Abstract
The Lys65 residue, in the fingers domain of HIV reverse transcriptase (RT), interacts in a sequence independent fashion with the incoming dNTP. Previously, we showed that a 5 amino acid deletion spanning Lys65 and a K65A substitution both enhanced the fidelity of dNTP insertion. We hypothesized that the Lys65 residue enhances dNTP misinsertion via interactions with the γ-phosphate of the incoming dNTP. We now examine this hypothesis in pre-steady state kinetic studies using wild type HIV-1 RT and two substitution mutants: K65A and K65R. The K65R mutation did not greatly increase misinsertion fidelity, but the K65A mutation led to higher incorporation fidelity. For a misinsertion to become a permanent error, it needs to be accompanied by the extension of the mispaired terminus thus formed. Both mutants, and the wild-type enzyme, discriminated against the mismatched primer at the catalytic step (kpol). Additionally, K65A and K65R mutants displayed a further decrease in mismatch extension efficiency, primarily at the level of dNTP binding. We employed hydroxyl radical footprinting to determine the position of the RT on the primer-template. The wild-type and Lys65 substituted enzymes occupied the same position at the primer terminus; the presence of a mismatched primer terminus caused all three enzymes to be displaced to a −2 position relative to the primer 3’ end. In the context of an efficiently extended mismatched terminus, the presence of the next complementary nucleotide overcame the displacement, resulting in a complex resembling the matched terminus. The results are consistent with the observed reduction in kpol in mispaired primer extension being due to the position of the enzyme at a mismatched terminus. Our work shows the influence of stabilizing interactions of Lys65 with the incoming dNTP in affecting two different aspects of polymerase fidelity.
Keywords: HIV-1 reverse transcriptase, polymerase fidelity, pre-steady state kinetics of HIV-1 RT, NRTI resistance mutation, site-specific foot-printing
Introduction
HIV-1 reverse transcriptase (RT) is required for replication of the HIV genome. HIV-1 RT shares its overall architecture with all other DNA polymerases; it resembles a right hand in shape, with fingers, palm and thumb subdomains 1. Although the polymerase has several features in common with high fidelity, replicative DNA polymerases (e.g., a sterically constrained active site, movement of the fingers domain following dNTP binding, and a lack of dependence on Watson-Crick hydrogen bonds for dNTP incorporation), it has a lower fidelity of DNA replication 2,3. This error-proneness is one of the major factors in the high level of sequence diversity observed within viral populations, from which drug- and immune-escape variants emerge swiftly.
A crucial event during viral DNA synthesis, at which the errors are introduced, involves the selection of correct dNTP for insertion at the growing viral DNA terminus. The incoming dNTP in the polymerization reaction is stabilized via direct interactions with RT residues in the dNTP binding site; hydrogen bonds are formed between Lys65 and the γ-phosphate of the incoming residue, Arg72 and the α and β-phosphate, and Gln151 with the sugar 3’-OH 4. These interactions are sequence independent; they do not require that the incoming dNTP be complementary to the template base. Following dNTP binding, the RT undergoes a conformational change, which is the rate-limiting step in catalysis 4–7. The rate-limiting step is further reduced in dNTP misinsertion or mismatch extension; this discrimination is hypothesized to be due to constraints imposed by the shape and flexibility of the dNTP binding pocket8–12. The RT dNTP binding pocket is sterically constrained by residues that interact directly with the dNTP, and by the template base, the primer 3’ terminus, the divalent cation co-factors and by stacking interactions between Arg72 and the incoming dNTP 4,9. The primer and template strand are themselves constrained by interactions with the RT. Polymerase fidelity is thus influenced by changes in either residues directly interacting with the incoming dNTP or altering the steric constraint of the dNTP binding pocket (reviewed in reference 13.)
The fingers subdomain of RT contains a small structural element, the β3-β4 loop, which projects into the dNTP-binding site of the enzyme. The loop contains several residues that are important in drug resistance and replication fidelity, including the lysine 65 residue that, as previously mentioned, forms a hydrogen bond with the incoming dNTP. We, and others, have previously demonstrated that deletion of the entire β3- β4 loop, or a K65A substitution 14,15, leads to an increased fidelity through an increase in dNTP selection stringency. The increase in stringency was hypothesized to occur because in the wild-type enzyme, a hydrogen bond between the K65 residue and the γ-phosphate of the incoming dNTP stabilizes the dNTP in the enzyme active site. In the absence of this interaction, the dNTP stabilization is reliant on other interactions, which are more sequence specific, leading to higher fidelity 14. In the present work, we have examined this hypothesis via the use of pre-steady state kinetics. Our results revealed that correct dNTP insertion was enhanced over incorrect for the K65A mutant via a decreased incorporation efficiency for the incorrect dNTP.
Insertion of an incorrect dNTP is only one aspect of enzyme fidelity. In order for a misinserted dNTP to become part of a DNA chain, the polymerase must be able to add a dNTP to the 3’-OH of the newly misinserted dNMP, extending the nascent DNA strand. HIV reverse transcriptase is very efficient at extending a DNA strand from a mismatched terminus in comparison to other DNA polymerases and reverse transcriptases including, for example, a replicative DNA polymerase 16 and other retroviral reverse transcriptases 17. However, even HIV RT is less efficient at extending a mismatched terminus than a fully matched terminus. Interestingly, the discrimination against a mismatched terminus appears to occur through a reduction in dNTP incorporation efficiency, rather than through a reduced efficiency in binding the primer-template with a mismatched terminus 18. In this report, we have used pre-steady state kinetics to investigate the role played by the Lys65 residue in mismatch extension fidelity. Our results show that both substitutions (Arg and Ala) at Lys65 led to decreased efficiency of mismatch extension, primarily through a decreased affinity for the incoming dNTP. Site specific hydroxyl radical footprinting of HIV-1 RT complexes with matched and mismatched template primers revealed that with a mismatched terminus the wild-type and mutant RTs are displaced to a −2 position with respect to the templating base, which is consistent with the observed reduction in kpol in mispair extension.
Results
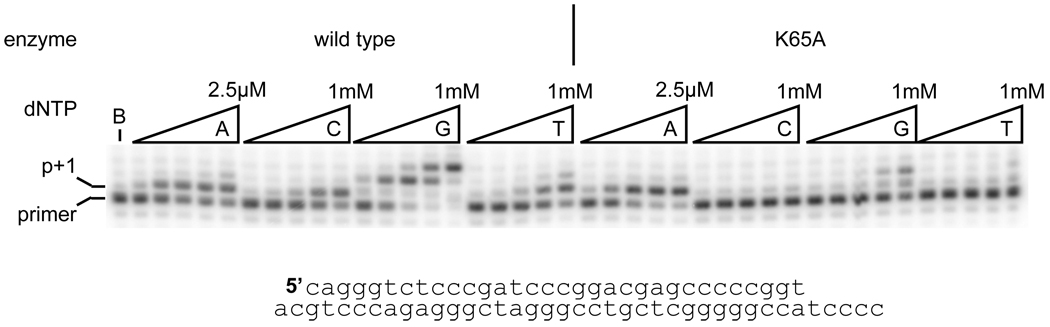
In order to examine the role of lysine 65 on replication fidelity, we employed recombinant, purified HIV-1 RT with either a K65R or K65A substitution, and compared them to the wild-type enzyme in fidelity assays. First, single-dNTP extension assays, in which a 32P 5’-labeled primer annealed to an unlabeled template oligonucleotide was extended in the presence of a single dNTP, were used to assess the effect of Lys65 substitutions on RT fidelity. Qualitative experiments showed, as previously demonstrated, that the K65A substitution resulted in an RT that was far less efficient at dNTP misinsertion than the wild-type enzyme (Figure 1), particularly in the misinsertion of dCMP and dTMP opposite a template thymine. In order to investigate the role that the K65 residue plays in this discrimination, pre-steady state single dNTP incorporation assays were performed.
Figure 1.
