
Materials and methods
Source of inoculum
The sample was obtained from the liquid phase of underground gas storage reservoir in the period of gas extraction as described by Ivanova et al. (2007). The dry weight of the sample was about 700 mg l−1. It contained the following minerals; Fe(II) (140 mg l−1), NH+4 (2.8 mg l−1), K+ (2.6 mg l−1), Na+ (18 mg l−1), Mg2+ (4.4 mg l−1), Ca2+ (27 mg l−1), NO−3 (<1 mg l−1), SO2−4 (3.6 mg l−1), and Cl− (57 mg l−1). The pH of the sample was 6.8. Although the initial temperature in the sampling place was around 60–65°C, after the injection of the cold gas (20–23°C), the temperature became around 37°C (personal communication, Dr. Anna Ivanova).
Culture medium
The culture medium for enrichment, isolation, and maintenance of strain An4 was prepared as described by Stams et al. (1993), except that (per)chlorate was tested without the addition of sulfide. Unless stated otherwise, all cultivations were carried out at 37°C. The cultures were routinely grown in 117-ml serum vials with butyl rubber stoppers and aluminum crimp seals. The vials contained 50-ml basal medium and a gas phase of 1.7-bar N2/CO2 (80/20%, v/v). Concentrated stock solutions of substrates were prepared anoxically, sterilized by filtration and added to the medium to final concentrations of 5–20 mM. The pH of the medium was 7. By varying the CO2 concentration in the headspace and adding a few drops of 0.1 N HCl or NaOH per vial, the pH of the medium could be adjusted within the range of 5.5–8.5. In all growth experiments in liquid medium, the inoculum size was 1% (v/v).
Enrichment and isolation of strain An4
The enrichment culture was cultivated in a bicarbonate-buffered medium containing methanol and perchlorate at 37°C. The culture was repeatedly transferred to fresh medium when methanol and perchlorate were consumed. Serial dilutions of the sample from the underground gas storage were prepared in liquid media containing 20 mM of methanol and 10 mM of perchlorate. The highest dilution showing growth at 37°C was used for further study. The culture was diluted in agar media (1.8%, w/v, Agar Noble) in the serum vials. Colonies from the highest dilution were picked, and serial agar and liquid dilutions were repeated until a pure culture was obtained. The pure culture was designated strain An4.
Cell morphology and purity were examined with a phase-contrast microscope. Gram-staining was carried out according to the standard procedure (Doetsch 1981).
Substrate utilization tests
The ability of strain An4 to metabolize substrates was tested in the bicarbonate-buffered medium. Substrates were added from sterile anoxic concentrated stock solutions. To test he use of potential electron acceptors on methanol, lactate, and fructose, sodium perchlorate (10 mM), sodium chlorate (10 mM), sodium sulfate (20 mM), sodium thiosulfate (20 mM), sodium sulfite (5 mM), FeCl3 (10 mM), Fe(III)-NTA (10 mM), Fe(III)-citrate (10 mM), anthraquinone-2,6-disulfonate (AQDS) (20 mM), and sodium nitrate (10 mM) were added to the medium. Fe(III)-NTA was prepared as described by Caldwell et al. (1999).
16S RNA sequence analysis and G+C content of DNA
For the phylogenetic characterization of strain An4, DNA was extracted and purified using the UltraClean Soil DNA kit (MoBio) according to the manufacturer’s instructions. PCR was performed with the bacterial primers 7f and 1510r (Lane 1991) by using the Taq DNA polymerase kit (Life Technologies) to amplify the bacterial 16S rRNA gene. The PCR products were purified with the Qiaquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Sequencing of the complete 16S rRNA gene was performed at Westburg Genomics (http://genomics.westburg.nl). A total of 1,386 nucleotides of the 16S rRNA gene were sequenced. The sequences were checked with the alignment programs of the ARB package (Ludwig et al. 2004), and a rooted neighbor-joining tree (Escherichia coli 8, D83536 positions 38–1427) was constructed using Dechloromonas hortensis (DSM 15637) as an outgroup. On-line similarity analysis of the 16S rRNA gene sequences was performed with the BLAST program at NCBI and EMBL databases.
The G+C content of the DNA was determined by using HPLC method described by Mesbah et al. (1989) at the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Genomic DNA was isolated according to the procedure that was described previously (Cashion et al. 1977).
Analytical methods
Most substrates were measured by HPLC as previously described (Stams et al. 1993). Gasses and alcohols were measured by gas chromatography (Balk et al. 2003; Henstra and Stams 2004). Perchlorate, chlorate, chloride, thiosulfate, nitrate, and sulfate were analyzed by a HPLC system equipped with an Ionpac AS9-SC column and an ED 40 electrochemical detector (Dionex, Sunnyvale, CA) (Scholten and Stams 1995). Perchlorate showed a very broad peak in the chromatogram. Therefore, the assessment of perchlorate concentration was semi-quantitative. Perchlorate consumption was quantified based on the increase in chloride formation. Chloride and chlorate measurements by HPLC were very accurate. Sulfide was analyzed by the method of Trüper and Schlegel (1964). The protein content of the cell extracts was determined according to the method of Bradford (1976) with bovine serum albumin as a standard.
