Abstract
Excessive alcohol use, a major cause of morbidity and mortality, is less well understood than other addictive disorders. Dopamine release in ventral striatum is a common element of drug reward, but alcohol has an unusually complex pharmacology, and humans vary greatly in their alcohol responses. This variation is related to genetic susceptibility for alcoholism, which contributes more than half of alcoholism risk. Here, we report that a functional OPRM1 A118G polymorphism is a major determinant of striatal dopamine responses to alcohol. Social drinkers recruited based on OPRM1 genotype were challenged in separate sessions with alcohol and placebo under pharmacokinetically controlled conditions, and examined for striatal dopamine release using positron emission tomography and [11C]-raclopride displacement. A striatal dopamine response to alcohol was restricted to carriers of the minor 118G allele. To directly establish the causal role of OPRM1 A118G variation, we generated two humanized mouse lines, carrying the respective human sequence variant. Brain microdialysis showed a four-fold greater peak dopamine response to an alcohol challenge in h/mOPRM1-118GG than in h/mOPRM1-118AA mice. OPRM1 A118G variation is a genetic determinant of dopamine responses to alcohol, a mechanism by which it likely modulates alcohol reward.
Keywords: alcohol, dopamine, opioids, reward, polymorphism, positron emission tomography
INTRODUCTION
Alcohol use is a major cause of disability worldwide 1, but available treatments have limited efficacy 2. Development of novel, mechanism-based pharmacotherapies will require an improved understanding of the neurobiology that underlies addictive properties of alcohol 3. Compared to other addictive drugs, alcohol has a complex pharmacology. Sedative, ataxic and anxiolytic alcohol effects are primarily mediated through GABA- and glutamate signaling. In contrast, rewarding properties of alcohol such as euphoria and psychomotor stimulation are thought to involve endogenous opioids and mesolimbic dopamine (DA). In response to alcohol, µ-opioid receptor (OPRM1) activation in the ventral tegmental area (VTA) suppresses the activity of inhibitory GABA-ergic interneurons, resulting in disinhibition of DA-neurons and DA release from their terminals in the ventral striatum 4. Accordingly, OPRM1 blockade is a treatment for alcohol dependence 3.
Humans vary substantially in their alcohol responses, and this variability is related to genetic susceptibility for alcohol use disorders, which accounts for more than half the disease risk in this condition 5,6. Striatal DA-release is a common element of drug reward 7, and alcohol-induced DA release has been shown both in rodents 8,9 and humans 10. There is, however, marked individual variation in alcohol-induced behavioral responses thought to be related to DA activation, such as psychomotor stimulation. Functional variation in opioid genes may contribute to this variation by modulating alcohol-induced DA release. A functional OPRM1 118G variant 11 confers enhanced subjective alcohol responses 12, and a functionally equivalent rhesus macaque 77G variant 13 confers enhanced alcohol-induced psychomotor stimulation 14. Despite this, it remains controversial whether the OPRM1 A118G polymorphism is a risk factor for alcohol use disorders 15.
Here, we asked whether OPRM1 A118G modulates striatal DA release in response to alcohol. We first carried out a human positron emission tomography (PET) study using displacement of the DA-D2 ligand [11C]-raclopride, an indirect measure of endogenous DA release 16, and found that 118G carriers had a markedly more vigorous striatal DA response to alcohol compared to subjects homozygous for the major 118A allele. In parallel, we isolated the influence of OPRM1 A118G variation by generating humanized mice, carrying the human exon 1 of the OPRM1 gene, either as the major 118A allele, or with the 118G SNP introduced through site directed mutagenesis. Direct microdialysis measures of the response to a rewarding dose of alcohol showed a four-fold higher peak DA response to the alcohol challenge.
MATERIALS AND METHODS
Additional details for all methods are provided in Supplementary Information.