Single dNTP incorporation by wild-type and K65A reverse transcriptases. 5’-end labeled primer, annealed to the template strand, was incubated with reverse transcriptase for 10 minutes in the presence of either the correct dNTP (dATP, 5, 25, 100, 500, 2500 nM) or incorrect dNTP (2.5, 10, 50, 250, 1000 µM).
Pre-steady state kinetics can be used to determine both the dNTP binding affinity (Kd) and the rate of catalysis (kpol), which, in HIV-1 RT, is limited by the conformational change required to align the incoming dNTP with the primer terminus. Nucleotide discrimination is due to changes in either, or both, Kd or kpol for incorrect dNTP incorporation, relative to that of correct dNTP. Selcectivity and fidelity of nucleotide insertion were assessed, as described in the legend to Table 1. Overall, the K65A mutation caused an increase in misinsertion fidelity of between 2 and 4-fold compared to the wild-type enzyme. Utilizing a template purine, a greater fold-reduction in catalytic efficiency (kpol/Kd) was observed for misinsertion of the pyrimidine nucleotides (dCMP and TMP) by K65A, as compared to misinsertion of dGMP (Table 1). Selection against the pyrimidine nucleotides was mostly at the level of dNTP binding; the effect of the K65A substitution was to increase the Kd of the incorrect dNTP (for dCTP, the Kd was increased 166-fold for wild type and 515-fold in K65A compared to the correct dNTP), with only a minor effect on kpol relative to the wild-type (Table 1).
Table 1.
Pre-steady state analysis of nucleotide misinsertion by wild-type and K65 substituted RT
| kpol Fold Change | Kd Fold Change | Fidelity Fold Change | |||||||
|---|---|---|---|---|---|---|---|---|---|
| dNTP | RT | kpol (s−1)a | from Correct | Kd (µM)a | from Correct | kpol/Kd (µM−1 s−1)a | Selectivityb | Fidelityc | from WT |
| dATP | WT | 56.85 ± 17.89 | 17.95 ± 2.33 | 3.13 ± 0.594 | |||||
| K65R | 20.45 ± 3.04 | 16.8 ± 3.25 | 1.22 ± 0.057 | ||||||
| K65A | 2.99 ± 0.990 | 6.96 ± 1.95 | 0.465 ± 0.276 | ||||||
| dGTP | WT | 0.854 ± 0.111 | 67 | 1491 ± 635.7 | 83 | 6.12×10−4 ± 1.89×10−4 | 5114 | 5361 | |
| K65R | 0.244 ± 0.021 | 84 | 1430 ± 140 | 85 | 1.71×10−4 ± 1.70×10−6 | 7135 | 7123 | 1.3 | |
| K65A | 0.02265 ± 0.00021 | 132 | 722.1 ± 33.23 | 104 | 3.13×10−5 ± 1.76×10−6 | 14856 | 14844 | 2.8 | |
| dCTP | WT | 0.2369 ± 0.0205 | 240 | 2980 ± 733.3 | 166 | 8.28×10−5 ± 2.73×10−5 | 37802 | 40265 | |
| K65R | 0.0272 ± 0.0032 | 751 | 833 ± 347 | 50 | 3.26×10−5 ± 1.41×10−6 | 37423 | 37441 | 0.9 | |
| K65A | 0.0109 ± 0.0013 | 274 | 3583 ± 1.41 | 515 | 3.06×10−6 ± 3.63×107 | 151961 | 153044 | 3.8 | |
| dTTP | WT | 0.2703 ± 0.0589 | 210 | 5478 ± 2463 | 305 | 4.94×10−5 ± 2.46×10−5 | 63360 | 63425 | |
| K65R | 0.0636 ± 0.0126 | 322 | 4909 ± 2238 | 292 | 1.29×10−5 ± 6.44×10−6 | 94574 | 94200 | 1.5 | |
| K65A | 0.0530 ± 0.0086 | 56 | 12700 ± 2879 | 1825 | 4.17×10−6 ± 1.16×10−6 | 111511 | 111482 | 1.8 |
Values represent averages and standard deviations from at least two independent experiments
Calculated as (kpol/Kd)correct/(kpol/Kd)incorrect
Calculated as [(kpol/Kd)correct + (kpol/Kd)incorrect]/(kpol/Kd)incorrect
For utilization of dGTP, the K65A substitution caused a greater fold change in kpol compared to wild-type. The fold change in kpol from correct dNTP insertion for WT was 67-fold and for K65A, it was 132-fold whilst the Kd fold change from correct dNTP was similar for the two enzymes (83-fold and 104-fold for wild-type and K65A respectively, see Table 1).
The K65R substitution had a less defined effect on misinsertion fidelity. The substitution did not alter affinity of the RT for the correct dNTP, and either had no effect (dGTP, dTTP) or increased affinity (dCTP) for incorrect dNTPs. The kpol was reduced compared to wild-type for both correct (approximately 2-fold) and incorrect (3 to 9-fold) dNTP incorporation, with a resultant modest effect on fidelity (defined by [(kpol/Kd)Correct + (kpol/Kd)incorrect]/(kpol/Kd)Incorrect). However, in absolute terms, the K65R RT is considerably less efficient at misinsertion compared to the wild-type enzyme (WT(kpol/Kd)Incorrect / K65R(kpol/Kd)Incorrect = 2.5 to 3.6-fold), largely due to the reduction in kpol (Table 1).
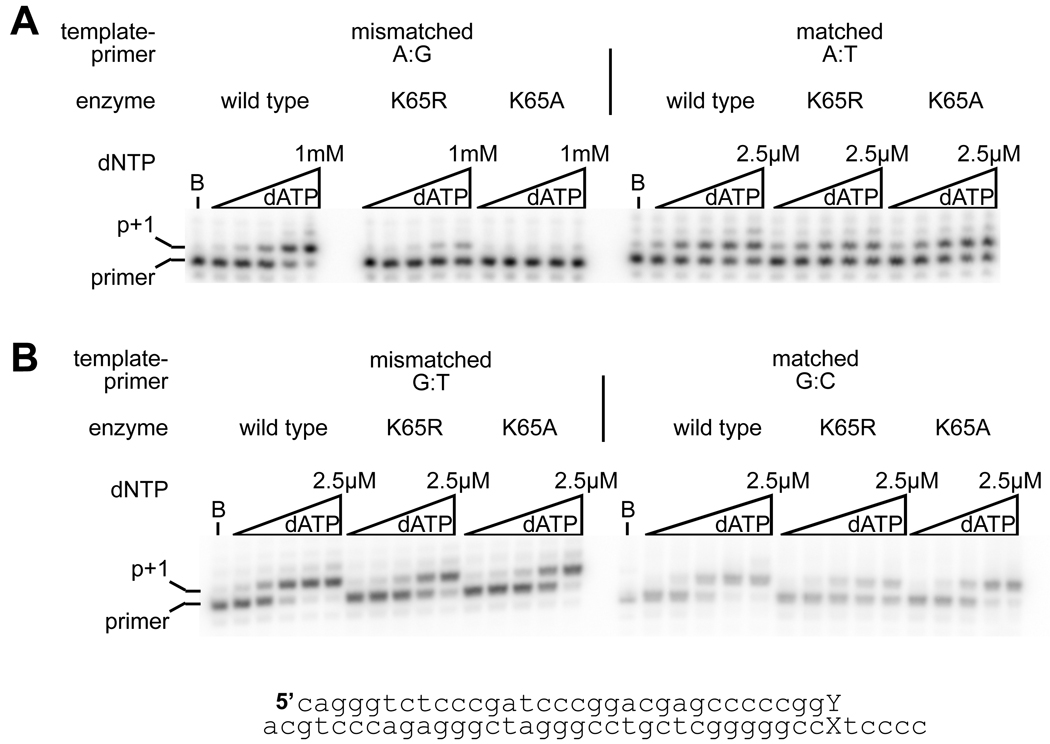
Next, the influence of the lysine 65 substitutions on mismatch extension was investigated by comparing the extension of radiolabeled DNA primers with matched and mismatched 3’ termini, annealed to DNA templates, in reactions containing dATP (the next complementary dNTP). Two different mismatched termini, A:G and G:T, were employed in these assays. The G:T base-pair is more similar to a canonical base-pair than the purine-purine A:G mismatch. Qualitatively, it was clear that both the conservative (K65R) and nonconservative (K65A) substitutions greatly reduced the efficiency of mismatch primer extension from an A:G mismatch (Figure 2), with K65A having a greater effect. Extension from the G:T mismatch was similar to extension from the matched terminus, and no qualitative difference could be seen between the three enzymes. Pre-steady state kinetic measurements were used to quantify the differences in extension, and it was demonstrated that from the A:G mismatch the K65R mutant did not alter kpol, and with the K65A mutant, the reduction in kpol was modest (4.5-fold compared to wild-type). However, the influence of the substitutions on the dNTP Kd was dramatic, leading to a decrease in affinity of 13-fold and 52-fold respectively for K65R and K65A (Table 2). From the more conservative G:T mismatch K65R and K65A again showed a small reduction in kpol compared to the wild-type enzyme. The nucleotide Kd was also increased for the K65A enzyme, but to a much more modest extent (2.5-fold compared to wild-type).