Growth was measured as the optical density at 600 nm (OD600). Uninoculated medium served as a reference. The results are representative of replicate experiments.
Enzyme assays
Cell extracts used for enzyme assays were obtained from cells grown in the medium supplemented with 20 mM of methanol and 10 mM of perchlorate. Chlorate, perchlorate, and nitrate reductase levels were measured with anoxic techniques in stoppered quartz cuvettes, by monitoring the oxidation of reduced methyl viologen (MV) at 578 nm and 30 and 37°C as it was described by Kengen et al. (1999). The assay mixture (1 ml) consisted of 50 mM Tris buffer (pH 7.5), 0.5 mM MV, and an appropriate amount of enzyme. The assay mixture was prereduced by a small amount of a dithionite solution (0.2 M) until an absorbance of 1.5 was reached, and then the reaction was started by the addition of 10 μl of chlorate (0.4 M) or perchlorate (0.4 M). Specific activities were calculated from the linear decrease in absorbance, with an extinction coefficient of 9.7 mM−1 for MV. Chlorite dismutase activity was determined by measuring the production of oxygen with a Clark-type oxygen electrode (van Ginkel et al. 1996).
Lipid analysis
Bacterial cultures of strain An4, Sporomusa ovata, and Sporomusa aerivorans grown on methanol in the bicarbonate-buffered medium were harvested by centrifugation (14,500 × g, 20 min, 4°C), and pellets were washed with distilled water. Lipids from lyophilized cells were extracted ultrasonically using a mixture of dichloromethane (DCM):methanol of 2:1 (v/v), and this procedure was repeated four times. The residue was saponified with 1 N KOH in 96% of methanol by refluxing for 1 h and subsequently neutralized and extracted with DCM. After addition of internal standards, the “free” and “bound” lipid extracts were methylated and silylated and subsequently analyzed by GC and GC-mass spectrometry (GC-MS).
Accession numbers
The 16S rRNA gene sequence of strain An4 has been deposited in the GenBank database under the accession number EF060193. The EMBL database accession numbers of the strain most closely related to strain An4 is S. ovata (DSM 2662) (98% sequence similarity).
Results
Isolation and morphological characterization
An enrichment culture that grew at 37°C in a perchlorate-containing bicarbonate-buffered medium with methanol as the growth substrate was obtained from an underground gas storage in Russia. The culture formed mainly acetate and chloride as products, but no methane was formed. A pure culture of perchlorate-reducing bacteria was obtained by using the agar and liquid dilution methods. The pure culture was designated strain An4 (=DSM 21435, =JCM 15643).
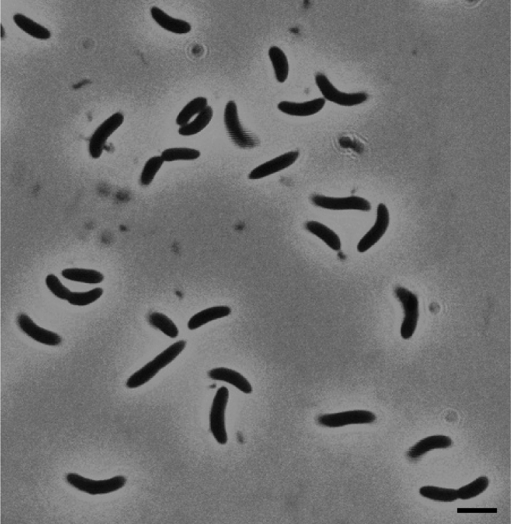
Strain An4 is a spore-forming bacterium, and the cells are straight to curved rods (Fig. 1). In the stationary growth-phase, they formed terminal endospores in club-shaped sporangia. When grown on methanol or fructose, the cells were 0.4–0.6 μm in diameter and 2–8 μm in length, and on H2/CO2, the cells were 0.4–0.6 μm in diameter and 6–8 μm in length. Spores located terminally, and cells stained Gram-negative.
Fig. 1.
Phase-contrast micrograph of strain An4. Bar 2.5 μm
16S rRNA analysis and G+C content
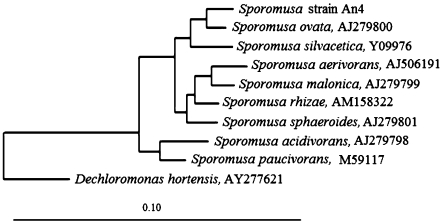
A phylogenetic analysis of the almost full-length 16S rRNA sequence (1,386 bases) revealed that strain An4 is a member of the genus Sporomusa within the family Acidaminococcaceae of the order Clostridiales of class “Clostridia” of the phylum Firmicutes of the domain Bacteria (Garrity and Holt 2000) (Fig. 2).
Fig. 2.