PET-study
Subjects were healthy male social drinkers, consuming less than 20 standard drinks per week. They were non-smokers who had never smoked, or had quit at least a year prior to enrolling in the study. Because the ventral striatum shrinks an average of 3.6 to 3.7% per decade 17, the age range was restricted to 21–45 years in order to reduce age-related variability. Furthermore, because rhesus macaque OPRM1 genotype predominantly modulates psychomotor responses to alcohol in males 14, and because recent human data indicate only borderline detectable striatal DA release in response to alcohol in females 18, the study population was restricted to males. Subject demographics are provided in table S1. Allele frequencies of OPRM1 118G in Caucasian populations are around 15% 19, and 118G homozygotes are rare. Subjects were recruited into two approximately equally sized groups: 1) individuals homozygous for the major 118A allele (118AA genotype; n=16) of the OPRM1 polymorphism; 2) individuals carrying one or two copies of the variant 118G allele (118AG or 118GG genotype; because no subject with the rare 118GG genotype was in fact found, this group is hereafter called 118AG; n=12) of the OPRM1 polymorphism. Genotyping was on DNA from whole blood. SNP rs1799971 (A/G) was genotyped using the assay-on-demand (assay ID: C_8950074_1) from Applied Biosystems (Foster City, CA, USA). The alleles were discriminated by post-PCR plate read on ABI PRISM® 7900HT Sequence Detector System (Applied Biosystems).
Because blood alcohol concentrations (BACs) after oral alcohol administration are highly variable 20, we used a physiologically based pharmacokinetic model, and intravenous (i.v.) alcohol infusion to achieve highly standardized brain alcohol exposure 21 successfully used in prior studies 20,22–27. Subjects underwent three infusion sessions, on separate days, separated by approximately 1 week.
A baseline session was carried out outside the PET scanner to ensure that subjects tolerated the alcohol infusion without experiencing nausea or marked sedation, and allow them to habituate to the procedure. All subjects received i.v. alcohol that raised BACs from 0 to 80 mg/dl within 15 min. This is a peak BAC commonly achieved by social drinkers, although the infusion procedure produced this level at a faster rate than is typically achieved with oral alcohol use. The exposure profile was chosen based on prior findings showing that it robustly activates the ventral striatum in a manner that can be detected by functional brain imaging 27. Subjective response to alcohol was measured using a modified Drug Effects Questionnaire [DEQ; 28] obtained before the beginning of the infusion and every 7.5 min minutes during the infusion. Breath alcohol concentrations (BrAC) were monitored every 7.5 min during the infusion, and every 30 min thereafter. Blood samples for BACs were collected before starting the infusion, and then at 15, 30 and 45 min.
In the second and third sessions, subjects received infusions of alcohol or placebo in counter-balanced order while undergoing a PET scan with [11C]-raclopride (GE Advance Tomograph, Waukesha, WI). An 8-min transmission scan was first acquired with 2 rotating rod sources for the purpose of attenuation correction. Next, a 10 mCi dose of [15O]-water was administered over 1 min through an IV line to assess regional perfusion and co-register the PET image with a brain MRI obtained from the subject during a separate visit. Following this, the infusion (6% v/v ethanol or normal saline) was started. At 15 min after the start of the infusion, a 12 mCi dose of [11C]-raclopride was administered over 1 min. Dynamic PET scans began with the [11C]-raclopride injection and continued for 90 min. A schematic of the PET sessions is shown in Figure 1D.