Figure 2.
Extension from a mismatched terminus by wild-type, K65R and K65A reverse transcriptases. 5’-end labeled primer with either a matched or non-complementary 3’ dNTP, was annealed to the template strand, and extended with reverse transcriptase. A. extension from a template:primer strand terminated with an A:G mismatch or an A:T base-pair. B. extension from G:T and G:C terminated template :primer. The 10-minute extension reactions were performed with dATP at 5, 25, 100, 500, 2500 nM for the matched terminus and G:T mismatch, and 2.5, 10, 50, 250, 1000 µM at the mismatched A:G terminus. The sequence of the template:primer utilized in these experiments is shown; the nucleotides that are changed are shown as an X (template nucleotide A or G) and a Y (primer nucleotide, C, G or T).
Table 2.
Pre-steady state kinetics of extension from a mismatched primer terminus
| Mismatch | RT | kpol (s−1)a | Kd (µM)a | kpol/Kd (µM−1 s−1)a | Mismatched Extension Fidelityb,c | Fold Change to Wild Type |
|---|---|---|---|---|---|---|
| A:G | WT | 1.06 ± 0.325 | 42.1 ± 5.37 | 0.0245 ± 0.0049 | 129 | |
| K65R | 1.45 ± 0.876 | 543.3 ± 45.11 | 0.0028 ± 0.0018 | 436 | 3 | |
| K65A | 0.239 ± 0.076 | 2175 ± 910.4 | 0.00011 ± 0.00006 | 3987 | 31 | |
| G:T | WT | 34.3 ± 10.6 | 369.9 ± 14.2 | 0.0923 ± 0.0250 | 35 | |
| K65R | 14.2 ± 0.025 | 251.9 ± 36.1 | 0.0571 ± 0.0083 | 22 | 0.6 | |
| K65A | 7.02 ± 0.264 | 934.0 ± 254.6 | 0.0078 ± 0.0024 | 60 | 1.7 |
Values represent averages and standard deviations from at least two independent experiments.
Calculated as [(kpol/Kd)matched + (kpol/Kd)mismatched]/(kpol/Kd)mismatched
Compared to (kpol/Kd)matched from Table 1
In a comparison between matched (A:T, Table 1) and mismatched primer termini (A:G, Table 2), it can be seen that the wild-type enzyme discriminates against the mismatched primer terminus largely on the basis of a reduced kpol; however, with the K65R and K65A substitutions the effect of increased Kd for dNTP (decreased affinity) becomes an increasingly important factor in the discrimination.
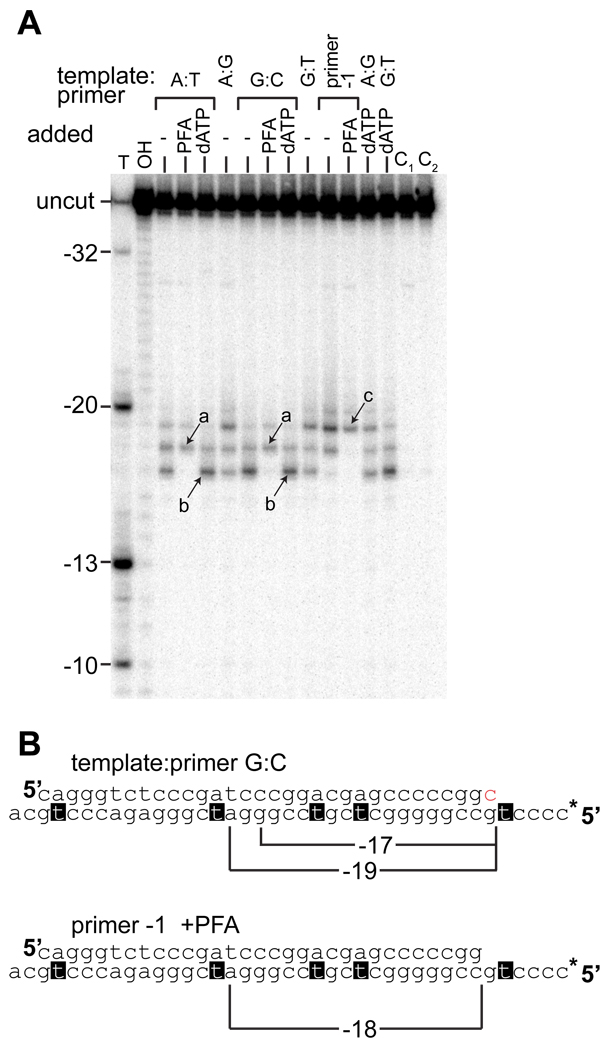
Site-specific footprinting, in which hydroxyl radicals are generated at the RNase H active site in the presence of Fe2+ 19, help identify the position of the reverse transcriptase on the primer-template. Comparison of a ladder generated by thymidine-specific cleavage of the template strand to the iron-dependent, RNase H directed cleavage of the template strand demonstrated that the products corresponded to cleavage at the −17, −18 and −19 positions, relative to the primer terminus (Fig. 3A and B). The specific position of the RT on the mismatched primer-template was identified by comparison to cleavage reactions performed under different conditions, such that the RT was locked into a particular position on the primer-template (Figure 3A). The post-translocation position of the RT on the primer-template was identified by Fe2+ directed footprinting on a primer blocked at the 3’-end with a dideoxynucleotide in the presence of the next complementary dNTP (Fig. 3A, band labeled “b”). The pre-translocation position (labeled “a” in Fig. 3A) was identified by footprinting in the presence of phosphonoformic acid (PFA), a non-hydrolyzable pyrophosphate analogue 20. Footprinting was also carried out with a primer shortened by a single dNTP at the 3’ end, both in the presence and absence of PFA. Using PFA, the pre-translocation complex on this template-primer was determined to lead to a −19 cleavage product. Control reactions demonstrated that the footprinting was dependent on the presence of both enzyme (Fig. 4) and Fe2+ (Figure 3A), and that the presence of the dideoxynucleotide on the blocked primer did not itself alter positioning compared to the deoxynucleotide terminated primer (result not shown).
Figure 3.
Position of RT on primer-template with matched or mismatched primer terminus by iron footprinting of the RNase H site on the radiolabeled template strand. A. Wild-type RT was incubated with different primer-templates, containing primer termini with the specified terminal base-pairs. 1 mM PFA or 1 mM dATP was added as indicated; in reactions containing dATP, the primer was terminated with a dideoxynucleotide. Cleavage was initiated by addition of 33 µM Fe(NH4)2(SO4)2. Products due to cleavage of RT primer-template pre-translocation and post-translocation complexes are identified by arrows and lettered a and b respectively. Cleavage products from a template-primer shortened by a single nucleotide at the 3’-end (primer −1, sequence shown in B) are shown in the presence and absence of PFA. The band representing a pre-translocation complex on this template:primer is indicated by an arrow and the label c. Control reactions C1 and C2 contained matched and mismatched primer termini, but Fe(NH4)2(SO4)2 was not added. The size of the cleavage products was determined by comparison to a ladder generated by thymidine-specific and nonspecific hydroxyl radical cleavage of the radiolabeled template strand (labeled T and OH respectively). The relative positions of the thymidines and the RNase H directed cleavage events are indicated in B. Likewise, the position of the cleavage products utilizing the primer −1 sequence are indicated. * on the primer-template sequence indicates the position of the radiolabeled 5’ terminus.