Dendrogram showing the position of strain An4 among the members of the genus Sporomusa. Phylogenetic analysis based on 16S rRNA gene sequences available from GenBank data bases. Bar evolutionary distance of 0.10
The closest relative of strain An4 is Sporomusa strain DR5 and S. ovata based on 16S rRNA sequence analysis (99% and 98% sequence similarity, respectively). Because strain DR5 was not deposited in a culture collection (personal communication, Dr. Peter H. Janssen), we could not study this strain for its physiology. Nevertheless, we were able to compare strain An4 with S. ovata (=DSM 2662) and S. aerivorans (=DSM 13326) in several physiological aspects (Table 1).
Table 1.
Characteristics of strain An4 and related representatives of the genus Sporomusa
| An 4 | S. ovata a | S. aerivorans b | S. silvacetica c | |
|---|---|---|---|---|
| Opt. temp(°C) | 37 | 34–39 | 30 | 25–30 |
| Temp. range | 20–40 | 15–45 | 19–35 | ND |
| Cell size (μm) | 0.5–1.3 × 0.8–3 | 0.7–1.0 × 1–5 | 1.3–7.0 × 0.6–0.7 | 3.5 × 0.7 |
| G+C content (mol%) | 42.6 | 41.3–42.2 | ND | 43 |
| pH range | 5.5–8.0 | 5.0–8.1 | 6.2–8.2 | 6.8 |
| Habitat | Underground gas storage | Silage | Termite gut | Forest soil |
| H2/SO2−4 | − | − | − | − |
| H2/CO2 | + | + | + | + |
| Formate | + | + | + | + |
| CO | + | + | ND | − |
| Methanol | + | + | + | + |
| Ethanol | + | + | + | + |
| n-Propanol | + | + | ND | ND |
| n-Butanol | + | + | ND | ND |
| 1,2-propanediol | − | ± | ND | ND |
| Glycerol | − | − | ND | + |
| 1,2-Butanediol | − | ND | ND | ND |
| 2,3-Butanediol | − | ND | ND | + |
| Pyruvate | + | + | + | + |
| DL-Lactate | + | + | + | + |
| Fumarate | ± | − | + | + |
| Succinate | + | − | + | − |
| Malate | ± | ND | + | ND |
| Glutamate | + | ND | − | ND |
| Citrate | − | − | + | − |
| Oxaloacetate | − | ND | + | − |
| Fructose | + | + | − | + |
| Glucose | − | − | − | − |
| Mannitol | − | − | + | − |
| Cellobiose | − | ND | ND | ND |
| Alanine | + | − | + | ND |
| Betaine | + | + | ND | + |
| Sarcosine | + | + | ND | ND |
| 3,4,5-Trimethoxy benzoate | + | − | + | |
| PCE with methanol | + | + | ND | ND |
| Nitrate reduction | + | + | − | − |
| (Per)chlorate reduction | + | − | − | ND |
| Sulfate reduction | − | − | − | − |
| Thiosulfate reduction | − | − | − | − |
| Sulfite reduction | − | − | − | − |
| Fe-NTA | − | ND | ND | ND |
The DNA base composition (G+C) of strain An4 was 42.6 mol%.
Growth and substrate utilization
Strain An4 grew between 20°C and 40°C; optimum growth occurred at 37°C. It grew optimally at pH 7. Strain An4 was able to grow on H2/CO2, CO, methanol, ethanol, glycerol, n-propanol, n-butanol, glycine, alanine, sarcosine, betaine, acetoin, choline, methyl amines, pyruvate, lactate, fructose, succinate, and glutamate (Table 1). Table 2 shows the stoichiometry and fermentation products of strain An4 grown on selected compounds. Strain An4 did not grow well on formate, but in the presence of (per)chlorate or nitrate, formate supported good growth. The following substrates were tested, but not utilized for growth in the absence or presence of perchlorate: glucose, cellobiose, ribose, oxaloacetate, butyrate, glycolate, glycerol, and citrate. When the isolate was grown with (per)chlorate, the organic substrates were mainly converted to CO2, while only small amounts of acetate were formed (Table 2). In the absence of (per)chlorate, the product was mainly acetate. The bacterium reduced perchlorate to chloride. The isolate was not able to respire with sulfate, thiosulfate, FeCl3, Fe(III)-NTA, Fe(III)-citrate, and AQDS. Strain An4 was also able to cometabolically dechlorinate tetrachloroethylene (PCE) to trichloroethylene during growth on methanol + CO2 in a similar way as it was previously described for S. ovata (Terzenbach and Blaut 1994).
Table 2.