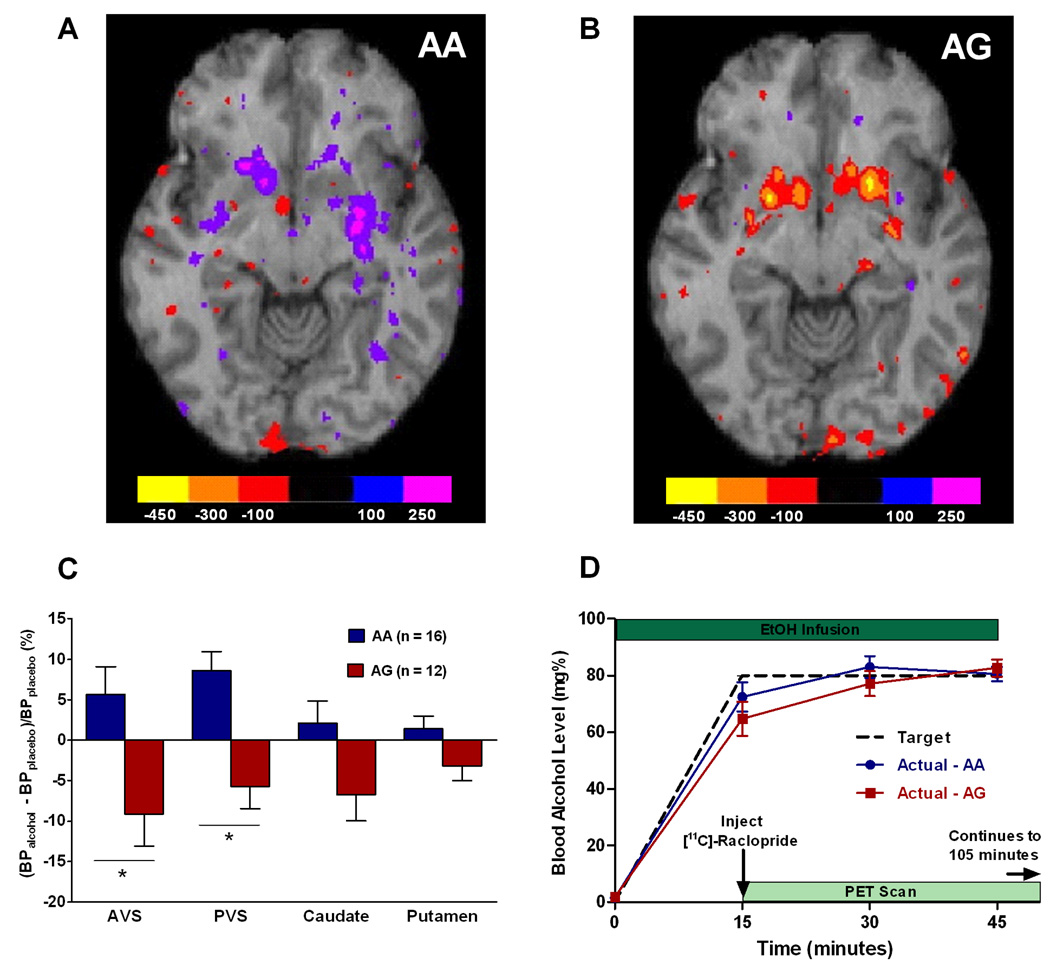
Fig. 1.
Human PET study. Axial view of group maps showing change of [11C]-raclopride binding potential (ΔBP; nCi/cc) between placebo and alcohol sessions in (A) AA individuals and (B) AG individuals. Color bars indicate corresponding ΔBP values. Reduction in raclopride binding is attributed to competition with dopamine released by the alcohol challenge; thus, a negative ΔBP indicates an increase in endogenous dopamine release. (C) Relative change in binding potential (%ΔBP) for [11C]-raclopride between alcohol and placebo sessions in four striatal regions of interest. Data are least square means (±SEM). Main genotype effect: p=0.006; *p<0.05 on post hoc tests within individual regions. AVS = anterior ventral striatum, PVS = posterior ventral striatum. (D) Schematic of PET sessions, and blood alcohol concentration profiles over time during the alcohol session (mean±SEM). There was no significant difference between genotypes (F[1,24]=0.51, p=0.48).
PET Analysis was based on a Region of Interest (ROI) methodology. A structural MR scan was co-registered with the subject’s PET scans. ROIs were drawn in the primary areas of post-synaptic dopamine release, namely the anterior ventral striatum, posterior ventral striatum, caudate and putamen (figure S1). The anterior ventral striatum was defined as striatum anterior and inferior to the anterior commisure. The posterior ventral striatum was defined as striatum posterior and inferior to the anterior commisure. The PET data from the placebo session were used as a measure of baseline raclopride binding potential, using the simplified reference tissue model 29. Percent change in binding potential (%ΔBP) following alcohol was calculated as (BPplacebo - BPalcohol)/( BPplacebo). Reduction in raclopride binding is attributed to competition with dopamine endogenously released by the alcohol challenge, and the percent change in binding potential has been shown to be proportional to the magnitude of dopamine release 16.
