SUMMARY
ATG12, an ubiquitin-like modifier required for macroautophagy, has a single known conjugation target, another autophagy regulator called ATG5. Here, we identify ATG3 as a substrate for ATG12 conjugation. ATG3 is the E2-like enzyme necessary for ATG8/LC3 lipidation during autophagy. ATG12-ATG3 complex formation requires ATG7 as the E1 enzyme and ATG3 autocatalytic activity as the E2, resulting in the covalent linkage of ATG12 onto a single lysine on ATG3. Surprisingly, disrupting ATG12 conjugation to ATG3 does not affect starvation-induced autophagy. Rather, the lack of ATG12-ATG3 complex formation produces an expansion in mitochondrial mass and inhibits cell death mediated by mitochondrial pathways. Overall, these results unveil a role for ATG12-ATG3 in mitochondrial homeostasis, and implicate the ATG12 conjugation system in cellular functions distinct from the early steps of autophagosome formation.
INTRODUCTION
Ubiquitin-like (UBL) protein conjugations, such as ubiquitination and sumoylation, influence diverse cell biological processes, including protein targeting, organelle trafficking, cell division, signal transduction, and transcription (Welchman et al., 2005). These post-translational modifications, which typically result in the covalent attachment of the UBL tag to the ε-amine group of lysine residues in target substrates, are highly dynamic, reversible, and tightly regulated by well-established biochemical cascades. First, the UBL is activated in an ATP dependent manner by an E1 activating enzyme in which the C-terminal glycine residue of the UBL moiety forms a high-energy thioester bond with a cysteine residue in the active site of the E1. Second, the activated UBL is transferred to an E2 conjugating enzyme via a trans-esterification reaction. Finally, the UBL is transferred onto a lysine of a target substrate; in many cases, this final transfer requires an E3 ligase enzyme (Kerscher et al., 2006). To date, two family members, ubiquitin and small ubiquitin-like modifier (SUMO), have received the greatest attention; nonetheless, other UBLs undoubtedly have undiscovered functions in biology and disease (Welchman et al., 2005).
Two UBLs, ATG12 and ATG8 (ATG stands for AuTophaGy regulator), play critical roles in macroautophagy (hereafter called autophagy), a tightly controlled lysosomal degradation process in which a cell digests its own proteins and organelles during starvation or stress (Levine and Kroemer, 2008; Ohsumi, 2001). Although ATG12 and ATG8 possess little primary sequence homology to ubiquitin, both contain an “ubiquitin superfold” and the C-terminal glycine required for isopeptide linkage (Hanada and Ohsumi, 2005; Sugawara et al., 2004; Suzuki et al., 2005). Importantly, the early steps of autophagosome formation require these two ubiquitin-like conjugation processes, which covalently attach ATG12 to the target protein ATG5 (Mizushima et al., 1998a; Mizushima et al., 1998b) and ATG8 (for which, microtubule associated protein light chain 3 (LC3) is a chief mammalian orthologue) to the lipid phosphotidylethanolamine (PE) (Ichimura et al., 2000; Kabeya et al., 2000). Both ATG8 and ATG12 are activated in an ATP-dependent process by a ubiquitin-like E1 activating enzyme, called ATG7 (Tanida et al., 1999). Subsequently, ATG12 is conjugated to ATG5 by ATG10, an E2-like conjugating enzyme (Nemoto et al., 2003), whereas ATG8/LC3 is conjugated to PE via another E2-like molecule, ATG3 (Tanida et al., 2002b; Yamada et al., 2007). Furthermore, recent biochemical evidence supports a model in which the ATG12-ATG5 complex possesses an E3-like activity for efficient PE-lipidation of ATG8 (Fujita et al., 2008; Hanada et al., 2007).
Unlike other UBLs, ATG12 is proposed to modify a single target, ATG5. However, it remains unclear why a complex energy-consuming enzymatic cascade has evolved to conjugate a single substrate. Furthermore, certain stimuli, namely mitochondrial outer membrane permeabilization (MOMP) and endoplasmic reticulum (ER) stress, produce an increase in ATG12 transcript or protein levels, but no concomitant rise in ATG5; such discordant regulation is enigmatic given that ATG12 is solely proposed to function as part of an 1:1 stoichiometric complex with ATG5 (Colell et al., 2007; Kouroku et al., 2007). One potential explanation for these previous findings is that, similar to other UBLs, ATG12 modifies additional substrates that regulate autophagy or other cellular functions.
Hence, we sought to test the hypothesis that ATG12 is not an isolated UBL modification confined to a single target. Here, we identify ATG3, the E2 enzyme necessary for ATG8/LC3 lipidation during autophagy, as a substrate for ATG12 conjugation. ATG12-ATG3 complex formation requires ATG7 as the E1-activating enzyme and an autocatalytic function of ATG3 as the E2, resulting in the covalent attachment of ATG12 onto a single lysine on ATG3. Remarkably, disrupting ATG12-ATG3 complex formation has no discernable effect on nonselective autophagy. Rather, upon disrupting ATG12 conjugation to ATG3, cells display increased mitochondrial mass and enhanced survival in response to agents that activate mitochondrial cell death pathways. Overall, these results demonstrate a previously unrecognized role for the ATG12-ATG3 complex in mitochondrial homeostasis and cell death. We propose that the ATG12 conjugation system directs cellular functions distinct from the early steps of autophagosome formation.
RESULTS
ATG12 covalently modifies multiple protein targets in addition to ATG5 in mammalian cells
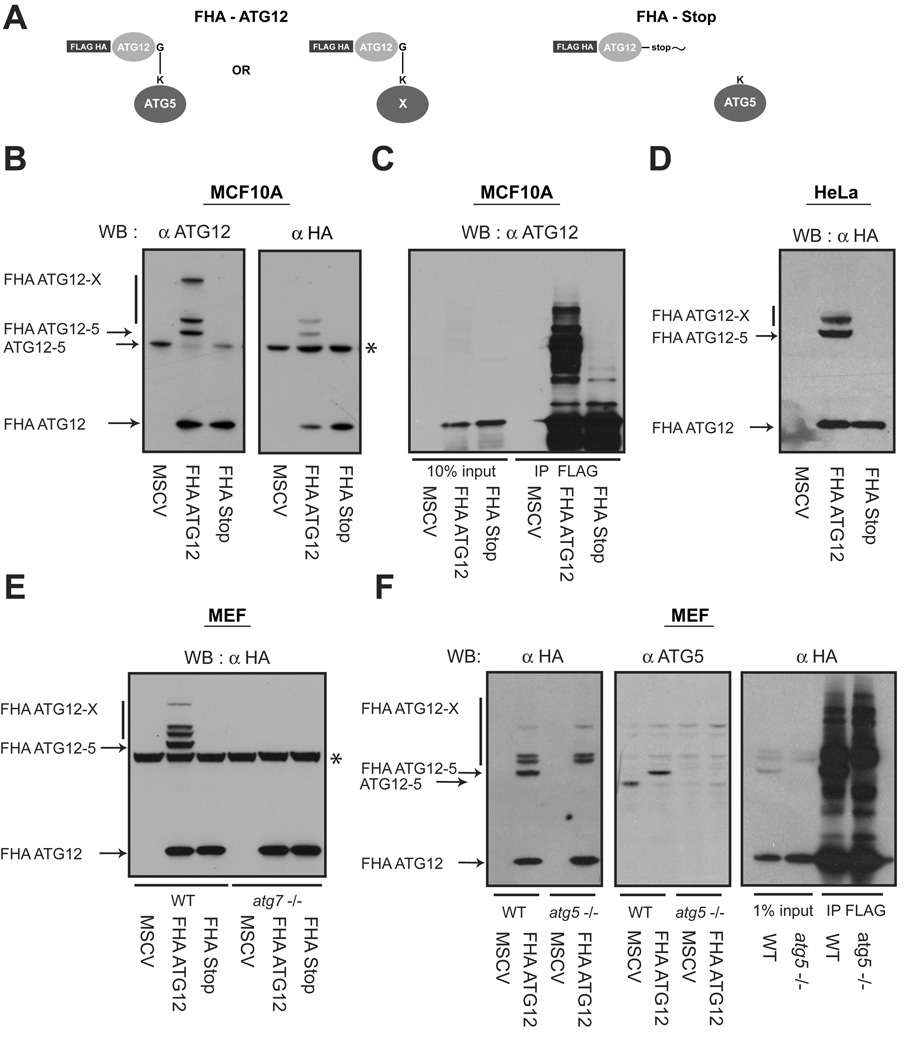
To test whether ATG12 covalently modifies protein targets in addition to ATG5, we created a retroviral construct encoding epitope-tagged mouse ATG12 (FHA-ATG12); as a negative control, we generated a conjugation incompetent version in which the C-terminal glycine in ATG12, required for the isopeptide linkage to target lysine residues, was replaced with a stop codon (FHA-Stop) (Figure 1A) (Mizushima et al., 1998b). Upon stable expression in MCF10A human mammary epithelial cells, we observed a slower migrating ATG12-ATG5 complex, consistent with epitope tag addition, as well as additional higher molecular weight conjugates in FHA-ATG12 expressing cells but not in FHA-Stop cells (Figure 1B). Importantly, these higher molecular weight proteins were observed using denaturing and reducing conditions, suggesting that ATG12 was covalently attached to multiple protein targets. Furthermore, upon α-FLAG immunoprecipitation, we observed numerous additional ATG12-X species that were unique to cells expressing conjugatable ATG12 (Figure 1C). Similar results were obtained upon stable FHA-ATG12 expression in HeLa cervical carcinoma cells, indicating that the ATG12-X proteins are present in multiple human cell types (Figure 1D).