Stoichiometry and fermentation products of strain An4 grown on selected substrates
| Product formed (mM) | |||||
|---|---|---|---|---|---|
| Substrate | (mM) | OD600 | Acetate | Propionate | Butyrate |
| H2/CO2 | 23.5 | 0.38 | 7.1 | ||
| Formate | 19.7 | 0.14 | 5.2 | ||
| CO | 22.1 | 0.22 | 5.5 | ||
| Methanol (+CO2) | 19.6 | 0.18 | 15.7 | ||
| Ethanol | 10.3 | 0.15 | 14.2 | ||
| n-Propanol | 18.9 | 0.19 | 11.1 | 17.8 | |
| n-Butanol | 21.3 | 0.18 | 11.7 | 18.8 | |
| Pyruvate | 19.8 | 0.18 | 21.7 | ||
| Succinate | 20.8 | 0.27 | 0.8 | 18.8 | |
| Glutamate | 19.9 | 0.23 | 20.3 | ||
| Fructose | 21.3 | 0.33 | 44 | ||
| Alanine | 10.4 | 0.21 | 18.4 | 0.3 | |
| Lactate | 21.2 | 0.17 | 41.3 | 0.2 | |
| Lactate + perchlorate | 9.2 | 0.46 | |||
| Lactate + chlorate | 9.1 | 0.34 | 4.5 | ||
During growth with perchlorate, the medium became pink due to the presence of resorufin formed from resazurin upon deoxygenation and when all the (per)chlorate was consumed, the pink color disappeared again.
Testing (per)chlorate reduction by other members of the genus Sporomusa
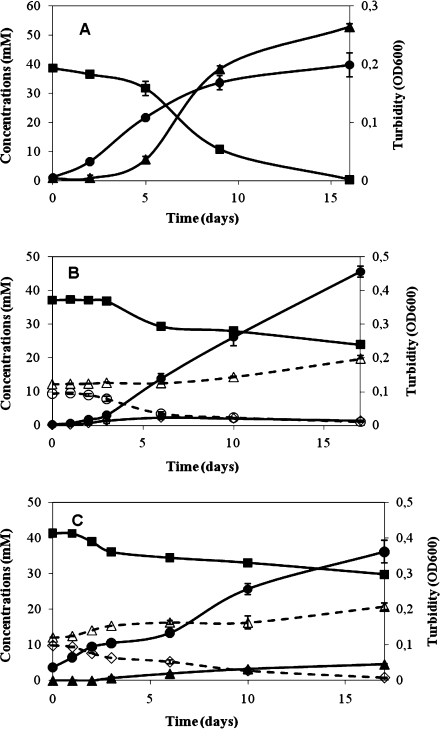
The ability of Sporomusa species, S. ovata (=DSM 2662) and S. aerivorans (=DSM 13326) to reduce perchlorate was tested either with lactate or with methanol. Both substrates are utilized by the two strains of the genus Sporomusa (Möller et al. 1984; Boga et al. 2003). However, S. ovata and S. aerivorans were not able to reduce perchlorate or chlorate to chloride.
Growth of these two strains in the medium with (per)chlorate did not result in a change of product formation with lactate as a substrate, which was the case for strain An4 (Fig. 3, Table 2). Transferring and adapting the strains several times to the medium containing perchlorate and lactate or methanol did not result in a substantial reduction of perchlorate to chloride. Although perchlorate and chlorate were reduced by the strain An4, lactate was not consumed completely. Probably, (per)chlorate utilization inhibited further fermentative growth with the remaining lactate.
Fig. 3.
Growth and lactate utilization by strain An4 in the absence of (per)chlorate (a) and in the presence of perchlorate (b) and chlorate (c). Bacterial growth was determined by measuring turbidity (OD600). Curves are labeled as follows: black square lactate, white circle perchlorate, white diamond chlorate; white triangle chloride, black triangle acetate, black circle OD,
Enzyme measurements
(Per)chlorate reductase activity could be demonstrated by using reduced MV as the artificial electron donor. The activity towards chlorate was approximately threefold higher than the activity towards perchlorate (Table 3). The pathway also appeared to involve the dismutation of chlorite yielding chloride and molecular oxygen.
Table 3.
The enzyme activities in cell extracts of strain An4 grown on methanol and perchlorate at two different temperatures
| 30°C | 37°C | |
|---|---|---|
| Perchlorate reductase | 0.45 ± 0.01 | 0.74 ± 0.00 |
| Chlorate reductase | 1.59 ± 0.04 | 2.19 ± 0.10 |
| Nitrate reductase | 0.74 ± 0.01 | 1.10 ± 0.01 |
| Chlorite dismutase | 15.16 ± 0.75 | 15.51 ± 2.90 |
Values are reported as 1 μmol of sodium perchlorate, sodium chlorate, or sodium nitrate reduced, and sodium chlorite dismutated per minute per milligram of cell protein
Lipids analysis
The fatty acid profile of strain An4 was not similar to S. ovata and S. aerivorans while grown on methanol (Table 4). Although the major fatty acids, iso-βOH-C13:0 and C16:1 Δ7, were present in all the strains, the composition of the remaining fatty acids was quite different.
Table 4.