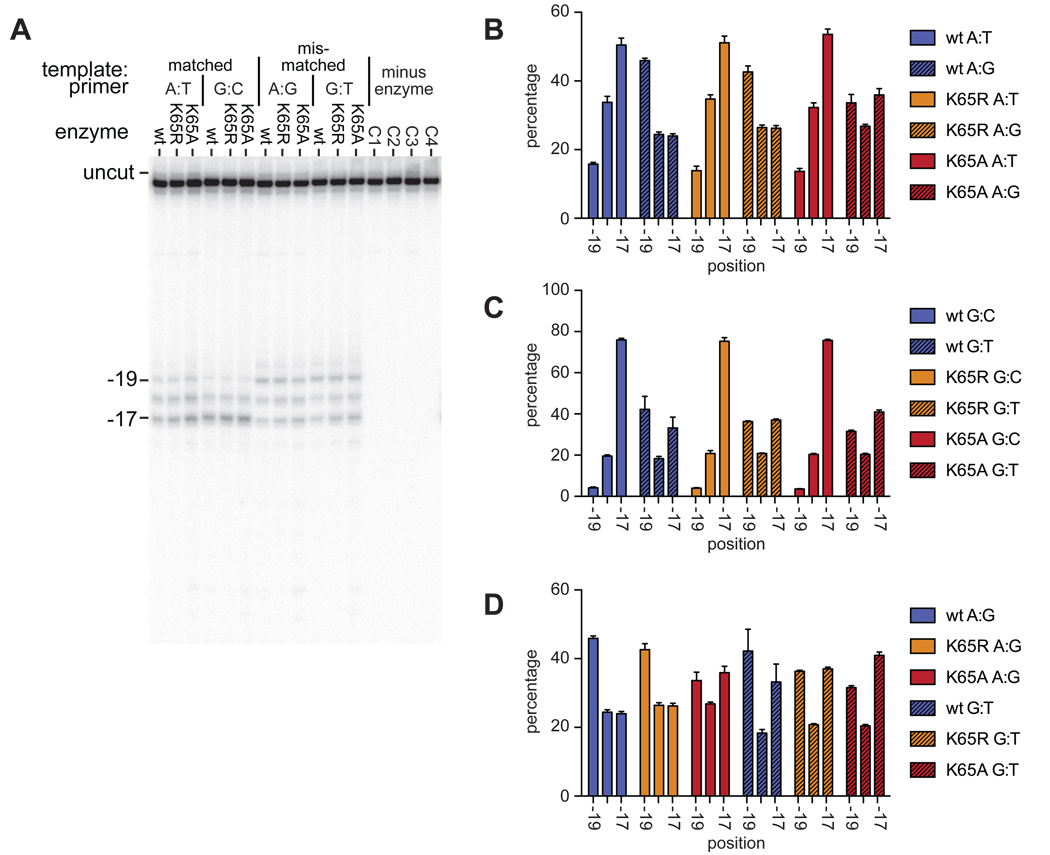
Figure 4.
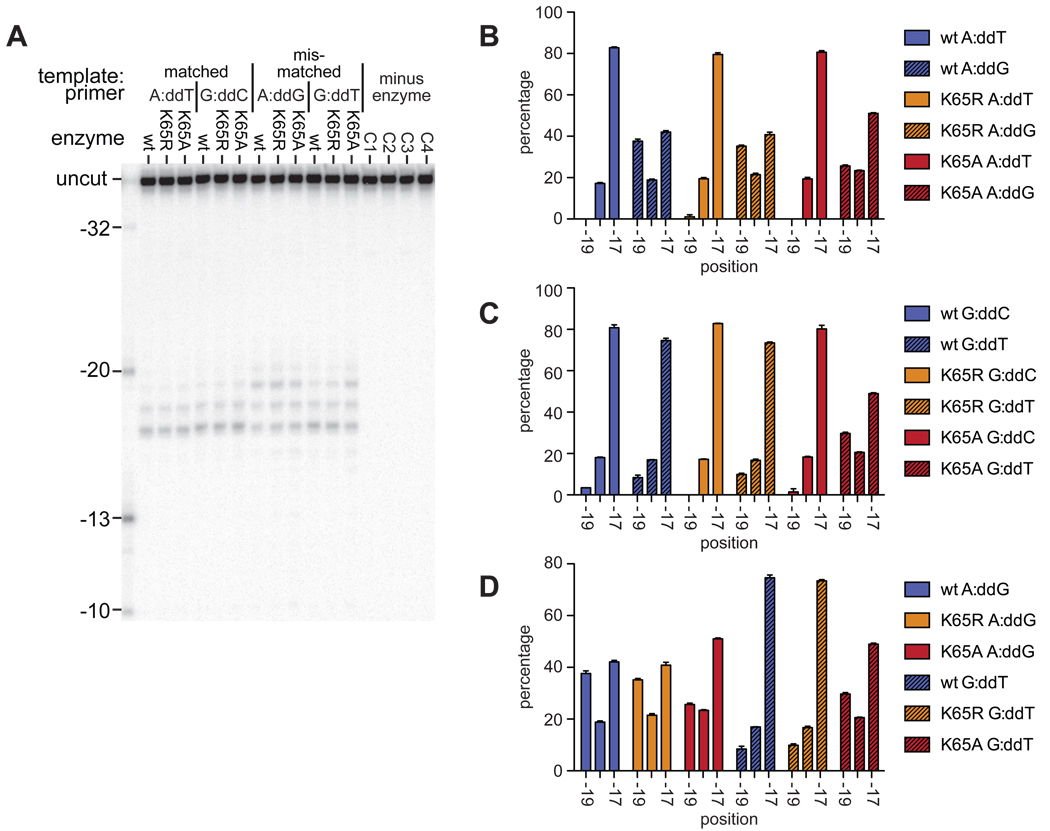
RNase H directed hydroxyl radical footprinting of wildtype and K65 substituted enzymes on matched and mismatched printer-templates. A. Representative gel showing the positions of Fe2+ dependent cleavage, relative to the primer 3’ end, with either matched (A:T, G:C) or mismatched (A:G, G:T) template-primer terminal base-pair. Control reactions C1–C4 contained each template:primer in turn incubated in the absence of enzyme. B, C and D. Graphical representation of position of wild-type and mutant RTs on template:primer with a template A, template G, and mismatched terminus respectively. Percentage of cleavage products is plotted against the position of the cleavage, which is relative to the 3’ terminus of the primer. Results are from at least three independent reactions, error bars show SEM.
Using primer-templates with the same sequence as those used in the pre-steady state kinetics assays, the position of the RT was shifted when the primer terminus was mismatched (Figures 3 and 4). It is clear that on a mismatched primer, the RT favored a position equivalent to the pre-translocation position on a primer shortened by a single dNTP. However, the positioning was not grossly altered in the K65A and K65R RT mutants compared to the wild-type (Figure 4). Additionally, the shift in position was seen with both the mismatched termini tested, A:G and G:T (Fig 4D), although with the A:G mismatch a greater proportion of −19 complexes were observed.