Figure 1. ATG12 covalently modifies multiple protein targets in mammalian cells.
(A) Schematic of ATG12 constructs: FHA-ATG12 is mouse ATG12 tandem tagged at the N-terminus with FLAG and HA epitopes. In FHA-Stop, the C-terminal glycine in ATG12 required for conjugation is replaced with a Stop codon. (B) MCF10A cells stably expressing empty vector (MSCV), FHA-ATG12 and FHA-Stop were lysed and immunoblotted with α-ATG12 and α-HA antibodies. Asterisk (*) indicates non-specific band during α-HA immunoblotting. (C) MCF10A cells stably expressing the indicated constructs were lysed and immunoprecipitated with α-FLAG; immune complexes were resolved using SDS-PAGE and immunoblotted with α-ATG12. (D) HeLa cells stably expressing the indicated constructs were lysed and immunoblotted with α-HA. (E) atg7+/+ (WT) and atg7−/− mouse embryonic fibroblasts (MEFs) stably expressing the indicated constructs were lysed and immunoblotted with α-HA. Asterisk (*) indicates nonspecific band. (F) Left: atg5+/+ (WT) and atg5−/− stably expressing the indicated constructs were lysed and immunoblotted with α-HA or α-ATG5. Right: WT and atg5−/− MEFs expressing FHA-ATG12 were lysed and immunoprecipitated with α-FLAG; immune complexes were resolved and immunoblotted with α-HA.
We next asked whether the E1-like enzyme ATG7 is required for the activation of ATG12 and its subsequent conjugation to these targets (Tanida et al., 1999; Tanida et al., 2001). We stably infected ATG7 deficient fibroblasts with FHA-ATG12 or FHA-Stop and found that all ATG12 conjugations were eliminated in the absence of ATG7 (Figure 1E). Thus, ATG12 requires ATG7 as an E1-activating enzyme to be conjugated to any of its multiple protein targets. Since ATG5 is the only known substrate of ATG12, we tested if the additional higher molecular weight ATG12-X proteins were unique ATG12-modified proteins versus multiple ATG12 conjugations onto the ATG12-ATG5 complex (Mizushima et al., 1998a; Mizushima et al., 1998b). To distinguish among these possibilities, we stably expressed FHA-ATG12 and FHA-Stop in both wild type (atg5+/+) and ATG5 null (atg5−/−) mouse embryonic fibroblasts (MEFs). Similar to our results in human cells, we observed numerous ATG12 conjugates in both atg5+/+ and −/− mouse fibroblasts. In atg5−/− cells, only the band corresponding to the ATG12-ATG5 protein complex disappeared; the remaining ATG12-X proteins continued to be produced, supporting that these species are unique conjugation targets that can be formed in the absence of ATG5 (Figure 1F).
ATG12 and ATG3 form a covalent complex
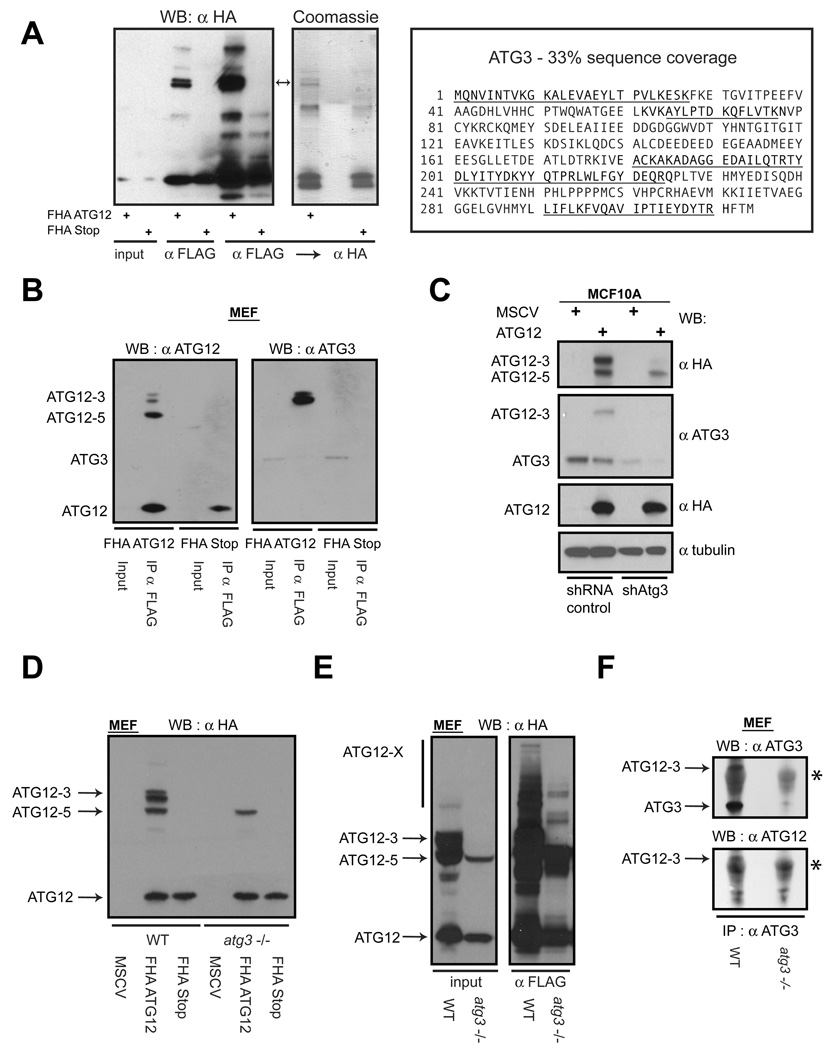
To identify putative ATG12-X proteins, lysates from atg5−/− fibroblasts stably expressing either FHA-ATG12 or FHA-Stop were subject to large-scale tandem affinity purification with α–FLAG and α–HA antibodies. Upon resolving the eluted proteins, each protein in the predominant doublet on a Coomassie Brilliant Blue stained gel (Figure 2A, asterisk) was trypsin-digested and subject to tandem mass spectrometry (MS/MS). Both proteins in this doublet were identified as ATG12 conjugated to ATG3 (Figure 2A). ATG3 is the E2-like enzyme responsible for ATG8/LC3 lipidation and has previously been demonstrated to interact with ATG12 (Tanida et al., 2002a; Tanida et al., 2002b).
Figure 2. ATG12 and ATG3 form a covalent complex.
(A) Lysates from atg5−/− MEFs stably expressing FHA-ATG12 or FHA-Stop were subject to affinity purification with α-FLAG followed by α-HA antibodies. Eluted proteins were resolved using SDS-PAGE and immunoblotted with α-HA (left) or stained with Coomassie (middle). Each protein in the indicated doublet (arrows) was individually subject to MS/MS analysis and both were identified as ATG3. Right: Amino acid sequence of mouse ATG3; underlined sequences correspond to independent peptides identified by mass spectrometry. (B) Lysates from MEFs expressing the indicated constructs were immunoprecipitated with α-FLAG; immune complexes (FLAG IP) were resolved using SDS-PAGE and immunoblotted with α-ATG12 or α-ATG3. (C) MCF10A cells stably expressing FHA-ATG12 were infected with lentiviruses encoding non-targeting control shRNA or shRNA targeted to ATG3 (shATG3). Lysates were immunoblotted as indicated. (D) Wild type and atg3−/− MEFs stably expressing the indicated constructs were lysed and subject to α-HA immunoblotting. (E) Wild type and atg3−/− MEFs stably expressing FHA-ATG12 were lysed and subject to α-FLAG immunoprecipitation; immune complexes were resolved and immunoblotted with α-HA. (F) Lysates from wild type and atg3−/− MEFs were immunoprecipated with α-ATG3; immune complexes were resolved using SDS-PAGE and subject to α-ATG3 or α-ATG12 immunoblotting. Asterisk (*) indicates immunoglobulin heavy chain. See also Figure S1.