PET data were examined for homogeneity of variance and analyzed using general linear models (GLM, Statistica 6.0, StatSoft, Tulsa, OK). For the %ΔBP, a repeated measures analysis of covariance (ANCOVA) was performed, with genotype and testing sequence as between subjects variables, region of interest as the within subjects variable, and drinking frequency (measured by the Lifetime Drinking History questionnaire 30) as the covariate. Subjective responses to alcohol were analyzed using repeated measures ANOVA with genotype and testing sequence as between subjects variables and time point as the within subjects variable. Post hoc comparisons were conducted using Newman-Kuel tests. Effect sizes were calculated using partial eta2 from Statistica, and Cohen’s f calculated using GPower 3.1.0 (Franz Faul, Universität Kiel, Germany).
Humanized mouse lines
At the OPRM1 locus, linkage disequilibrium (LD) exists between OPRM1 A118G and other markers 31. We therefore generated two humanized mouse lines, where the OPRM1 exon 1 was replaced by the corresponding human sequence. One of the lines, h/mOPRM-118AA, is homozygous for the major human 118A allele. For the other (h/mOPRM1–118GG), site directed mutagenesis was used to introduce a G in position 118. Thus, the lines are genetically identical, with the exception of the A → G substitution (Figure 2A–C). The lines are on a C57BL/6 background, where robust voluntary alcohol consumption is observed 32.
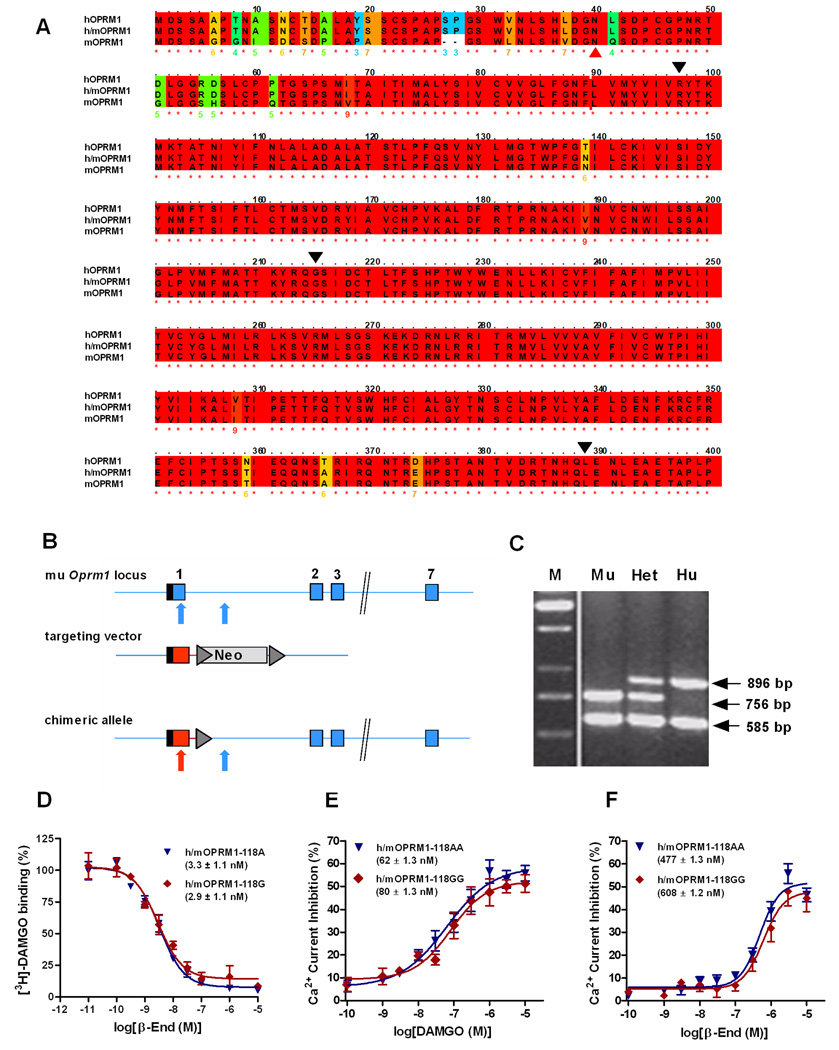
Fig. 2.