Following binding of the next complementary nucleotide, the enzyme is in a post-translocation complex. The ability of the wild-type and mutant RTs to enter the post-translocation complex was measured on both matched and mismatched primer termini (Figure 5), using primers in which the 3’ terminus is blocked with a dideoxynucleotide, and in the presence of the next complementary nucleotide (dATP). Under these conditions, the enzymes were found to occupy a predominantly −17 position on the matched termini. On the G:T mismatched termini the wild type and K65R enzymes were also mostly in a −17 position; however, this effect was less pronounced with the K65A substitution. When the template:primer contained the more extreme A:G mismatch, all three enzymes were present at positions −19, −18 and −17 (Fig. 4D).
Figure 5.
RNase H directed hydroxyl radical footprinting of wildtype and K65 substituted enzymes on matched and mismatched printer-templates, in the presence of the next complementary nucleotide (1 mM dATP). Primers were terminated with dideoxynucleotide. Reactions and labeling as for Figure. 5.
Discussion
The Lys65 residue has been implicated in misinsertion fidelity 14,15,21. Using a forward mutation assay, in which substitution, deletion and insertion mutations were measured, the K65R mutation was found to increase RT fidelity by 8-fold through a decrease in all three types of error 21. Steady state experiments suggested that the increased fidelity in RT with Lys65 substitutions was due to a reduced binding of incorrectly base-paired dNTPs in the active site; in the absence of the interaction between Lys65 and the incoming dNTP, polymerization is dependent on Watson-Crick hydrogen bonds between the incoming and templating bases 14. In addition to the role that Lys65 plays in the binding of the incoming dNTP, it also has a role in maintaining the dNTP and residues around the active site in the correct conformation for efficient catalysis 22.
Discrimination against misinsertion of pyrimidine dNTPs (with a pyrimidine template base) by K65A RT is due to a decrease in kpol and increased Kd; the decrease in dNTP binding affinity is the largest factor. However, reduced misinsertion of dGMP with a template thymidine is predominantly through a reduction in kpol. The loss of the interaction between Lys65 and the γ-phosphate of the incoming dNTP in the K65A substituted RT will result in a greater dependency on other interactions in the dNTP-binding pocket to stabilize binding of the incoming nucleotide. With a template pyrimidine, it may be expected that an incoming purine will be more able to form these interactions with the RT, even in the absence of complementary Watson-Crick hydrogen bond formation, than an incoming pyrimidine. Thus, stable binding of the incorrect dNTP in the active site would be less dependent on the hydrogen bond between the γ-phosphate of the incoming dNTP and Lys65, and the K65A substitution has less effect on Kd. However the change in kpol caused by the K65A substitution suggests that after binding dGTP, the rate of the conformational change preceding the chemistry step is reduced, possibly due to the improper alignment of the incorrect dNTP with the template base.
As expected, the K65R substitution has no, or little, effect on the binding of either a correct or incorrect dNTP; one of the guanidinium nitrogens of the Arg65 residue is positioned to coordinate the incoming dNTP similarly to Lys65 in the wild type. The K65R mutation has a significant effect on kpol for misinsertion. The recently solved ternary structure of K65R RT in complex with template-primer and an incoming dATP suggests that the K65R substitution locks the Arg72 side-chain into a particular orientation via a stacking interaction, and it was hypothesized that the structural constraint imposed by the K65R/Arg72 interaction could be involved in the higher fidelity of the K65R mutation 22. The Arg72 side chain is extremely important in catalysis, interacting with the incoming dNTP through stacking interactions with the base, and hydrogen bond formation with the α-phosphate 4. In the context of misinsertion, the Arg72, with less freedom of movement due to the K65R substitution, may be less able to coordinate the dNTP for optimal catalysis, hence reducing the kpol. Similarly, K65R results in resistance to dideoxynucleotide inhibitors 23, and this has been shown to be due to a reduction in kpol 24,25; the K65R mutation also allows binding of dNDPs by RT, but the wild-type residue is required for incorporation 26.
In order for a misincorporated dNTP to be “fixed” within a nascent DNA strand, the polymerase has to be capable of extending the mismatched primer terminus that results from such a misinsertion 16. In addition to the frequency of misinsertion, the efficiency of mismatch primer extension is an important aspect of DNA replication fidelity. Both the K65A and K65R mutations lead to a decrease in mismatch extension efficiency compared to the wild-type enzyme. The difference in fidelity caused by the substitutions is almost entirely at the level of dNTP binding; on the purine-purine mismatched terminus, the K65R and K65A substitutions increase the dNTP Kd, compared to the wild-type, approximately 13-fold and 52-fold respectively. However, when extension of matched and mismatched primer termini are compared, it is clear that the wild-type enzyme discriminates against the mismatched primer on the basis of kpol rather than Kd. The reduction in kpol in the wild-type enzyme on a mismatched primer has been previously reported, and it was suggested that, depending on sequence context, this reflects a change in the limiting step of catalysis 18. Mismatch extension may be limited by the rate of the chemical step, rather than the preceding conformational change as is the case for the wild-type enzyme at a matched primer terminus 5,27.
The K65R mutation has previously been demonstrated to result in an 8-fold increase in base substitution fidelity using a forward mutation assay 21. A high proportion of the substitutions observed in that assay occurred at homopolymeric runs, hypothesized to be due to increased pausing of the reverse transcriptase at homopolymeric sequences. The proportion of errors made by the wild type enzyme at homopolymeric runs was higher than that of the K65R. This dynamic effect of sequence context on the misinsertion event cannot easily be analyzed by pre-steady state kinetics.
K65R reduces sensitivity of HIV to tenofovir and ddNTPs, but is rarely selected as a result of drug treatment. Several studies have shown that patient-derived or recombinant virus containing the K65R mutation has a lower replicative capacity than wild-type in cell culture (from ~20% to 60% that of wild type) 28–30, but conflicting results have also been reported, showing that the K65R mutation does not grossly impact viral replicative capacity in cell culture, in either single- or multiple cycles of infection 23,25. The K65R mutation results in a decrease in kpol for the correct, natural dNTPs28, but this does not seem to be sufficient to explain the lack of selection in vivo. It has been suggested that a biased reduction in efficiency in dNTP incorporation, particularly affecting purine utilization 28, or a reduction in processivity at low dNTP concentrations 30, reduces the viral fitness. However, the results presented here suggest other possibilities for a reduction in fitness. Although K65R results in an increase in misinsertion fidelity, there is an additional decrease in mismatch extension from some termini, which is due to a 10-fold increase in dNTP Kd compared to the wild-type. Thus, following a misinsertion, the K65R substituted RT is less capable of extending the nascent strand than the wild-type enzyme, presumably reducing the overall replication efficiency and therefore viral fitness. This effect is clearly influenced by the particular base-pair at the mismatched terminus; by comparison, at a G:T mismatch the affinity of the K65R substituted RT for the next nucleotide was increased.
The wild-type RT does not discriminate against a mispaired primer terminus through decreased primer-template binding, but rather through incorporation of the incoming dNTPs. However, mutant RTs that have non-aromatic substitutions of residues that directly interact with the primer-template (Trp24 and Phe61) do reduce mismatched primer-template binding and additionally reduce the rate with which a mismatched terminus is extended 31,32. V148I and Q151N substitutions in reverse transcriptase also increase mismatch extension fidelity, producing mismatch extension efficiencies that approach the misinsertion efficiency 33; in contrast, the wild-type enzyme is 1–2 orders of magnitude more efficient in primer misextension than dNTP misinsertion 18. The Arg72 residue, which like Lys65 is involved in dNTP binding, has also been implicated in both misinsertion and mismatch extension fidelity by playing a role in stabilizing the binding of incorrect dNTPs in the RT active site 34. The Y115W substitution also reduces dNTP binding affinity and decreases primer mismatch extension, although this effect can be somewhat mitigated by the presence of a primer-grip substitution, M230I 35. In RTs containing V75I, a secondary mutation associated with mutations conferring resistance to both ddNTPs and NNRTIs, there is an increase in incorporation fidelity and, to a greater degree, a decrease in efficiency of mismatch extension. The mechanism through which V75I acts is less clear, as the residue does not interact directly with the incoming dNTP, but does interact with the template overhang 36.