In α-FLAG immunoprecipitates prepared from MEFs expressing FHA-ATG12, we detected the 65kD doublet using antibodies against either ATG12 or ATG3, confirming that this protein corresponded to ectopic FHA-ATG12 conjugated to endogenous ATG3 (Figure 2B). Remarkably, this doublet collapsed to a single protein when treated with calf intestinal phosphatase, suggesting that the ATG12-ATG3 complex undergoes phosphorylation (Figure S1). Furthermore, MCF10A cells stably expressing shRNA against ATG3 exhibited specific depletion of the ATG12-ATG3 complex, relative to a non-targeting shRNA control, whereas the ATG12-ATG5 complex continued to be produced (Figure 2C). Finally, the ATG12-ATG3 complex was not observed in atg3−/− MEFs expressing FHA-ATG12, in contrast to wild type controls (Figure 2D), whereas ATG12-ATG5 and other higher molecular weight species, corresponding to other ATG12-X conjugates, persisted in atg3−/− cells (Figure 2E) (Sou et al., 2008). Finally, to assess if the ATG12-ATG3 complex was present at endogenous expression levels, protein extracts were immunoprecipitated with α-ATG3 antibodies. We detected a 51kd protein complex in wild type fibroblasts that immunoblotted with both α-ATG3 and α-ATG12 antibodies; this complex was not present in protein lysates prepared from atg3−/− MEFs (Figure 2F). Overall, these data support that ATG12 forms a covalent complex with ATG3 and that this complex represents the second (with ATG12-ATG5 being the first) of several potential ATG12 conjugation targets.
Auto-conjugation of ATG12 onto a single lysine on ATG3
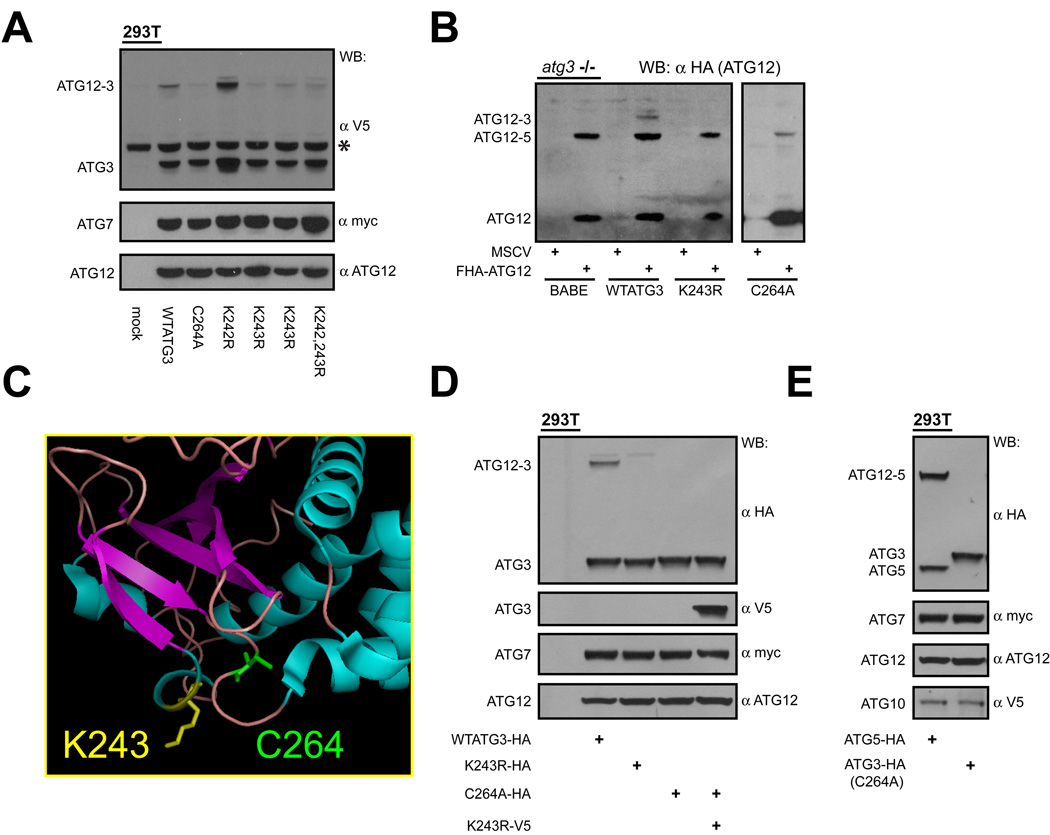
Similar to other UBLs, ATG12 is covalently attached to the ε-amine group of a lysine residue of its target (Kerscher et al., 2006; Mizushima et al., 1998a). To elucidate potential functions of the ATG12-ATG3 complex, we sought to identify the target lysine(s) in ATG3 required for ATG12 conjugation. First, we compared the amino acid sequence of ATG3 from multiple species and found five lysines to be highly conserved (Figure S2A). Using site-directed mutagenesis, we systematically eliminated each lysine, or groups of lysines, and transiently co-expressed each mutant version of ATG3 along with ATG12 and ATG7 in HEK293T cells (Figure 3A and Figure S2B). Using this strategy, we reconstituted ATG12-ATG3 complex formation and identified lysine 243 (K243) as the primary lysine required for ATG12 conjugation. Upon mutation of this single lysine to arginine, complex formation was virtually eliminated. Remarkably, mutating an adjacent lysine at position 242 of ATG3 had no effect on ATG12 conjugation. In contrast, mutating the ATG3 catalytic cysteine to alanine (C264A) potently inhibited formation of the ATG12-ATG3 complex (Figure 3A) (Tanida et al., 2002b). To corroborate these results, we stably complemented ATG3 deficient fibroblasts with epitope-tagged wild type (WT) ATG3, the non-conjugatable ATG3 mutant K243R and the catalytically inactive mutant C264A (Sou et al., 2008). Unlike cells reconstituted with WTATG3, ATG12-ATG3 complex formation was not observed in cells co-expressing FHA-ATG12 and either K243R or C264A (Figure 3B).
Figure 3. Auto-conjugation of ATG12 onto a single lysine of ATG3.
(A) HEK293T cells expressing YFP-ATG12, Myc-tagged ATG7, and V5-tagged WTATG3 or the indicated ATG3 mutants. (B) atg3−/− MEFs stably reconstituted with empty vector (BABE), wild type ATG3, or the indicated ATG3 mutants, were then transduced with empty vector (MSCV) or FHA-ATG12. Lysates were α-HA immunoblotted to detect ATG12-conjugated proteins. (C) Crystal structure of yeast ATG3 with labeled α-helices (blue) and β-sheets (purple). Positions of the conserved catalytic cysteine (C264 in mouse ATG3, green) and the principal lysine conjugated with ATG12 (K243, yellow) are shown. Diagram prepared using PyMOL. (D) 293T cells transfected with YFP-ATG12, myc-tagged ATG7 and mutants of either HA or V5-tagged ATG3, as indicated. (E) HEK293T cells transfected with Myc-tagged ATG7, V5-tagged ATG10, YFP-ATG12 and either HA-tagged ATG5 or ATG3 C264A; catalytically inactive ATG3 (C264A) was used to distinguish the E2 activity of ATG10 from ATG3 during ATG12-ATG3 formation. See also Figure S2.
Since ATG12-ATG3 formation required ATG3 catalytic activity, we hypothesized that an autocatalytic activity of ATG3 served as the E2 enzyme for this conjugation. Several E2 enzymes undergo self-ubiquitination (Gwozd et al., 1995; Machida et al., 2006; Walter et al., 2001). Typically, these are cis-acting intramolecular reactions, in which the target lysine of the E2 enzyme lies in close proximity to the active site cysteine; upon inspection of its tertiary structure, this also appeared to be the case for ATG3 (Figure 3C) (Yamada et al., 2007). To test whether ATG12 conjugation to ATG3 could occur in trans, we co-expressed two distinct tagged versions of ATG3, along with ATG12 and ATG7, in 293T cells. This first was catalytically inactive ATG3 (C264A) possessing the target lysine, while the second was catalytically active ATG3 that lacked the target lysine (K243R). In contrast to WTATG3, no ATG12-ATG3 complex was formed upon co-expressing these two mutant forms of ATG3, supporting that ATG12 conjugation to ATG3 was a cis-acting reaction (Figure 3D). Finally, we assessed if ATG10, the E2 enzyme responsible for ATG12 conjugation to ATG5, contributed to the formation of ATG12-ATG3. However, ATG10 was unable to mediate ATG12-ATG3 complex formation, and the expression of catalytically inactive ATG10 (C165A) was unable to dominantly inhibit the auto-conjugation of ATG12 onto ATG3 (Figure 3E and Figure S2C). Overall, these results indicate that ATG3 auto-catalyzes the conjugation of ATG12 onto a single conserved lysine residue (K243) of this E2-like enzyme required for autophagy.