Fatty acid composition of strain An4 in comparison to related species
| Fatty acids | An4 | S. ovata | S. aerivorans |
|---|---|---|---|
| Iso-C11:0 | 2.3 | 1.2 | 1.1 |
| βOH-C11 | – | 1.3 | 0.9 |
| C14:0 | 0.5 | 1.2 | 1.1 |
| βOH-C12 | 1.5 | 7.3 | 10.5 |
| Iso-C15:1 Δ7 + 9 | 0.9 | – | – |
| Iso-C15:0 | 4.3 | 0.7 | 0.8 |
| C15:1 Δ7 | 1.3 | 11.6 | 7.3 |
| Iso-βOH-C13:0 | 26.5 | 8.0 | 8.6 |
| βOH-C13:1 | – | 1.1 | 0.8 |
| C15:0 | 0.9 | 3.8 | 1.5 |
| Iso-C16:1 Δ7 | 1.4 | 0.4 | – |
| Iso-C16:0 | 0.7 | – | – |
| βOH-C13:0 | – | 4.1 | 2.0 |
| C16:1 Δ7 | 7.0 | 20.6 | 27.5 |
| C16:1 Δ9 | 1.9 | 2.2 | 4.0 |
| C16:1 Δ11 | 0.6 | 0.6 | 0.5 |
| C16:0 | 4.0 | 7.2 | 7.1 |
| Iso-C17:1 Δ7 | 22.3 | 1.7 | 2.0 |
| Iso-C17:1 Δ9 | 5.6 | 0.6 | 1.0 |
| Anteiso-C17:1 Δ7 | 3.1 | – | – |
| Iso-C17:0 | 6.3 | – | – |
| Anteiso-C17:0 | 1.4 | – | – |
| C17:1 Δ7 | 2.0 | 6.1 | 2.7 |
| C17:1 Δ9 | 1.4 | 12.6 | 10.2 |
| C17:1 Δ11 | – | 1.5 | 1.1 |
| C17:0 | 0.5 | 1.7 | 0.7 |
| C18:1 Δ9 | 2.1 | 3.4 | 5.5 |
| C18:1 Δ11 | 0.5 | 1.0 | 2.5 |
| C18:0 | 1.2 | 0.6 | 0.7 |
| Iso-C19:1 Δ9 | 0.7 | – | – |
Values are percentages of total quantified fatty acids
– Not detected
Discussion
A Sporomusa strain An4 that is capable of reducing (per)chlorate to chloride was obtained from an underground gas storage in Russia. The genus Sporomusa was created in 1984 (Möller et al. 1984) to accommodate a number of strains of anaerobic acetogenic bacteria distinguished by having a Gram-negative cell wall and an ability to form endospores, two properties whose coincidence is uncommon in Microbiology. In general, the members of the genus Sporomusa can utilize a wide variety of electron donors. Sporomusa has been isolated from a large variety of habitats including anoxic sediments of freshwater rivers, lakes, creeks, and ditches (Möller et al. 1984; Hermann et al. 1987; Dehning et al. 1989; Sass et al. 1998), and anoxic soil from rice paddy microcosms (Rosencrantz et al. 1999), from soils and silage (Möller et al. 1984; Kuhner et al. 1997), from sugar beet factory and distillery wastewater (Möller et al. 1984; Ollivier et al. 1985), from horse and cattle dung (Möller et al. 1984), from the guts of wood-feeding termites (Breznak and Switzer 1986; Breznak et al. 1988), and soil-feeding termites (Boga et al. 2003). All Sporomusa species have been described to gain energy for growth by the reduction of carbon dioxide to acetate. The reduction of CO2 to acetate occurs via acetogenesis, and acetogens utilize the acetyl-CoA pathway as a terminal electron-accepting energy-conserving CO2-fixing process (Drake et al. 2006; Drake 2009).
Perchlorate and chlorate reductase activities of strain An4 were active at 30 to 37°C. Strain An4 was also able to reduce nitrate, and nitrate reductase activity was detected (Table 3). Currently, we do not know the genes encoding (per)chlorate reduction and chlorite dismutation. Although microbial nitrate reductases have activity with chlorate, it is not known whether perchlorate is a substrate as well (Shanmugam et al. 1992; Bender et al. 2005). We tested and found that S. ovata was also able to reduce nitrate, but it did not reduce perchlorate.
More than 50 dissimilatory (per)chlorate-reducing bacteria are now in pure culture, and this number continues to increase (i.e., Bruce et al. 1999; Coates et al. 1999; Wolterink et al. 2002; Weelink et al. 2008). (Per)chlorate-reducing bacteria have been isolated from a broad variety of environments, including contaminated soils and sediments (Rikken et al. 1996; Wallace et al. 1996; Bruce et al. 1999; Michaelidou et al. 2000; Coates and Achenbach 2004; Weelink et al. 2008). The bacteria described to date belonged mostly to the Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Epsilonproteobacteria subclasses of the Proteobacteria, but the majority are in the Betaproteobacteria subclass and are members of the genus Dechloromonas or Azospira (formerly Dechlorosoma) (Coates et al. 1999; Achenbach et al. 2001). The isolation of strain An4 suggests that many other bacteria might be capable of (per)chlorate reduction than have been considered up to now.
Acknowledgments
We thank Anna Ivanova (Institute of Microbiology of the Russian Academy of Sciences in Moscow) for kindly providing the sample that was used in this study.