Site-specific Fe2+-dependent hydroxyl radical generation at the RNase H active site has been used to identify the position of RT on a primer-template 19, and has been used to demonstrate that positioning is altered by a mismatched primer terminus37. The Fe2+ induced cleavage pattern that we observe here is somewhat different to that reported previously 37, in which only two cleavage products corresponding to 17 and 18 dNTPs from the primer terminus were reported using a matched primer-terminus. The precise positioning of the RNase H site relative to the primer terminus is likely sequence-dependent; several cleavage products have been reported using this assay on other primer-templates 38. More intriguingly, although the previous report with the wild-type enzyme and a mismatched primer demonstrated a difference in positioning compared to the matched primer, the difference was in the opposite direction from what we observe here 37; the results in this report demonstrate that on a mismatched primer the RT can occupy a position corresponding to a pre-translocation complex on a primer shortened by 1 nucleotide at the 3’ end (Fig. 3), whereas in the previous report a mismatched primer terminus increased the proportion of the RT in a post-translocation complex. This may be due to differences in the sequences of the primer-templates utilized, or differences in the primer length; as the double-stranded primer-template in the previous report was only 18 dNTPs in length, that may have prevented the RT from sliding to an upstream position corresponding to that observed here.
At a matched primer terminus, the RT was found predominantly in a −17 position, corresponding to the post-translocation complex. The presence of either the A:G or G:T mismatch caused a shift in this position, such that between 30 and 40% of the cleavage occurred at the −19 position, corresponding to a −2 pre-translocation complex. There was little difference in the extent of this shift between the A:G and G:T mismatches; the change in position on the template:primer did not reflect the difference in efficiency of extension from the two mismatched termini. However, utilizing a dideoxynucleotide blocked primer, and in the presence of the next complementary nucleotide, the RT was found predominantly in the post-translocation complex (−17 cleavage product) with both matched primer termini, and with the G:T mismatch (Fig 5). However, with an A:G mismatch, the RT was still found distributed between the −19 and −17 positions. This suggests that the deformation of the DNA structure induced by the A:G mismatch is sufficient to prevent the RT from sliding into the position that it would occupy when a nucleotide is bound in the active site. In contrast, with a less drastic mismatch, in the presence of the next complementary nucleotide, the RT can occupy a similar position to that at a matched terminus.
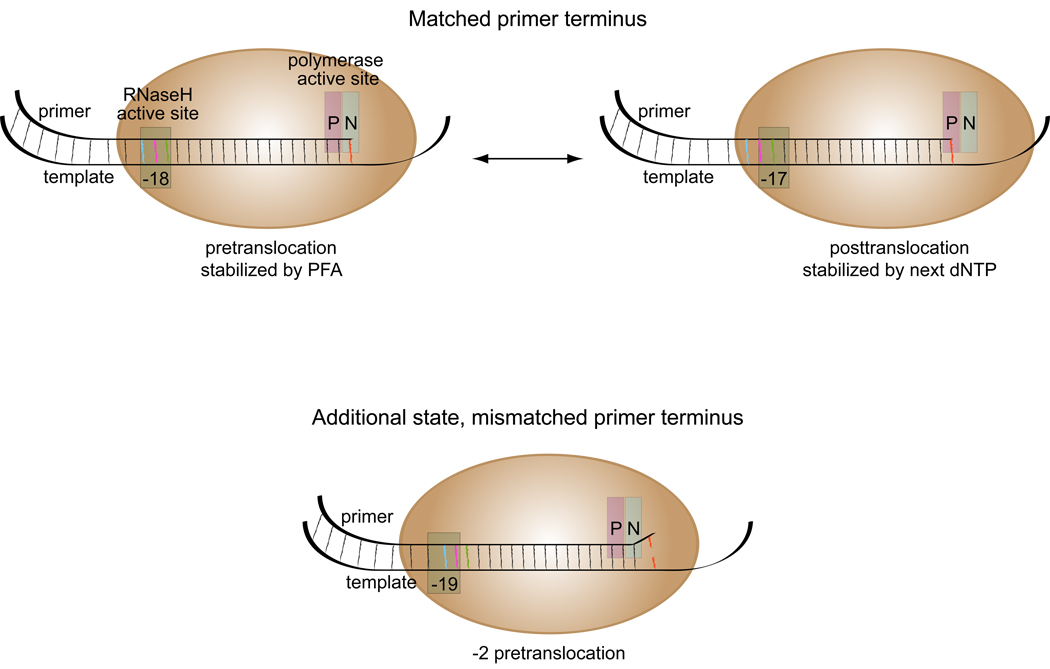
The overall pattern of cleavage is similar for the wild-type and Lys65 substituted RTs, suggesting that the observed change in positioning of the RT plays a role in discrimination between matched and mismatched termini for all three of the RTs analyzed. The additional increase in mismatch extension fidelity displayed by RT with Lys65 substitutions is not due to any further change in position relative to the template-primer. This would suggest that the change in position correlates with the reduced kpol that all three enzymes display on a mismatched primer. As shown in the model depicted in Fig. 6, the mismatched primer termini may reduce occupancy by the RT at the primer terminus. With the RT in the −2 pre-translocation position, the incoming dNTP cannot be bound, which would result in a non-productive complex, and a reduction in the measured kpol.
Figure 6.
Diagrammatic representation of reverse transcriptase on matched and mismatched primer termini. Positions of cleavage relative to the 3’-termini of the primer are marked at the RNase H active site. The P (polymerization) and N (dNTP binding) – sites of the polymerase active site are indicated. With a matched primer terminus, the RT can be present in either a pre- or posttranslocated complex; with a mismatched primer terminus, an additional −2 pre-translocation complex was observed.
Materials and Methods
RT expression and purification
Plasmids encoding RT with substitutions at position 65 were a kind gift of Professor Parniak. The constructs contained the HIV-1 RT and protease sequences; following induction of expression, RT was cleaved to the p66 and p51 subunits by the coexpressed protease, so the mutation was contained in both subunits. Expression and purification were essentially as described14. Briefly, the RT was expressed in DH5α F’LacIq (codonplus), and the RT heterodimers were purified via an N-terminal hexahistidine tag. Following initial purification with a Ni2+-Sepharose column, the enzyme was desalted and purified through two ion exchange chromatography steps; the flow-through from a DEAE column was collected and applied to a Mono-S column, and eluted with a NaCl gradient. All purification steps were carried out at room temperature using an Äkta Explorer (Amersham Biosciences). All preparations were found to be free of nucleases.
Oligonucleotide preparation and purification
Oligonucleotides (sequences shown in Fig. 2) were synthesized by Integrated DNA Technologies (Coralville, Iowa, USA), and purified on a 10% denaturing acrylamide gel. For blocked primers, oligonucleotide (15 µM) was incubated at 37°C for 30 minutes in a reaction containing 1 mM dideoxynucleotide, 0.2 mg/ml BSA, 1×TdT buffer (as supplied by the manufacturer) and 20 units Terminal deoxynucleotidyl transferase (TdT; USB Corp, Ohio, USA) in 100µl. Enzyme was heat inactivated, and the oligonucleotides were phenol/chloroform extracted, ethanol precipitated and purified by size exclusion on a p30 Micro Bio-Spin column (Biorad). For the pre-steady state kinetic analysis, DNA oligonucleotides were synthesized on an Applied Biosystems 380A DNA synthesizer (DNA synthesis facility, Yale University) and purified using denaturing polyacrylamide gel electrophoresis (20% acrylamide, 8 M urea). Primer strands of the DNA/DNA duplex was 5'-radiolabeled with T4 polynucleotide kinase (New England Biolabs) as previously described1. The DNA/DNA duplex was formed by annealing a 1:1.4 molar ratio of primer to template at 90°C for 5 min, 50°C for 10 min, and 0°C for 10 min. The duplex mixtures were then analyzed by nondenaturing polyacrylamide gel electrophoresis (15%) to ensure complete annealing. The concentrations of oligonucleotides were determined by UV absorbance at 260 nm using calculated extinction coefficients.