Starvation-induced autophagy remains intact upon disrupting ATG12 conjugation to ATG3
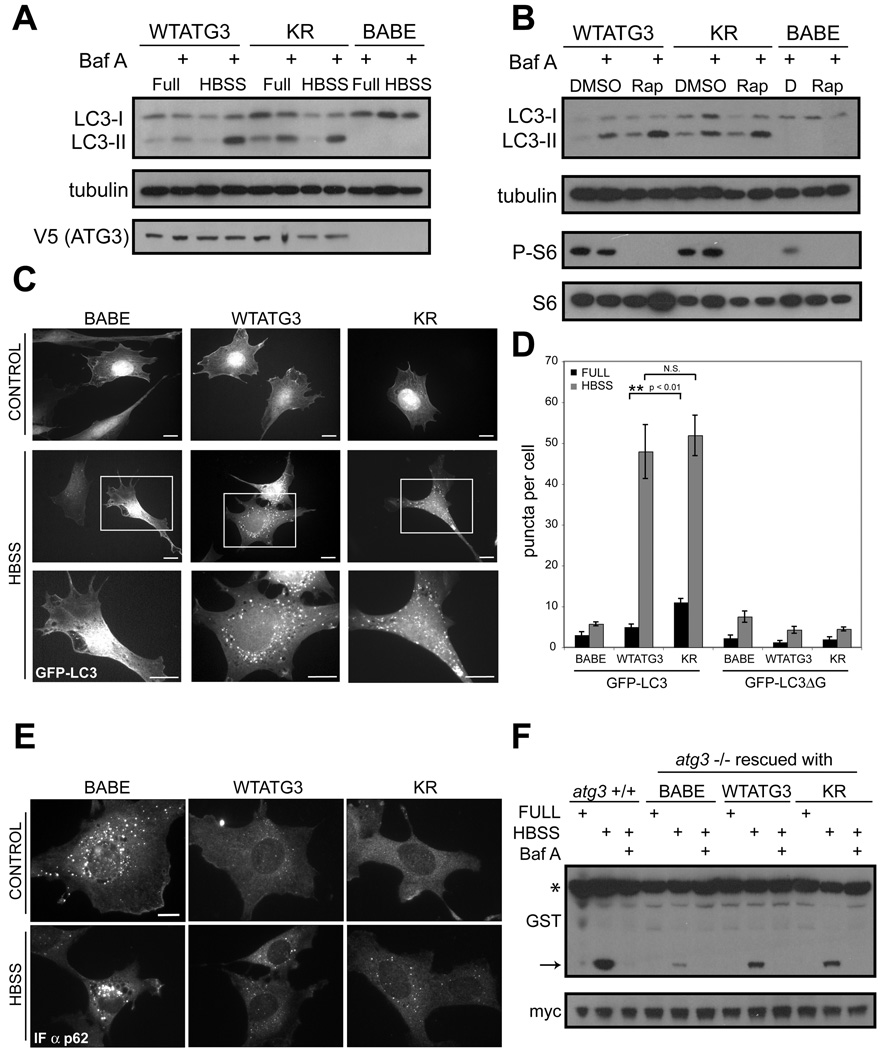
We next tested if ATG12-ATG3 modulates known ATG3 biological activities, namely PE-lipidation of ATG8/LC3 and autophagosome formation (Machida et al., 2006; Sou et al., 2008). For these experiments, we reconstituted atg3−/− fibroblasts with a control empty vector (BABE), wild type ATG3 (WTATG3), or the K243R mutant (KR) that was incapable of ATG12-ATG3 complex formation (Figure 3B and Figure S3A). In response to nutrient starvation using Hank’s buffered saline (HBSS) or rapamycin-mediated mTORC1 inactivation, we found equivalent levels of LC3 lipidation (LC3-II) between WT and KR and efficient LC3-II turnover in the lysosome (Figure 4A–B). We also observed no significant differences in the number or morphology of GFP-LC3 puncta, a widely utilized marker for autophagosome formation, upon HBSS starvation, although we did note an increase in punctate GFP-LC3 in KR cells cultured in nutrient rich conditions (Figure 4C–D and Figure S3B) (Kabeya et al., 2000). Furthermore, WTATG3 and KR cells exhibited no differences in the lipidation of other mammalian ATG8 orthologues, including GABARAP and GATE-16 (Figure S3C).
Figure 4. Starvation-induced autophagy remains intact upon disrupting ATG12 conjugation to ATG3.
Stable pools of atg3−/− fibroblasts expressing an empty vector (BABE), wild type mouse ATG3 (WTATG3) or mutant ATG3 unable to be conjugated by ATG12 (KR) were used for experiments as indicated. (A and B) Cells were grown in complete media, starved in Hank’s buffered salt solution (HBSS) for 4h, or treated with 10nM rapamycin (B) for 6h. Cells were lysed and immunoblotted with indicated antibodies. Phosphorylated ribosomal S6 (P-S6) was used to verify rapamycin-mediated mTORC1 inhibition. When indicated, bafilomycin A (BafA, 10nM) was added to cells 1h prior to lysis. (C) Indicated cells types expressing GFP-LC3 were grown in complete media (control) or HBSS-starved for 4h; boxed areas from center panels are enlarged below. Bars, 25µm. (D) Quantification of punctate GFP-LC3 or GFP-LC3ΔG (mean +/−SEM puncta per cell). (E) Indicated cell types grown in complete media (control) or HBSS-starved for 4h, and then fixed and immunostained with α-p62 antibody. Bar, 25µm. (F) Indicated cell types were transfected with a GST-BHMT fusion construct, HBSS-starved for 6h, lysed and immunoblotted with α-GST. Asterisk (*) indicates full-length GST-BHMT and arrow indicates cleaved BHMT produced in autolysosomes. When indicated, bafilomycin A (BafA, 10nM) was used to inhibit lysosomal function. α-Myc was used to detect GFP-myc (expressed from an IRES sequence) to control for transfection efficiency (Dennis and Mercer, 2009). See also Figure S3.
To extend these results, we evaluated the accumulation of p62SQSTM, an ubiquitin-binding scaffold protein selectively degraded by autophagy, via immunofluorescence (Bjorkoy et al., 2005). In both nutrient-rich and starvation conditions, p62 cytoplasmic bodies amassed in autophagy-deficient cells lacking ATG3; this accumulation was profoundly decreased in cells rescued with either WTATG3 or the KR mutant (Figure 4E). Finally, we assessed the processing of an ectopically-expressed autophagy cargo protein betaine homocysteine methyltransferase (BHMT) (Dennis and Mercer, 2009). Efficient proteolytic cleavage of BHMT during HBSS starvation required both ATG3 and lysosomal function; this autophagy dependent cleavage was restored at equivalent levels upon rescue with either WTATG3 or KR (Figure 4F). Overall, these data support that disrupting ATG12 conjugation to ATG3 does not impair ATG8/LC3 lipidation, autophagosome formation or autophagic proteolysis in response to nutrient starvation.