This work was supported by the Darwin Center for Biogeology of The Netherlands Organization for Scientific Research (NWO) and also partly supported by Award No KUK-C1-017-12, made by King Abdullah University of Science and Technology (KAUST). This is publication no. 4770 of the Netherlands Institute of Ecology (NIOO-KNAW).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The GenBank accession number for the 16S rRNA sequence of Sporomusa sp. is EF060193
References
- Achenbach LA, Michaelidou U, Bruce RA, Fryman J, Coates JD. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int J Syst Evol Microbiol. 2001;51:527–533. doi: 10.1099/00207713-51-2-527. [DOI] [PubMed] [Google Scholar]
- Attaway H, Smith M. Reduction of perchlorate by an anaerobic enrichment culture. J Ind Microbiol. 1993;12:408–412. doi: 10.1007/BF01569673. [DOI] [Google Scholar]
- Balk M, Weijma J, Friedrich MW, Stams AJM. Methanol utilization by a novel thermophilic homoacetogenic bacterium, Moorella mulderi sp. nov., isolated from a bioreactor. Arch Microbiol. 2003;179:315–320. doi: 10.1007/s00203-003-0523-x. [DOI] [PubMed] [Google Scholar]
- Balk M, van Gelder T, Weelink SA, Stams AJM. (Per)chlorate reduction by the thermophilic bacterium Moorella perchloratireducens sp. nov., isolated from underground gas storage. Appl Environ Microbiol. 2008;74:403–409. doi: 10.1128/AEM.01743-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender KS, Shang C, Chakraborty R, Belchik SM, Coates JD, Achenbach LA. Identification, characterization, and classification of genes encoding perchlorate reductase. J Bacteriol. 2005;187:5090–5096. doi: 10.1128/JB.187.15.5090-5096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boga HI, Ludwig W, Brune A. Sporomusa aerivorans sp. nov., and oxygen-reducing homoacetogenic bacterium from the gut of a soil-feeding termite. Int J Syst Evol Microbiol. 2003;53:1397–1404. doi: 10.1099/ijs.0.02534-0. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Breznak JA, Switzer JM. Acetate synthesis from H2 plus CO2 by termite gut microbes. Appl Environ Microbiol. 1986;52:623–630. doi: 10.1128/aem.52.4.623-630.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breznak JA, Switzer JM, Seitz H-J. Sporomusa termitida sp. nov., an H2/CO2-utilizing acetogen isolated from termites. Arch Microbiol. 1988;150:282–288. doi: 10.1007/BF00407793. [DOI] [Google Scholar]
- Bruce RA, Achenbach LA, Coates JD. Reduction of (per)chlorate by a novel organism isolated from paper mill waste. Environ Microbiol. 1999;1:319–329. doi: 10.1046/j.1462-2920.1999.00042.x. [DOI] [PubMed] [Google Scholar]
- Caldwell ME, Tanner RS, Suflita JM. Microbial metabolism of benzene and the oxidation of ferrous iron under anaerobic conditions: implications for bioremediation. Anaerobe. 1999;5:595–603. doi: 10.1006/anae.1999.0193. [DOI] [Google Scholar]
- Cashion P, Holder-Franklin MA, McCully J, Franklin M. A rapid method for the base ratio determination of bacterial DNA. Anal Biochem. 1977;81:461–466. doi: 10.1016/0003-2697(77)90720-5. [DOI] [PubMed] [Google Scholar]
- Coates JD, Achenbach LA. Microbial perchlorate reduction: rocket-fueled metabolism. Nat Rev Microbiol. 2004;2:569–580. doi: 10.1038/nrmicro926. [DOI] [PubMed] [Google Scholar]
- Coates JD, Michaelidou U, Bruce RA, O’Connor SM, Crespi JN, Achenbach LA. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl Environ Microbiol. 1999;65:5234–5241. doi: 10.1128/aem.65.12.5234-5241.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta PK, Kirk AB, Dyke JV, Ohira S. Intake of iodine and perchlorate and excretion in human milk. Environ Sci Technol. 2008;42(21):8115–8121. doi: 10.1021/es801549w. [DOI] [PubMed] [Google Scholar]
- Dehning I, Stieb M, Schink B. Sporomusa malonica sp. nov., a homoacetogenic bacterium growing by decarboxylation of malonate or succinate. Arch Microbiol. 1989;151:421–426. doi: 10.1007/BF00416601. [DOI] [Google Scholar]
- Doetsch RN. Determinative methods of light microscopy. In: Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB, editors. Manual of methods for general bacteriology. Washington, DC: American Society for Microbiology; 1981. pp. 21–33. [Google Scholar]
- Drake HL. Sporomusa. In: De Vos P, Garrity GM, Jones D, Frieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB, editors. Bergey’s manual of systematic bacteriology, Vol. 3. 2. New York: Springer-Verlag; 2009. pp. 1112–1116. [Google Scholar]