Qualitative single dNTP insertion assays
5’ 32P end-labeled primer was annealed to the template oligonucleotide. The enzyme and primer-template were incubated at 37°C for 10 minutes in a reaction mix containing 10 nM RT, 100 nM primer-template 50 mM Tris-HCl, pH 8.0, 50 mM KCl, 6 mM MgCl2, 1 mM DTT, 0.1 mg/ml ultrapure BSA (Ambion) and one dNTP, at a concentration specified in the figure legend. The reaction products were separated on a denaturing 10% polyacrylamide sequencing gel, and exposed to a phosphoimager screen.
Rapid Quench Experiments
Rapid chemical quench experiments with a KinTek Instruments Model RQF-3 rapid-quench-flow apparatus were performed as previously described 5,39. Single-turnover pre-steady state kinetic experiments were used to examine incorporation of the next dNTP into a DNA/DNA duplex at 37°C. These are conducted under conditions in which enzyme is in excess over radiolabeled oligonucleotide to allow one to examine chemical catalysis during a single enzyme turnover40. Values for Kd and kpol were determined for incorporation of correct and incorrect dNTPs on a matched DNA/DNA primer/template and for incorporation of correct dNTPs on a DNA/DNA primer/template with a mismatched primer 3'-end. The reactions were carried out by rapidly mixing a solution containing the preincubated complex of 250 nM HIV-1 RT (active site concentration) and 5'-end labeled 50 nM DNA/DNA duplex with a solution of 10 mM Mg2+ and varying concentrations of dNTP in the presence of 50 mM Tris-HCl and 50 mM NaCl at pH 7.8. For dNTP misinsertion experiments, the correct dNTP (dATP) ranged from 100 nM to 50 µM and incorrect dNTPs (dCTP, dGTP, and dTTP) ranged from 100 µM to 10 mM. For mismatched extension experiments, the correct dNTP (dATP) ranged from 5 µM to 1.5 mM. Polymerization was quenched with 0.3 M EDTA at specific time intervals. For dNTP misinsertion experiments, the time points ranged from 0.01 to 2 sec for correct dNTP and from 0.5 sec to 30 min for incorrect dNTPs. For mismatched extension experiments, time points ranged from 0.01 sec to 120 min. All concentrations refer to the final concentration after mixing. In the cases where long reaction times were required, manual quench experiments were performed by mixing the two solutions (RT preincubated with primer/template and dNTP with MgCl2) were mixed and incubated at 37°C, and at regular time intervals, 5 µl of the reaction solution was removed and quenched with EDTA. Products were analyzed by sequencing gel analysis (20% polyacrylamide gel, 8M urea, 1× TBE running buffer) and quantitated using a Bio-Rad Molecular Imager FX (Bio-Rad Laboratories, Inc., Hercules, CA).
Data Analysis
Data was fit by nonlinear regression using the program KaleidaGraph (Synergy Software, Reading, PA). Single-turnover incorporation experiments were fit to a single-exponential equation: [product] = A[1 − exp(−kobsdt)], where A is the amplitude of product formation and kobsd is the observed rate at a specific substrate concentration. The dissociation constant (Kd) of the substrate (dNTP) binding to the complex of RT and primer/template during dNMP incorporation was calculated by fitting observed rate constants at different concentrations of dNTP to the hyperbolic equation kobsd = kpol[dNTP]/(Kd + [dNTP]), where kpol is the maximum first-order rate constant for dNMP incorporation, [dNTP] is the corresponding concentration of dNTP, and Kd is the equilibrium dissociation constant for the productive interaction of dNTP with the RT and primer/template complex.
Fe2+-dependent RNase H site footprinting
Reactions, containing 40 nM enzyme, 50 mM Tris-HCl, pH 8.0, 50 mM KCl, 5 mM DTT, 1 mM MgCl2, 0.1 mg/ml ultrapure BSA and 2.5 nM primer-template (5’-32P labeled template strand annealed to a 2-fold molar excess of unlabeled primer strand) were incubated for 5 minutes at 37°C. Phosphonoformic acid (PFA, Foscarnet) (1 mM) or dATP (1 mM) were added as indicated, and incubated for a further 5 minutes, before cleavage was initiated by addition of 33 µM Fe(NH4)2(SO4)2. The cleavage reaction was allowed to proceed for 2 minutes, and was terminated by the addition of 5 volumes of stop mix (95% formamide, 15 mM EDTA, 0.2 mg/ml bromophenol blue). Cleavage reactions were separated on a denaturing 15% polyacrylamide sequencing gel. Product size was determined by comparison to 5’ end labeled template cleaved to generate thymidine specific and single-nucleotide ladders. The thymidine specific ladder was generated by mixing 25 fmol of 5’ 32P end-labeled template oligonucleotide with an equal volume (20 µl) of 1 mM KMnO4 in 3M tetraethylammonium chloride 1, and incubating for 10 minutes at room temperature. Following ethanol precipitation, the modified nucleotides were cleaved by treatment with 10% piperidine at 90°C for 30 minutes, ethanol precipitated, and resuspended in formamide stop mix (as above). The single nucleotide ladder was generated using the Fenton reaction; 50 fmol of endlabeled oligonucleotide was incubated at room temperature for 2.5 minutes in a reaction containing 1 mM ascorbate, 0.03% H2O2, and 10 µM Fe(II)-EDTA, as described1.
Acknowledgements
The research described in this report was supported by NIH grant R37AI030861 to V.R.P and NIH Grant GM49551 to K.S.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 2.Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 3.Roberts JD, Bebenek K, Kunkel TA. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 4.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 5.Kati WM, Johnson KA, Jerva LF, Anderson KS. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 6.Rittinger K, Divita G, Goody RS. Human immunodeficiency virus reverse transcriptase substrate-induced conformational changes and the mechanism of inhibition by nonnucleoside inhibitors. Proc Natl Acad Sci U S A. 1995;92:8046–8049. doi: 10.1073/pnas.92.17.8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reardon JE. Human immunodeficiency virus reverse transcriptase: steady-state and pre-steady-state kinetics of nucleotide incorporation. Biochemistry. 1992;31:4473–4479. doi: 10.1021/bi00133a013. [DOI] [PubMed] [Google Scholar]
- 8.Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 9.Harris D, Kaushik N, Pandey PK, Yadav PN, Pandey VN. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J Biol Chem. 1998;273:33624–33634. doi: 10.1074/jbc.273.50.33624. [DOI] [PubMed] [Google Scholar]
- 10.Moran S, Ren RX, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc Natl Acad Sci U S A. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Post CB, Ray WJ. Reexamination of induced fit as a determinant of substrate specificity in enzymatic reactions. Biochemistry. 1995;34:15881–15885. doi: 10.1021/bi00049a001. [DOI] [PubMed] [Google Scholar]
- 12.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 13.Menéndez-Arias L. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog Nucleic Acid Res Mol Biol. 2002;71:91–147. doi: 10.1016/s0079-6603(02)71042-8. [DOI] [PubMed] [Google Scholar]
- 14.Garforth SJ, Kim TW, Parniak MA, Kool ET, Prasad VR. Site-directed mutagenesis in the fingers subdomain of HIV-1 reverse transcriptase reveals a specific role for the beta3-beta4 hairpin loop in dNTP selection. J Mol Biol. 2007;365:38–49. doi: 10.1016/j.jmb.2006.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sluis-Cremer N, Arion D, Kaushik N, Lim H, Parniak MA. Mutational analysis of Lys65 of HIV-1 reverse transcriptase. Biochem J. 2000;348(Pt 1):77–82. [PMC free article] [PubMed] [Google Scholar]