Cells expressing non-conjugatable ATG3 (KR) exhibit increased mitochondrial mass and a fragmented mitochondrial morphology
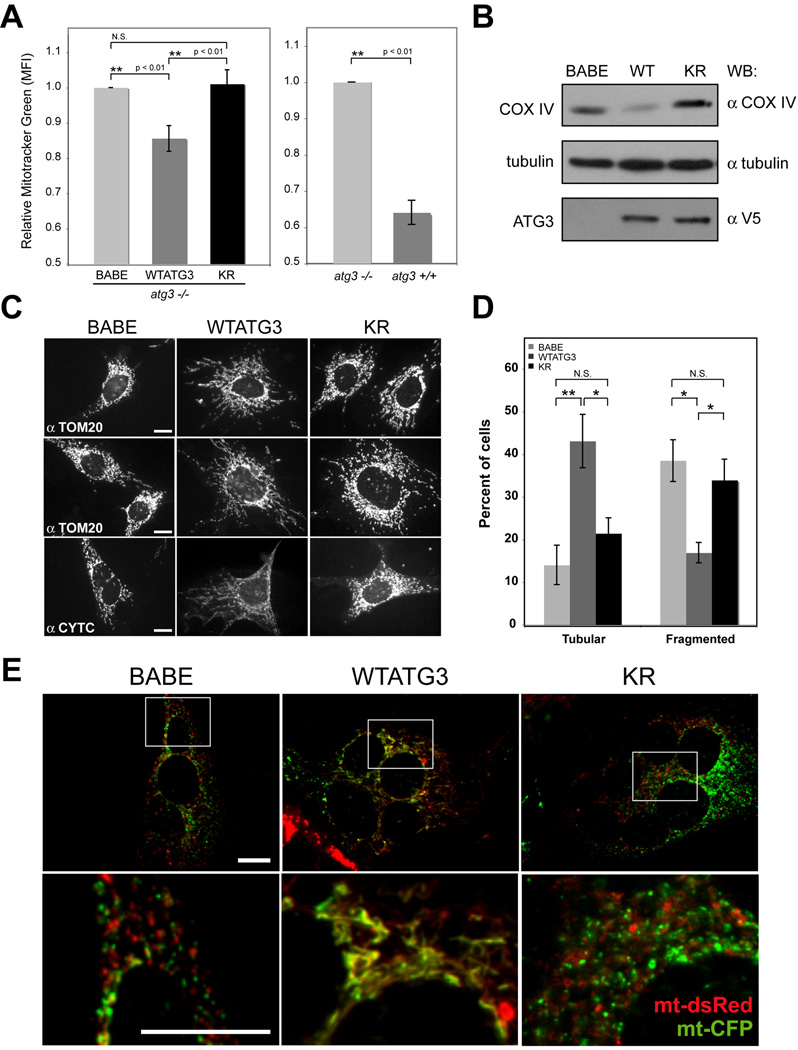
Recent studies support that autophagy is required to maintain mitochondrial homeostasis (Chen and Chan, 2009; Lemasters, 2005). Flow cytometric analysis for Mitotracker® Green (MTG), a mitochondria specific intravital dye, demonstrated that cells reconstituted with WTATG3 exhibit an approximately 15% reduction in total mitochondrial mass compared to atg3−/− (BABE) controls; similarly, atg3+/+ cells possessed reduced MTG staining compared to atg3−/− cells. In contrast, the mitochondrial mass of cells reconstituted with KR was unchanged relative to atg3−/− (BABE) cells (Figure 5A). Protein levels of the mitochondrial inner membrane protein, cytochrome c oxidase subunit IV (COX IV) corroborated these differences in MTG staining (Figure 5B).
Figure 5. Effects of disrupting ATG12 conjugation to ATG3 on mitochondrial mass and morphology.
(A) Left: Mitotracker Green (MTG) fluorescence intensity (mean+/−SEM from 5 experiments) for the indicated cell types relative to atg3−/− cells expressing empty vector (BABE). Statistical significance calculated using ANOVA, followed by Tukey's HSD test. Right: MTG fluorescence intensity (mean+/−SEM from 3 experiments) for atg3+/+ cells relative to atg3−/− cells. (B) Lysates from indicated cell types were immunoblotted with α-COX IV, α-tubulin and α-V5. (C) Indicated cell types were immunostained with TOM20 (top and middle) or cytochrome c (bottom) antibodies. (D) Percent of cells with purely fragmented/round morphology or purely tubular morphology was quantified from TOM20-immunostained images. Results are the mean+/−SEM from 5 experiments, where at least 250 cells were scored per condition for each individual experiment. Statistical significance calculated using ANOVA, followed by Tukey's HSD test. (E) Cells expressing either mitochondria-targeted dsRed (mt-dsRed, red) or CFP (mt-CFP, green) were hybridized using PEG to assess mitochondrial fusion activity. Representative merged images of cell hybrids from each cell type are shown; the colocalization of these signals (yellow) within cell hybrids indicates mitochondrial fusion (Chen et al., 2003). Each bottom panel is an enlargement of the boxed inset in the corresponding panel above. Bar, 25µm. See also Figures S4 and S5.
We next evaluated mitochondrial morphology in the various cell types by immunostaining for TOM20, a mitochondrial import receptor located in the outer membrane, and cytochrome c, located in the inner membrane. In both ATG3 null (BABE) and KR cultures, numerous cells possessed fragmented and round mitochondria, whereas in WTATG3 cells, the mitochondria predominantly formed tubular networks (Figure 5C). Because the mitochondria in individual cells in these cultures exhibited a range of morphologies (Figure S4A), we enumerated cells from each condition that showed purely fragmented/round versus purely tubular mitochondrial morphology. We confirmed a significant increase in cells possessing fragmented mitochondria in ATG3 null and KR cells compared to WTATG3 cells, and a corresponding decrease in cells with purely tubular mitochondria (Figure 5D). In contrast, we observed no obvious morphological differences in other organelles, such as the endoplasmic reticulum, peroxisomes, or Golgi apparatus (Figure S4B).
Fragmentation can arise as an early consequence of declining mitochondrial function or cell viability (Youle and Karbowski, 2005). Hence, to determine whether the excess mitochondria in KR cells were depolarized, we co-stained cells with Mitotracker® Red (MTR), whose accumulation in mitochondria depends on intact membrane potential, and MTG, which labels the lipid membranes of all mitochondria (Tal et al., 2009). A small population of ATG3 null cells had depolarized mitochondria (MTR negative, MTG positive), which was not significantly reduced upon rescue with either WTATG3 or KR (Figure S4C). We also performed growth curves for the three cell types and found no overt differences in proliferation or viability (Figure S4D).
Alternatively, defects in mitochondrial fusion also promote fragmentation (Chen and Chan, 2009). Accordingly, we evaluated mitochondrial fusion activity more directly using a polyethylene glycol (PEG) fusion assay (Chen et al., 2003). For these assays, either mitochondria-targeted red (mt-dsRed) or cyan (mt-CFP) fluorescent proteins were ectopically expressed in each cell type; subsequently, hybrids between the differentially labeled populations were scored for mitochondrial fusion. While we observed robust mitochondrial fusion among hybrids derived from WTATG3 cells, such events were reduced in both ATG3 null and KR-expressing hybrids, indicating decreased fusion activity (Figure 5E and Figure S5A–B). Altogether, these results support that disrupting ATG12 conjugation to ATG3 produces significant increases in mitochondrial mass and fragmentation, which correlates with a reduction in mitochondrial fusion activity. Most importantly, these phenotypes arise in cells capable of robust autophagy, indicating that the effects of the ATG12-ATG3 complex on mitochondria are distinct and separable from the well-established functions of ATG3 in ATG8/LC3 lipidation and autophagosome formation.