Targeting strategy and functional confirmation for humanized OPRM1 mouse lines. (A) Alignment of human (hOPRM1), mouse (mOPRM1) and humanized (h/mOPRM1) protein sequences. Black triangles indicate exon boundaries. Red triangle points to site of the N40D substitution, corresponding to A118G allelic variation. Color-coding of background indicates degree of conservation, with red indicating full human – mouse homology, and blue non-conservative substitution. As can be seen, replacement of mouse exon 1 with the corresponding human sequence yields a receptor that is highly homologous to the human protein. (B) Exon 1 of the murine OPRM1 locus including additional 400 bp of intron 1 was replaced by the corresponding human sequence, containing either A or G in position 118. Blue and red colors represent mouse and human sequences, respectively. Boxes: respective exons of the OPRM1 coding region. Hatched region: 5’-UTR. Triangle: frt site for flox recombinase. Neo: neomycin selection marker. Arrows point to position of the human and mouse specific PCR primers. (C) PCR based identification of the human (Hu), mouse (Mu) and heterozygote (Het) genotypes. M: Marker lane. (D) [3H]-DAMGO binding to respective humanized receptor transiently expressed in CHO cells. Data are means (±SEM) of triplicates, and mean (±SEM) IC50 values for the respective genotypes are indicated. Non-linear curve-fitting established that no significant genotypes differences were present (E – F) Concentration-response curves for DAMGO and β-endorphin (β-End) in trigeminal ganglion (TG) neurons isolated from h/mOPRM1 118AA or 118GG mice. Data points represent the mean (±SEM) of the agonist-mediated Ca2+ current inhibition. Mean EC50 values (±SEM) for the respective genotypes are indicated. Similar to the binding experiment, no genotype differences were found.
Generation of the h/mOPRM1–118AA and h/mOPRM1–118GG lines is detailed in Supplementary Information. Insertion of sequence corresponding to human OPRM1 exon 1 is shown in Fig. 2C. Determination of the respective h/mOPRM1–118 genotype was carried out using the PCR method described for the human PET study. First generation h/mOPRM1–118AA and GG mice obtained from a cross of the founder lines were assessed with regard to overall health and behavior using an established testing battery 33. Table S2 shows that there were no genotype differences, with the exception of some excessive running in GG subjects upon introduction into the novel environment, and a modest but statistically significant increase in locomotion in h/m118AA males compared to h/m118GG males. Male mice aged 13–16 weeks were used for the binding, signaling experiments, and male mice aged 24 – 32 weeks were used for the microdialysis experiments.
Receptor binding studies
Membrane binding was assessed in preparations from CHO-K1 cells transfected with OPRM1 receptor cDNA and incubated with 2nM [3H]-D-Ala2, N-MePhe4, Gly-ol]-enkephalin (DAMGO; 37Ci/mmol; NEN, Boston, MA) and nonradioactive naloxone, morphine, and β-endorphin (β-End) ranging from 0.01nM to 1000 nM. The mixture was incubated at room temperature, filtered through Whatman GF/B filters (FP-100, Brandel), washed with ice-cold Tris-HCl, and assayed for radioactivity by liquid scintillation. Data were analyzed by nonlinear analysis using the Prism computer program (GraphPad Software, Inc.).
Autoradiographic binding was studied on 20 µm coronal cryosections as described 34, using 5 nM [3H]DAMGO (48.9 Ci/mmol, NEN). Non-specific binding was determined from adjacent sections in the presence of the radioligand plus 10 µM naloxone. Radiolabeled, dried tissue sections and [3H]microscales (Amersham) were apposed to tritium-sensitive screens (Fujifilm imaging plates), and images were obtained using a Fujifilm BAS 5000 PhosphorImager.