- Drake HL, Küsel K, Matthies C. Acetogenic prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes, Volume 2. 3. New York: Springer-Verlag; 2006. pp. 354–420. [Google Scholar]
- Dyke JV, Ito K, Obitsu T, Hisamatsu Y, Dasgupta PK, Blount BC. Perchlorate in dairy milk. Comparison of Japan versus the United States. Environ Sci Technol. 2007;41(1):88–92. doi: 10.1021/es061429e. [DOI] [PubMed] [Google Scholar]
- Garrity GM, Holt JG. An overview of the road map to the manual. In: Garrity GM, Holt JG, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer Verlag; 2000. pp. 1–19. [Google Scholar]
- Giblin TL, Herman DC, Frankenberger WT. Removal of perchlorate from ground water by hydrogen-utilizing bacteria. J Environ Qual. 2000;29:1057–1062. doi: 10.2134/jeq2000.00472425002900040004x. [DOI] [Google Scholar]
- Henstra AM, Stams AJM. Novel physiological features of Carboxydothermus hydrogenoformans and Thermoterrabacterium ferrireducens. Appl Environ Microbiol. 2004;70:7236–7240. doi: 10.1128/AEM.70.12.7236-7240.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman DC, Frankenberger WT. Bacterial reduction of perchlorate and nitrate in water. J Environ Qual. 1999;28:1018–1024. doi: 10.2134/jeq1999.00472425002800030036x. [DOI] [Google Scholar]
- Hermann M, Popoff MR, Sebald M. Sporomusa paucivorans sp. nov., a methylotrophic bacterium that forms acetic acid from hydrogen and carbon dioxide. Int J Syst Bacteriol. 1987;37:93–101. doi: 10.1099/00207713-37-2-93. [DOI] [Google Scholar]
- Hogue C. Rocket-fueled river. Chem Eng News. 2003;81:37–46. [Google Scholar]
- Ivanova AE, Borzenkov IA, Tarasov AL, Milekhina EI, Belyaev SS. A microbiological study of An Underground gas storage in the process of gas extraction. Microbiology. 2007;76:461–468. doi: 10.1134/S0026261707040121. [DOI] [PubMed] [Google Scholar]
- Kengen SWM, Rikken GB, Hagen WR, van Ginkel CG, Stams AJM. Purification and characterization of (per)chlorate reductase from the chlorate-respiring strain GR-1. J Bacteriol. 1999;181:6706–6711. doi: 10.1128/jb.181.21.6706-6711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk AB, Martinelango PK, Tian K, Dutta A, Smith EE, Dasgupta PK. Perchlorate and iodide in dairy and milk. Environ Sci Technol. 2005;39:2011–2017. doi: 10.1021/es048118t. [DOI] [PubMed] [Google Scholar]
- Kuhner CH, Frank C, Griesshammer A, Schmittroth M, Acker G, Gossner A, Drake HL. Sporomusa silvacetica sp. nov., an acetogenic bacterium isolated from aggregated forest soil. Int J Syst Bacteriol. 1997;47:352–358. doi: 10.1099/00207713-47-2-352. [DOI] [PubMed] [Google Scholar]
- Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester: Wiley; 1991. pp. 115–175. [Google Scholar]
- Logan BE. A review of chlorate- and perchlorate-respiring microorganisms. Bioremediat J. 1998;2:69–79. doi: 10.1080/10889869891214222. [DOI] [Google Scholar]
- Logan BE. Assessing the outlook for perchlorate remediation. Environ Sci Technol. 2001;35:482A–487A. doi: 10.1021/es012564o. [DOI] [PubMed] [Google Scholar]
- Logan BE, Zhang HS, Mulvaney P, Milner MG, Head IM, Unz RF. Kinetics of perchlorate- and chlorate-respiring bacteria. Appl Environ Microbiol. 2001;67:2499–2506. doi: 10.1128/AEM.67.6.2499-2506.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah M, Premachandran U, Whitman W. Precise measurement of the G+C content of deoxyribonucleic acid by high performans liquid chromatography. Int J Syst Bact. 1989;39:159–167. doi: 10.1099/00207713-39-2-159. [DOI] [Google Scholar]
- Michaelidou U, Achenbach LA, Coates JD. Isolation and characterization of two novel (per)chlorate-reducing bacteria from swine waste lagoons. In: Urbansky ET, editor. Perchlorate in the environment. New York: Kluwer/Plenum Publishers; 2000. pp. 271–283. [Google Scholar]
- Möller B, Ossmer R, Howard BH, Gottschalk G, Hippe H. Sporomusa, a new genus of Gram-negative anaerobic-bacteria including Sporomusa sphaeroides spec. nov. and Sporomusa ovata spec. nov. Arch Microbiol. 1984;139:388–396. doi: 10.1007/BF00408385. [DOI] [Google Scholar]
- Motzer WE. Perchlorate: problems, detection, and solutions. Environ Forensics. 2001;2:301–311. doi: 10.1006/enfo.2001.0059. [DOI] [Google Scholar]
- Ollivier B, Cordruwisch R, Lombardo A, Garcia JL. Isolation and characterization of Sporomusa acidovorans sp. nov., a methylotrophic homoacetogenic bacterium. Arch Microbiol. 1985;142:307–310. doi: 10.1007/BF00693409. [DOI] [Google Scholar]