Effects of CCCP uncoupling on mitochondria in cells expressing non-conjugatable ATG3 (KR)
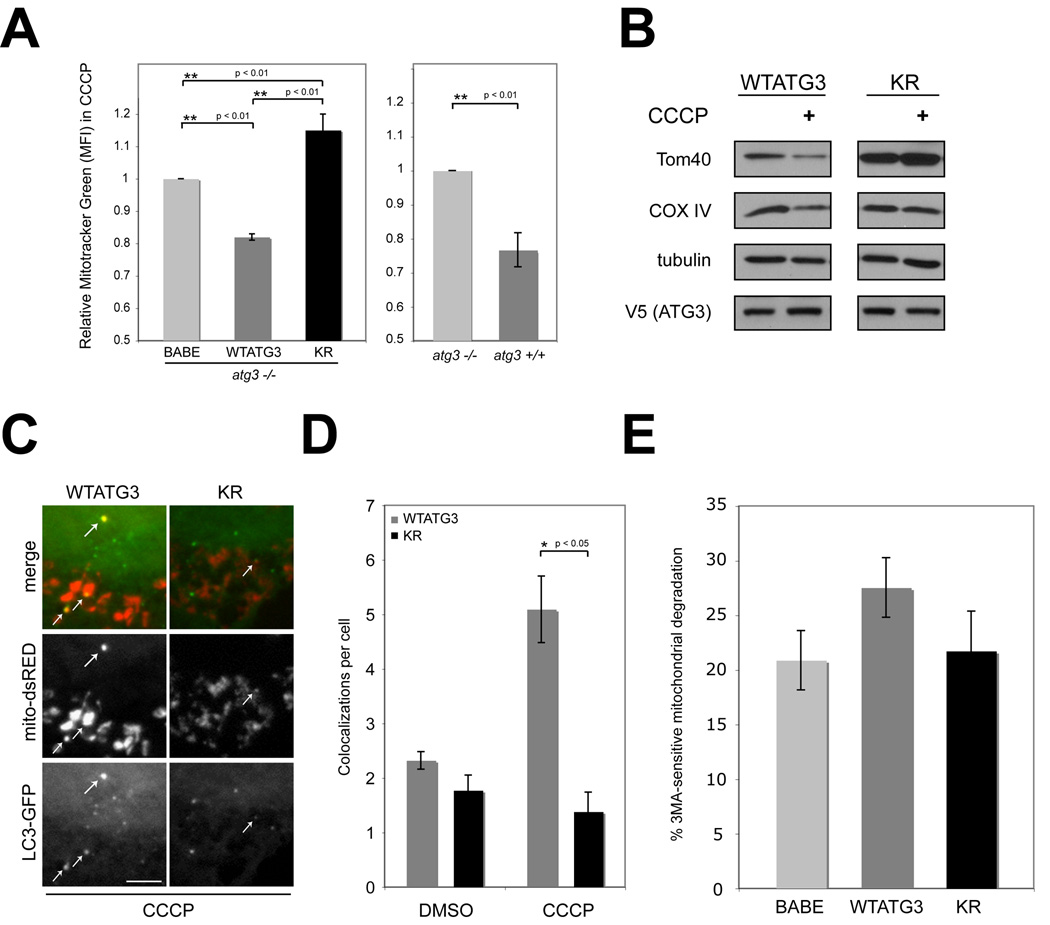
We next sought to clarify how the ATG12-ATG3 complex regulates mitochondrial homeostasis by stressing cells with carbonyl cyanide m-chlorophenylhydrazone (CCCP), a proton ionophore that uncouples mitochondria. CCCP-induced depolarization elicits a multifaceted cellular response that includes an overall increase in mitochondrial biogenesis coupled to the autophagic degradation of damaged mitochondria, termed mitophagy. (Lemasters, 2005; Narendra et al., 2008; Rohas et al., 2007). Indeed, CCCP treatment resulted in increased MTG intensity compared to DMSO-treated controls in all cell types, corroborating that overall mitochondrial mass increased during chemical uncoupling (Rohas et al., 2007). Notably, this CCCP-induced increase in MTG staining intensity was more pronounced in both ATG3 null (BABE, 27% increase) and KR cells (25% increase) in comparison to WTATG3 cells (14% increase) (Figure S6A). When directly compared to ATG3−/− (BABE) cells, CCCP-treated WTATG3 cells displayed 20% lower MTG staining intensity; similar differences were observed between atg3+/+ and atg3−/− cells. In contrast, KR cells possessed a 15% and 35% increase in mitochondria compared to BABE and WTATG3 cells, respectively (Figure 6A). TOM20 immunostaining of CCCP-treated cultures also supported that mitochondria were more abundant in ATG3 null and KR cells compared to WTATG3 cultures (Figure S6B). These results indicate that ATG12 conjugation to ATG3 restricts the expansion of mitochondrial mass during CCCP-induced depolarization.
Figure 6. Effects of CCCP treatment on mitochondria in cells expressing a non-conjugatable ATG3 mutant (KR).
(A) Left: Cells treated with 10 µM CCCP for 24h and stained with MTG. MTG fluorescence intensity (mean+/−SEM from 8 experiments) relative to atg3−/− cells expressing empty vector (BABE) is shown. Statistical significance calculated using ANOVA, followed by Tukey's HSD test. Right: MTG fluorescence intensity (mean+/−SEM from 3 experiments) for CCCP-treated atg3+/+ cells relative to atg3−/− cells. (B) Cells were CCCP-treated as indicated, lysed and immunoblotted with antibodies against the resident mitochondrial proteins, TOM40 and COX IV, V5 and tubulin (loading control). (C) Indicated cell types expressing GFP-LC3 were transfected with mito-dsRed and treated with 10µM CCCP for 24h. White arrows indicate colocalization of GFP-LC3 and mito-dsRed. Bar, 5µm (D) Quantification of mito-dsRed and GFP-LC3 colocalizations per cell (mean+/−SEM). (E) 3MA-sensitive mitochondrial degradation (mean+/−SEM from 5 experiments) during CCCP treatment. See also Figure S6.
We next assessed the protein levels of two mitochondrial resident proteins, COX IV and TOM40 and found that, compared to WTATG3, KR cells had higher levels of both proteins in control as well as CCCP-treated cultures. Interestingly, in WTATG3 cells, both proteins were slightly reduced during CCCP treatment, whereas in KR cells, these levels remained unchanged, suggesting that the rate of mitochondrial degradation may be reduced in cells lacking the ATG12-ATG3 complex (Figure 6B). As a result, we sought to more closely analyze if KR cells exhibited decreased mitophagy. We first compared the rates of CCCP-induced autophagosome induction in WTATG3 and KR cells, but found no significant differences in punctate GFP-LC3 between these cells following CCCP treatment (Figure S6C–D). To measure mitochondrial targeting to autophagosomes during CCCP treatment, we assessed the level of colocalization between mito-dsRed and GFP-LC3 (Figure 6C). Importantly, the number of colocalizations was significantly higher in WTATG3 cells compared to KR cells (Figure 6C and D). Hence, upon loss of ATG12 conjugation to ATG3, mitochondrial targeting to autophagosomes is reduced.
To further assess if mitophagy was impaired in KR cells, we tested if 3-methyladenine (3MA), a pharmacological autophagy inhibitor, was able to augment mitochondrial mass during CCCP-treatment (Seglen and Gordon, 1982). In WTATG3 cells, we observed a 27.5 % increase in mitochondrial mass upon treatment with CCCP+3MA compared to CCCP alone, which we attributed to 3MA-sensitive mitochondrial degradation. To control for the non-specific effects of 3MA, which can promote significant protein degradation in an autophagy-independent manner (Mizushima et al., 2001), we tested the effects of 3MA on atg3−/− (BABE) cells; 3MA-sensitive mitochondrial degradation in these autophagy-incompetent cells was reduced to 20% (Figure 6E). Similar to atg3−/− (BABE) cells, 3MA-sensitive mitochondrial degradation in KR cells was decreased to 22%. Overall, these results support that ATG12 conjugation to ATG3 facilitates mitochondrial degradation in the presence of CCCP. Importantly, this function of ATG12-ATG3 is entirely separable from the ability of ATG3 to induce autophagosome formation during mitochondrial depolarization.
Cells lacking ATG12-ATG3 exhibit reduced cell death mediated by mitochondrial pathways
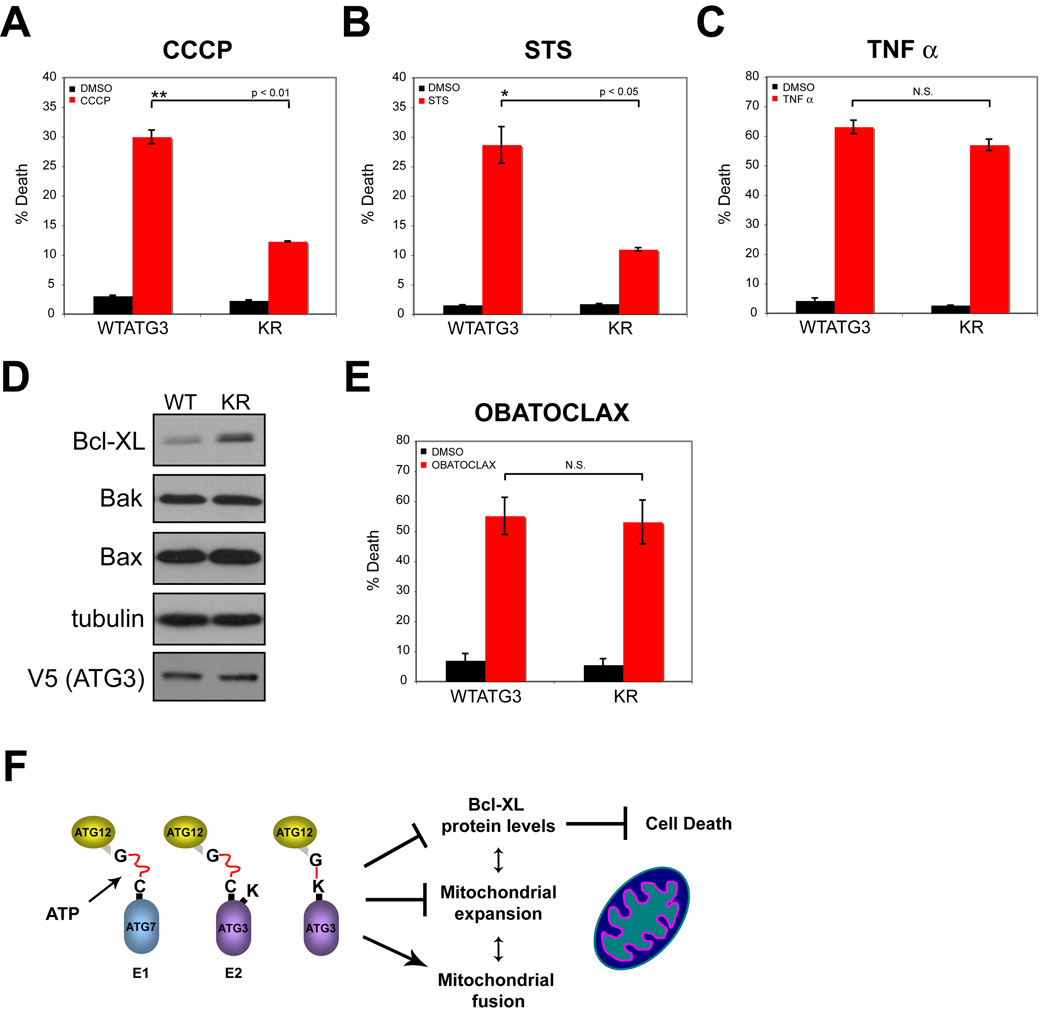
Mitochondria play critical roles in many forms of cell death, including apoptosis mediated by the intrinsic pathway (Brenner and Mak, 2009). Accordingly, we determined how the ATG12-ATG3 complex influenced cell death. First, we compared the effects of CCCP on cell death in WTATG3 and KR cells; similar to other mitochondrial uncoupling agents, high doses of CCCP induce death that can be inhibited by anti-apoptotic Bcl-2 family members (de Graaf et al., 2004). In response to CCCP treatment, KR cells exhibited a 2-fold reduction in death compared to WTATG3 controls (Figure 7A). Similarly, cell death was reduced in KR cells treated with staurosporine, a pan-kinase inhibitor that potently activates the intrinsic apoptosis pathway (Figure 7B). We also observed reduced cleavage of the executioner caspase, caspase-3, in KR cells in response to both agents, supporting that apoptosis was reduced compared to WTATG3 (data not shown). In contrast, both cell types exhibited robust and equivalent levels of cell death in response to Tumor Necrosis Factor-α (TNF–α). TNF-α induces apoptosis via the extrinsic pathway, in which death receptors directly activate executioner caspases independently of mitochondrial pathways (Figure 7C) (Brenner and Mak, 2009). Thus, ATG12 conjugation to ATG3 sensitizes cells to death downstream of mitochondrial pathways, but has no effect on death-receptor mediated apoptosis.