Agonist-induced GTP[γ -35S] activity was evaluated as described 35 on 20 µm coronal sections, by incubating GTP[γ-35S] (0.04 nM) with 3 µM DAMGO in assay buffer at 25°C for 2 hr. Agonist studies were also performed in the presence of the antagonist naloxone to verify the receptor specificity of G-protein activation. Basal activity was assessed in each experiment with GDP in the absence of agonist, and nonspecific binding was evaluated in the presence of 10 µM unlabeled GTP[γS] without GDP. Slides were rinsed, dried and exposed to tritium-sensitive screens (Fujifilm imaging plates).
Images were analysed using MCID Image Analysis Software (Imaging Research Inc., UK). Regions of interest were defined by anatomical landmarks 36. Agonist-stimulated activity in brain sections was calculated by subtracting the optical density in basal sections (incubated with GDP alone) from that of agonist-stimulated sections; results were expressed as percentage of basal activity.
Electrophysiology
Enzymatically dissociated trigeminal ganglion neurons were used. Ca2+ currents were recorded using whole-cell patch-clamping as described 37. Currents were acquired with the Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA), filtered, and cell membrane capacitance and pipette series resistance were electronically compensated. The concentration-response relationships were determined by sequential application of the respective agonist in increasing concentrations. Two to three different concentrations were used with each cell in order to minimize desensitization. The results were pooled and the concentration-response curves were fit to the Hill equation. Data and statistical analyses were performed with Igor Pro (Lake Oswego, OR) and Prism 4.0 (GraphPad Software, Inc.) software packages, respectively.
Microdialysis
Under isofluorane anaesthesia, mice were implanted with microdialysis probes as previously described 38, aimed at the nucleus accumbens (NAc), and secured to the skull using a skull screw and cement. Histologically verified placement within the NAc for each subject is shown in Figure 3A. During a 12h postsurgical recovery period, probes were slowly perfused with artificial cerebrospinal fluid. The perfusion flow rate was then increased to 0.6µL/min for 2h to reach an interstitial equilibrium before sample collection. Dialysate fractions were subsequently collected, during the dark cycle, at 10min intervals during a 60min baseline period and, after an injection of EtOH (2g/kg, ip), during a 120min post-injection period. Samples were analyzed for DA and 5-HT using high-performance liquid chromatography coupled with electrochemical detection (HPLC-ECD).
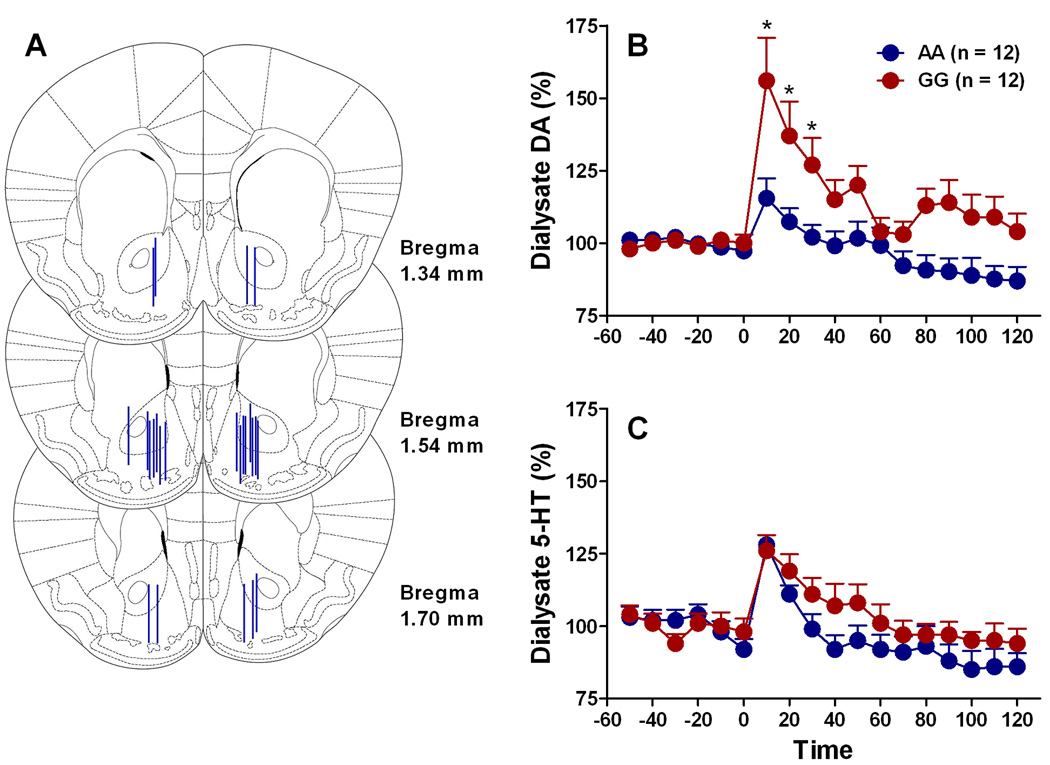
Fig. 3.