- Renner R. Perchlorate-tainted wells spur government action. Environ Sci Technol. 1998;9:210A. doi: 10.1021/es983489t. [DOI] [PubMed] [Google Scholar]
- Rikken GB, Kroon AGM, van Ginkel CG. Transformation of (per)chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl Microbiol Biotechnol. 1996;45:420–426. doi: 10.1007/s002530050707. [DOI] [Google Scholar]
- Rosencrantz D, Rainey FA, Janssen PH. Culturable populations of Sporomusa spp. and Desulfovibrio spp. in the anoxic bulk soil of flooded rice microcosms. Appl Environ Microbiol. 1999;65:3526–3533. doi: 10.1128/aem.65.8.3526-3533.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass H, Wieringa E, Cypionka H, Babenzien HD, Overmann J. High genetic and physiological diversity of sulfate-reducing bacteria isolated from an oligotrophic lake sediment. Arch Microbiol. 1998;170:243–251. doi: 10.1007/s002030050639. [DOI] [PubMed] [Google Scholar]
- Scholten JC, Stams AJM. The effect of sulphate and nitrate on methane formation in a freshwater sediment. Antonie Leeuwenhoek. 1995;68:309–315. doi: 10.1007/BF00874141. [DOI] [PubMed] [Google Scholar]
- Shanmugam KT, Stewart V, Gunsalus RP, Boxer DH, Cole JA, Chippaux M, DeMoss JA, Giordano G, Lin ECC, Rajagopalan KV. Proposed nomenclature for the genes involved in molybdenum metabolism in Escherichia coli and Salmonella typhimurium. Mol Microbiol. 1992;6:3452–3454. doi: 10.1111/j.1365-2958.1992.tb02215.x. [DOI] [PubMed] [Google Scholar]
- Stams AJM, van Dijk JB, Dijkema C, Plugge CM. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Appl Environ Microbiol. 1993;59:1114–1119. doi: 10.1128/aem.59.4.1114-1119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzenbach DP, Blaut M. Transformation of tetrachloroethylene to trichloroethylene by homoacetogenic bacteria. FEMS Microbiol Lett. 1994;123:213–218. doi: 10.1111/j.1574-6968.1994.tb07224.x. [DOI] [PubMed] [Google Scholar]
- Thrash JC, Pollock J, Torok T, Coates JD. Description of the novel perchlorate-reducing bacteria Dechlorobacter hydrogenophilus gen. nov., sp. nov. and Propionivibrio militaris, sp. nov. Appl Microbiol Biotechnol. 2010;86:335–343. doi: 10.1007/s00253-009-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trüper HG, Schlegel HG. Sulphur metabolism in Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Antonie Leeuwenhoek. 1964;30:225–238. doi: 10.1007/BF02046728. [DOI] [PubMed] [Google Scholar]
- Urbansky ET. Perchlorate chemistry: implications for analysis and remediation. Bioremediat J. 1998;2:81–95. doi: 10.1080/10889869891214231. [DOI] [Google Scholar]
- Urbansky ET, Schock MR. Issues in managing the risks associated with perchlorate in drinking water. J Environ Manag. 1999;56:79–95. doi: 10.1006/jema.1999.0274. [DOI] [Google Scholar]
- van Ginkel CG, Rikken GB, Kroon AGM, Kengen SWM. Purification and characterization of a chlorite dismutase: a novel oxygen generating enzyme. Arch Microbiol. 1996;166:321–326. doi: 10.1007/s002030050390. [DOI] [PubMed] [Google Scholar]
- Wallace W, Ward T, Breen A, Attaway H. Identification of an anaerobic bacterium which reduces perchlorate and chlorate as Wolinella succinogenes. J Ind Microbiol. 1996;16:68–72. doi: 10.1007/BF01569924. [DOI] [Google Scholar]
- Weelink SAB, Tan NCG, ten Broeke H, van den Kieboom C, van Doesburg W, Langenhoff AAM, Gerritse J, Junca H, Stams AJM. Isolation and characterization of Alicycliphilus denitrificans strain BC that grows on benzene with chlorate as electron acceptor. Appl Environ Microbiol. 2008;74:6672–6681. doi: 10.1128/AEM.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolterink AFWM, Jonker AB, Kengen SW, Stams AJ. Pseudomonas chloritidismutans sp. nov., a non-denitrifying, chlorate-reducing bacterium. Int J Syst Evol Microbiol. 2002;52:2183–2190. doi: 10.1099/ijs.0.02102-0. [DOI] [PubMed] [Google Scholar]
- Wolterink AWFM, Kim S, Muusse M, Kim IS, Roholl PJ, van Ginkel CG, Stams AJM, Kengen SW. Dechloromonas hortensis sp. nov. and strain ASK-1, two novel(per)chlorate-reducing bacteria, and taxonomic description of strain GR-1. Int J Syst Evol Microbiol. 2005;55:2063–2068. doi: 10.1099/ijs.0.63404-0. [DOI] [PubMed] [Google Scholar]
- Wu J, Unz RF, Zhang H, Logan BE. Persistence of perchlorate and the relative numbers of perchlorate- and chlorate-respiring microorganisms in natural waters, soils, and wastewater. Bioremediat J. 2001;5:119–130. doi: 10.1080/20018891079230. [DOI] [Google Scholar]