Figure 7. Cells lacking ATG12-ATG3 exhibit decreased cell death mediated by mitochondrial pathways.
Cells were treated for 24h with the following agents: (A) 100 µM CCCP; (B) 100nM staurosporine; and (C) 20ng/mL TNF-α + 2.5µg/mL cycloheximide. Percent cell death (mean+/−SEM) was assayed by propidium iodide uptake using flow cytometry. (D) Lysates prepared from WTATG3 and KR cells were immunoblotted for the indicated markers. (E) Cells were treated for 24h with 500 nM obatoclax. Cell death (mean +/−SEM) was quantified using trypan blue exclusion. Statistical significance calculated using t test. (F) Summary model of results.
Anti-apoptotic members of the Bcl-2 family, such as Bcl-xL, function as potent inhibitors of cell death; these proteins are mainly located on mitochondria, which is vital for their pro-survival functions (Brenner and Mak, 2009). Increased Bcl-xL protein levels were present in KR cells compared to WTATG3, whereas no differences in the protein levels of the pro-apoptotic Bcl-2 family members, Bax and Bak, were observed (Figure 7D). Accordingly, we predicted that both WTATG3 and KR cells would be sensitive to the potent chemical inhibition of anti-apoptotic Bcl-2 family members. To test this hypothesis, we utilized obatoclax, a small molecule BH3 mimetic that antagonizes multiple anti-apoptotic Bcl-2 family proteins, including Bcl-xL, Bcl-2 and Mcl-1 (Nguyen et al., 2007). As a single agent, obatoclax was able to robustly induce cell death in both WTATG3 and KR cells; most importantly, we found no significant differences in obatoclax-mediated death between these two cell types (Figure 7E). In conclusion, cells lacking ATG12-ATG3 exhibit reduced cell death mediated by mitochondrial pathways; this protection correlates with an increase in anti-apoptotic Bcl-2 proteins secondary to the lack of ATG12 conjugation to ATG3.
DISCUSSION
Since the initial discovery of the ATG12 ubiquitin-like conjugation system, ATG5 remains as the only known target of ATG12 (Mizushima et al., 1998a; Mizushima et al., 1998b). Here, we provide evidence that multiple ATG12 substrates exist and identify ATG3, the E2 that mediates ATG8/LC3 lipidation during autophagy, as a target for ATG12 conjugation (Tanida et al., 2002b). Since this complex links two components of the autophagy conjugation machinery, we reasoned that ATG12 modification of ATG3 would primarily regulate the early steps of autophagosome formation (Ohsumi, 2001). However, our results argue against this hypothesis. Disrupting ATG12 conjugation to ATG3 has no discernable effect on LC3/ATG8 lipidation or nonselective autophagy in response to three different stresses—nutrient starvation, rapamycin-mediated mTOR inhibition and CCCP-induced mitochondrial depolarization. Instead, the most obvious consequences of disrupting the ATG12-ATG3 complex are increased mitochondrial mass, fragmentation of the mitochondrial network, and resistance to cell death mediated by mitochondrial pathways (Figure 7F).
Mitochondrial homeostasis requires the careful balance and integration of numerous processes, namely fission, fusion, biogenesis and degradation (Chen and Chan, 2009). Although we observe decreased levels of mitophagy upon disrupting ATG12-ATG3 complex formation, we do not believe our results can be solely explained by the defective autophagy of damaged mitochondria. First, the reduction in mitophagy in cells lacking ATG12-ATG3 is modest. Second, the increase in mitochondrial mass in KR cells is not accompanied by mitochondrial depolarization, suggesting that the excess mitochondria are functional. Third, during CCCP-induced uncoupling, we have uncovered that mitochondrial mass is actually higher in autophagy-competent KR cells than in ATG3 null cells, which are autophagy-deficient. Based on these results, we hypothesize that the ATG12-ATG3 complex may also restrict mitochondrial expansion using mitophagy-independent mechanisms. Notably, other proteins, like Parkin, have versatile effects on mitochondria. In addition to promoting mitophagy, Parkin has been found to control mitochondrial dynamics and to induce biogenesis; the precise interrelationships between these diverse biological outcomes remain unclear (Deng et al., 2008; Kuroda et al., 2006; Lutz et al., 2009; Narendra et al., 2008). Notably, we have uncovered increased basal levels of Bcl-XL in KR cells, which may contribute to changes in both mitochondrial mass and morphology. Recent work in neurons demonstrates that Bcl-XL plays a vital role in mitochondrial homeostasis, and is able to increase mitochondrial fission, fusion and biomass (Berman et al., 2009). Future studies are required to more precisely dissect how this complex between two ATGs uniquely affects mitochondrial expansion and morphology, as well as to identify potential interconnections with the mitochondrial fission and fusion machinery.
Surprisingly, despite possessing fragmented mitochondrial morphology, cells lacking ATG12-ATG3 exhibit resistance to death mediated by mitochondrial pathways. Overall, the effects of mitochondrial fission and fusion on cell death are variable and context dependent (Suen et al., 2008). We hypothesize that the protection found in KR cells may result, at least in part, from increased basal levels of anti-apoptotic Bcl-2 family proteins. In support, specific and potent antagonism of anti-apoptotic Bcl-2 proteins causes equivalent amounts of cell death in WTATG3 and KR cells. A future challenge will be to discern which of these dramatic perturbations in mitochondrial morphology and function in KR cells is causal, since mitochondrial homeostasis, dynamics, and cell death are in delicate equilibrium (Figure 7F).
Importantly, our results indicate that the effects of the ATG12-ATG3 complex on mitochondrial homeostasis and cell death are unique functions that can be completely separated from the established roles of either ATG in autophagosome formation. Other studies demonstrate that pleiotropic roles for ATGs beyond autophagy may exist; for example, recent work delineates a macroautophagy-independent mechanism of LC3-recruitment to the mammalian phagosome, which is dependent on Beclin-1 and ATG5 (Sanjuan et al., 2007). Moreover, our studies support that ATG12, like ubiquitin and SUMO, may serve as a broad-based ubiquitin-like protein conjugation with far-reaching implications in biology and human disease. Accordingly, we are actively pursuing the identification and validation of additional substrates subject to “12-ylation.”
EXPERIMENTAL PROCEDURES
Materials
Dr. Noburu Mizushima (Tokyo Medical and Dental University) provided atg5+/+ and atg5−/− MEFs and Dr. Masaaki Komatsu (Juntendo University) provided atg7+/+, atg7−/−, atg3+/+ and atg3−/− MEFs. Fibroblasts and HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS, penicillin and streptomycin. A peptide corresponding to the N-terminus common to human, mouse and rat MAP1LC3 was used to create α-LC3 rabbit polyclonal antibody (Fung et al., 2008). See supplemental methods for commercial antibodies and chemicals.