Striatal DA response to alcohol (2g/kg) in the humanized mouse lines, determined by brain microdialysis. (A) Individual probe placements in ventral striatum / Nucleus Accumbens. (B) Dialysate DA levels in h/mOPRM1-118AA and h/mOPRM1-118GG mice. After confirmation of no group differences in baseline concentrations, the data were converted to the percent change from the average baseline concentration obtained prior to pretreatment. Alcohol was given at time point 0. Differential response was indicated by a genotype x time interaction (p<0.005); differences at individual data points as determined by post hoc test are indicated by *p<0.05 (C) In contrast to the DA response, the 5-HT response to the alcohol dose did not differ between genotypes.
After confirmation by ANOVA of no group differences in baseline DA or 5-HT concentrations, data were converted to percent change from the average baseline concentration obtained prior to pretreatment. Within- and between-groups analyses were performed by two-way ANOVA with repeated measures over time, with separate analyses performed for the DA and 5-HT data.
RESULTS
Human PET-study
BACs at the end of the alcohol infusion during the PET session were, on average, within 1 mg/dl of the target level, confirming the high level of accuracy and precision of the method. There were no differences between BACs achieved in the AA and AG groups (ANOVA: F[1, 24]=0.287, p = 0.597). Baseline binding potential (BP) for [11C]-raclopride, as measured on the saline infusion session, also did not differ between genotypes (AA: 3535.8±107.8; AG: 3722.7±124.5 nCi/cc, mean±SEM; F[1,26]=1.3, p=0.27), making pre-existing differences in D2 receptor densities unlikely.
Across the four striatal subregions of interest (anterior ventral striatum, posterior ventral striatum, caudate and Putamen, figure S1), the change in BP (ΔBP) attributable to the alcohol challenge [(BPplacebo - BPalcohol)/(BPplacebo )] differed markedly as a function of genotype, with AG subjects consistently showing signs of greater DA-release than AA individuals (main genotype effect: F[1,22]=9.3, p=0.006). Genotype accounted for 29.7% of the variance in response, measured as partial eta2, translating into an effect size of Cohen's f=0.65, i.e. between “moderate” – “large”. There was also a significant genotype x region interaction (F[3,66]=3.8, p=0.01); accordingly, post-hoc tests showed that the main genotype effect was predominantly driven by significant differences in the anterior and posterior ventral striatum, regions most closely associated with drug reward (Figure 1A–C). The highest response to alcohol was observed in the anterior ventral striatum of the AG group, where ΔBP was approximately 9%. This is more than half of the response seen with amphetamine 16, a drug that potently enhances synaptic DA by acting on the DA transporter. This relative potency is in agreement with direct measures obtained by brain microdialysis in experimental animals 9,16. In contrast, AA subjects did not show an alcohol-induced displacement of the radioligand in any region. Inclusion of the BAC at 45 min as a covariate in the statistical analysis of the %ΔBP data did not influence the main effect of genotype (F[1,19]=9.7, p=0.0057). The BAC at 45 min per se showed a trend for a significant effect on the %ΔBP (F[1,19]=3.86, p=0.064).