- 16.Perrino FW, Preston BD, Sandell LL, Loeb LA. Extension of mismatched 3' termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 1989;86:8343–8347. doi: 10.1073/pnas.86.21.8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakhanashvili M, Hizi A. Fidelity of the RNA-dependent DNA synthesis exhibited by the reverse transcriptases of human immunodeficiency virus types 1 and 2 and of murine leukemia virus: mispair extension frequencies. Biochemistry. 1992;31:9393–9398. doi: 10.1021/bi00154a010. [DOI] [PubMed] [Google Scholar]
- 18.Zinnen S, Hsieh JC, Modrich P. Misincorporation and mispaired primer extension by human immunodeficiency virus reverse transcriptase. J Biol Chem. 1994;269:24195–24202. [PubMed] [Google Scholar]
- 19.Götte M, Maier G, Gross HJ, Heumann H. Localization of the active site of HIV-1 reverse transcriptase-associated RNase H domain on a DNA template using site-specific generated hydroxyl radicals. J Biol Chem. 1998;273:10139–10146. doi: 10.1074/jbc.273.17.10139. [DOI] [PubMed] [Google Scholar]
- 20.Marchand B, Tchesnokov EP, Götte M. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J Biol Chem. 2007;282:3337–3346. doi: 10.1074/jbc.M607710200. [DOI] [PubMed] [Google Scholar]
- 21.Shah FS, Curr KA, Hamburgh ME, Parniak M, Mitsuya H, Arnez JG, Prasad VR. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate and error specificity of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 2000;275:27037–27044. doi: 10.1074/jbc.M002881200. [DOI] [PubMed] [Google Scholar]
- 22.Das K, Bandwar RP, White KL, Feng JY, Sarafianos SG, Tuske S, Tu X, Clark AD, Boyer PL, Hou X, Gaffney BL, Jones RA, Miller MD, Hughes SH, Arnold E. Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. J Biol Chem. 2009;284:35092–35100. doi: 10.1074/jbc.M109.022525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Z, Gao Q, Fang H, Salomon H, Parniak MA, Goldberg E, Cameron J, Wainberg MA. Identification of a mutation at codon 65 in the IKKK motif of reverse transcriptase that encodes human immunodeficiency virus resistance to 2',3'-dideoxycytidine and 2',3'-dideoxy-3'-thiacytidine. Antimicrob Agents Chemother. 1994;38:275–281. doi: 10.1128/aac.38.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sluis-Cremer N, Sheen CW, Zelina S, Argoti Torres PS, Parikh UM, Mellors JW. Molecular Mechanism by which K70E in HIV-1 Reverse Transcriptase Confers Resistance to Nucleoside Reverse Transcriptase Inhibitors. Antimicrob Agents Chemother. 2007;51:48–53. doi: 10.1128/AAC.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deval J, Navarro JM, Selmi B, Courcambeck J, Boretto J, Halfon P, Garrido-Urbani S, Sire J, Canard B. A loss of viral replicative capacity correlates with altered DNA polymerization kinetics by the human immunodeficiency virus reverse transcriptase bearing the K65R and L74V dideoxynucleoside resistance substitutions. J Biol Chem. 2004;279:25489–25496. doi: 10.1074/jbc.M313534200. [DOI] [PubMed] [Google Scholar]
- 26.Garforth SJ, Parniak MA, Prasad VR. Utilization of a deoxynucleoside diphosphate substrate by HIV reverse transcriptase. PLoS ONE. 2008;3:e2074. doi: 10.1371/journal.pone.0002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh JC, Zinnen S, Modrich P. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J Biol Chem. 1993;268:24607–24613. [PubMed] [Google Scholar]
- 28.Deval J, White KL, Miller MD, Parkin NT, Courcambeck J, Halfon P, Selmi B, Boretto J, Canard B. Mechanistic basis for reduced viral and enzymatic fitness of HIV-1 reverse transcriptase containing both K65R and M184V mutations. J Biol Chem. 2004;279:509–516. doi: 10.1074/jbc.M308806200. [DOI] [PubMed] [Google Scholar]
- 29.Weber J, Chakraborty B, Weberova J, Miller MD, Quiñones-Mateu ME. Diminished replicative fitness of primary human immunodeficiency virus type 1 isolates harboring the K65R mutation. J Clin Microbiol. 2005;43:1395–1400. doi: 10.1128/JCM.43.3.1395-1400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White KL, Margot NA, Wrin T, Petropoulos CJ, Miller MD, Naeger LK. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184V and their effects on enzyme function and viral replication capacity. Antimicrob Agents Chemother. 2002;46:3437–3446. doi: 10.1128/AAC.46.11.3437-3446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fisher TS, Prasad VR. Substitutions of Phe61 located in the vicinity of template 5'-overhang influence polymerase fidelity and nucleoside analog sensitivity of HIV-1 reverse transcriptase. J Biol Chem. 2002;277:22345–22352. doi: 10.1074/jbc.M200282200. [DOI] [PubMed] [Google Scholar]
- 32.Agopian A, Depollier J, Lionne C, Divita G. p66 Trp24 and Phe61 Are Essential for Accurate Association of HIV-1 Reverse Transcriptase with Primer/Template. J Mol Biol. 2007;373:127–140. doi: 10.1016/j.jmb.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 33.Weiss KK, Chen R, Skasko M, Reynolds HM, Lee K, Bambara RA, Mansky LM, Kim B. A role for dNTP binding of human immunodeficiency virus type 1 reverse transcriptase in viral mutagenesis. Biochemistry. 2004;43:4490–4500. doi: 10.1021/bi035258r. [DOI] [PubMed] [Google Scholar]
- 34.Lewis DA, Bebenek K, Beard WA, Wilson SH, Kunkel TA. Uniquely altered DNA replication fidelity conferred by an amino acid change in the nucleotide binding pocket of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 1999;274:32924–32930. doi: 10.1074/jbc.274.46.32924. [DOI] [PubMed] [Google Scholar]
- 35.Gutiérrez-Rivas M, Menéndez-Arias L. A mutation in the primer grip region of HIV-1 reverse transcriptase that confers reduced fidelity of DNA synthesis. Nucleic Acids Res. 2001;29:4963–4972. doi: 10.1093/nar/29.24.4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matamoros T, Kim B, Menéndez-Arias L. Mechanistic insights into the role of Val75 of HIV-1 reverse transcriptase in misinsertion and mispair extension fidelity of DNA synthesis. J Mol Biol. 2008;375:1234–1248. doi: 10.1016/j.jmb.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 37.Marchand B, Götte M. Impact of the translocational equilibrium of HIV-1 reverse transcriptase on the efficiency of mismatch extensions and the excision of mispaired nucleotides. Int J Biochem Cell Biol. 2004;36:1823–1835. doi: 10.1016/j.biocel.2004.02.029. [DOI] [PubMed] [Google Scholar]
- 38.Dash C, Scarth BJ, Badorrek C, Götte M, Le Grice SF. Examining the ribonuclease H primer grip of HIV-1 reverse transcriptase by charge neutralization of RNA/DNA hybrids. Nucleic Acids Res. 2008;36:6363–6371. doi: 10.1093/nar/gkn678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerr SG, Anderson KS. Pre-steady-state kinetic characterization of wild type and 3'-azido-3'-deoxythymidine (AZT) resistant human immunodeficiency virus type 1 reverse transcriptase: implication of RNA directed DNA polymerization in the mechanism of AZT resistance. Biochemistry. 1997;36:14064–14070. doi: 10.1021/bi9713862. [DOI] [PubMed] [Google Scholar]
- 40.Johnson KA. The Enzymes. Academic Press; 1992. Transient-state kinetic analysis of enzyme reaction pathways; pp. 1–61. [Google Scholar]
- 41.Roberts E, Deeble VJ, Woods CG, Taylor GR. Potassium permanganate and tetraethylammonium chloride are a safe and effective substitute for osmium tetroxide in solid-phase fluorescent chemical cleavage of mismatch. Nucleic Acids Res. 1997;25:3377–3378. doi: 10.1093/nar/25.16.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dixon WJ, Hayes JJ, Levin JR, Weidner MF, Dombroski BA, Tullius TD, Sauer RT. Methods in Enzymology Protein 3- DNA Interactions. Academic Press; 1991. Hydroxyl radical footprinting; pp. 380–413. [DOI] [PubMed] [Google Scholar]