Generation of stable pools
cDNAs in this study are described in the supplemental methods. For retroviral transduction, VSV-G-pseudotyped retroviruses were generated, and cells were infected and selected as previously described (Debnath et al., 2003). Following infection and drug selection, early passage stable pools (maximum of 6–8 passages) were utilized for experiments to avoid clonal selection or drift.
Mass spectrometry
Lysates were sequentially immunoprecipitated with α-FLAG M2 affinity gel and monoclonal α-HA conjugated to agarose and eluted with 3× FLAG peptide or HA peptide, respectively (Sigma). The final eluate was separated by SDS-PAGE and visualized with Novex Colloidal Blue Stain Kit (Invitrogen). Bands of interest were in-gel tryptically digested following destain, and subject to tandem mass spectrometry (MS/MS) using electrospray ionization (ESI) and a quadropole quadropole time-of-flight (QqTOF) mass spectrometer located in the UCSF Mass Spectroscopy Core (QStarXL).
Immunofluorescence
Immunofluorescent staining was carried out as previously described (Debnath et al., 2003) with the following modifications during fixation and permeabilization. Fibroblasts were fixed with 2% paraformaldehyde at 4C for 10 min, followed by cold methanol (−20C) for 10 min and then permeabilized for 10 min at 20C with 0.5% Triton X-100 in PBS.
Microscopy
Widefield immunofluorescence imaging was performed using the 63× (1.4 NA) or 100× (1.3 NA) objectives of a Zeiss Axiovert 200 microscope equipped with a Spot RT camera (Diagnostics Instruments) and mercury lamp; images were acquired using Metamorph (v6.0) software (Molecular Devices). Confocal analysis was performed using the 60× (1.4 NA) objective of a Nikon C1si Spectral Confocal System equipped with an argon laser (488 line) and two solid-state diodes (405 and 546 lines). Images were color-combined in Metamorph (v6.0) and arranged in Adobe Photoshop (v7.0).
Analysis of punctate GFP-LC3
Fibroblasts expressing GFP-LC3 were grown overnight on fibronectin-coated coverslips prior to HBSS starvation. Cells were fixed with 4% paraformaldehyde, washed with PBS, mounted using Immunomount (Thermo), and analyzed by widefield immunofluorescent microscopy as described above; images were acquired and punctate GFP-LC3 was quantified using Metamorph (v6.0) software. At least 250 cells from 4 independent experiments were enumerated. For analysis of GFP-LC3 colocation with mitochondria, cells expressing GFP-LC3 were transiently transfected with mito-dsRed, plated onto fibronectin-coated coverslips, and treated with 10 µM CCCP for 24h. Cells were fixed, washed and imaged as above and colocalization, defined as the complete overlap of a GFP-LC3 punctum with mito-dsRed, was quantified per cell; at least 50 cells from 3 independent experiments were enumerated.
Flow cytometry for mitochondrial mass
As indicated, cells were grown in full media or subject to the indicated treatments for 24h. Cells were stained with 100 nM Mitotracker® Green (MTG) for 25 min at 37C. Cells were trypsinized, collected by centrifugation, washed twice in PBS and analyzed using a FACSCalibur (BD) and CellQuest Pro software. When indicated, cells were co-stained with 100 nM Mitotracker® Red CMXRos (MTR). For assays of 3MA-sensitive mitochondrial degradation during CCCP treatment, cells were treated for 24h with 10 µM CCCP in the presence or absence of 5mM 3MA and then stained with MTG as described. 3MA sensitive degradation was calculated as follows:
Cell death assays
Cells were treated with death agents for 24h. Cells were then trypsinized, pooled with detached cells (floaters), pelleted by centrifugation, PBS washed, and stained with propidium iodide (0.5 µg/mL) in PBS for 10 min at 20C. Cells were analyzed using a FACSCalibur (BD) and CellQuest Pro software. Obatoclax treated cells were analyzed using trypan blue exclusion because nonspecific fluorescence associated with drug treatment technically interfered with PI detection by flow cytometry.
Statistics
Experimental groups were compared using t test for pairwise comparisons or ANOVA (followed by Tukey's HSD test). For all experiments, statistical significance indicated as follows: N.S., nonsignificant; * p<0.05; ** p<0.01; *** p<0.001.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Noboru Mizushima, Masaaki Komatsu and Patrick Dennis for generously providing reagents, and Drs. Gerard Evan, Don Ganem, Abul Abbas and Feroz Papa for critically reading the manuscript. Confocal microscopy was performed in the Biological Imaging Development Center at UCSF. Grant support to JD includes the NIH (RO1CA126792, KO8CA098419), a Culpeper Medical Scholar Award (Partnership For Cures), an AACR-Genentech BioOncology Award, a HHMI Physician-Scientist Early Career Award, and a Stewart Family Trust Award. LR was a Genentech/Sandler Graduate Student Fellow and EF was an HHMI Summer Undergraduate Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Berman SB, Chen YB, Qi B, McCaffery JM, Rucker EB, 3rd, Goebbels S, Nave KA, Arnold BA, Jonas EA, Pineda FJ, Hardwick JM. Bcl-x L increases mitochondrial fission, fusion, and biomass in neurons. J Cell Biol. 2009;184:707–719. doi: 10.1083/jcb.200809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D, Mak TW. Mitochondrial cell death effectors. Curr Opin Cell Biol. 2009 doi: 10.1016/j.ceb.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A, Waterhouse NJ, Li CW, et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- de Graaf AO, van den Heuvel LP, Dijkman HB, de Abreu RA, Birkenkamp KU, de Witte T, van der Reijden BA, Smeitink JA, Jansen JH. Bcl-2 prevents loss of mitochondria in CCCP-induced apoptosis. Exp Cell Res. 2004;299:533–540. doi: 10.1016/j.yexcr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci U S A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PB, Mercer CA. The GST-BHMT assay and related assays for autophagy. Methods Enzymol. 2009;452:97–118. doi: 10.1016/S0076-6879(08)03607-0. [DOI] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Fukuda M, Noda T, Yoshimori T. The Atg16L Complex Specifies the Site of LC3 Lipidation for Membrane Biogenesis in Autophagy. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of Autophagy during Extracellular Matrix Detachment Promotes Cell Survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwozd CS, Arnason TG, Cook WJ, Chau V, Ellison MJ. The yeast UBC4 ubiquitin conjugating enzyme monoubiquitinates itself in vivo: evidence for an E2-E2 homointeraction. Biochemistry. 1995;34:6296–6302. doi: 10.1021/bi00019a006. [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hanada T, Ohsumi Y. Structure-function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy. 2005;1:110–118. doi: 10.4161/auto.1.2.1858. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- Kuroda Y, Mitsui T, Kunishige M, Shono M, Akaike M, Azuma H, Matsumoto T. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum Mol Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, Lammermann K, Brunner B, Kurz-Drexler A, Vogel F, Reichert AS, et al. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998a;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998b;273:33889–33892. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto T, Tanida I, Tanida-Miyake E, Minematsu-Ikeguchi N, Yokota M, Ohsumi M, Ueno T, Kominami E. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J Biol Chem. 2003;278:39517–39526. doi: 10.1074/jbc.m300550200. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Marcellus RC, Roulston A, Watson M, Serfass L, Murthy Madiraju SR, Goulet D, Viallet J, Belec L, Billot X, et al. Small molecule obatoclax (GX15-070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc Natl Acad Sci U S A. 2007;104:19512–19517. doi: 10.1073/pnas.0709443104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Rohas LM, St-Pierre J, Uldry M, Jager S, Handschin C, Spiegelman BM. A fundamental system of cellular energy homeostasis regulated by PGC-1alpha. Proc Natl Acad Sci U S A. 2007;104:7933–7938. doi: 10.1073/pnas.0702683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004;9:611–618. doi: 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy. 2005;1:119–126. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci U S A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Nishitani T, Nemoto T, Ueno T, Kominami E. Mammalian Apg12p, but not the Apg12p.Apg5p conjugate, facilitates LC3 processing. Biochem Biophys Res Commun. 2002a;296:1164–1170. doi: 10.1016/s0006-291x(02)02057-0. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002b;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- Walter J, Urban J, Volkwein C, Sommer T. Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p. Embo J. 2001;20:3124–3131. doi: 10.1093/emboj/20.12.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Yamada Y, Suzuki NN, Hanada T, Ichimura Y, Kumeta H, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J Biol Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat Rev Mol Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.