Similar to other imaging genetic studies 39,40, subjective ratings showed higher variability than the biological measure. The only effect detected was a genotype x time interaction, with AG subjects reporting lower levels of intoxication than AA subjects at the later time points, suggestive of faster acute tolerance development (see Supplementary Materials and Figure S2 E).
Characterization of humanized receptors and mouse lines
When expressed in CHO cells, both humanized receptors showed binding of the classical OPRM1 selective ligand [3H]-DAMGO, displaced by the endogenous OPRM1 agonist β-End with expected low nanomolar IC50, and with no difference between genotypes (F[4,46]=1.3, p=0.3; Fig. 2D). Signaling, assessed as modulation of Ca2+ currents in acutely isolated superior trigeminal ganglion neurons, was also normal in response to both DAMGO and β-End, once again without any genotype differences (DAMGO: F[4,12]= 1.8, p=0.2; β-End: F[4,12]=1.2, p=0.4; Fig 2E–F). Finally, [3H]-DAMGO binding on mouse brain sections did not show genotype differences in receptor densities across a number of brain regions examined, including ventral and dorsal striatum and the ventral tegmental area (VTA; Figure S5).
DA responses to alcohol in humanized mouse lines
Microdialysis probes placement in ventral striatum / Nucleus Accumbens is shown in Figure 3A. Baseline DA levels did not differ between genotypes (F[1,22]= 6.9 × 10−6, n.s.) and were 2.42±0.4 nM for h/mOPRM1–118AA and 2.43±0.6 nM for h/mOPRM1–118GG mice, respectively (n=12/group). Alcohol injection (2g/kg, a dose that robustly induces conditioned place preference in C57BL/6 mice) increased dialysate DA in both lines (main effect: F[6,132]=12.4; p<0.0001), However, response differed markedly between genotypes (main genotype effect: F[1,22]=7.2; p<0.05; genotype x time interaction: F[6,132]= 12.4; p<0.005; Figure 3B). Peak DA increase was nearly 4-fold greater in GG mice than in AA animals. The genotype effect was specific for DA release induced by alcohol; although alcohol also significantly increased 5-HT (main effect: F[6,132]=15.6; p<0.0001), this was not affected by genotype (main genotype effect: F[1,22]=3.6; n.s; genotype x time interaction: F[6,132]=0.92; n.s.; Figure 3C).
DISCUSSION
Taken together, our data strongly support a causal role of the OPRM1 118G allele to confer a more vigorous DA response to alcohol in the ventral striatum. These findings may be related to observations of differential subjective alcohol effects as a function of OPRM1 A118G genotype in humans 12, and of markedly increased psychomotor responses to alcohol in rhesus males carrying the functionally equivalent 77G variant 14. Interestingly, we also found that subjective feelings of intoxication over time differed as a function of genotype. Carriers of the 118G allele showed signs of rapid acute tolerance, similar to that previously described in men at genetic risk for alcoholism 41. The use of a reverse-translational approach allowed us to effectively isolate the influence of the human 118G variant from that of other polymorphisms with which it might be in LD.
Excessive activation of brain reward systems by alcohol in OPRM1 118G carriers might be expected to confer susceptibility for alcohol use disorders. Although we in fact found evidence in support of this notion in an ethnically homogenous Caucasian sample 42, others have not. One study found that other polymorphisms within the same haplotype block, but not A118G, were associated with diagnoses of substance dependence 31, while a meta-analysis of all available data for A118G was negative 43. This apparent discrepancy closely parallels recent findings with the 5-HT transporter gene promoter length polymorphism (5-HTTLPR). A robust role of 5-HTTLPR variation has been found using an imaging based endophenotype that is closely related to negative affect 39,44, and these findings are paralleled by experimental data in a non-human primate model 45,46. Nevertheless, findings from 5-HTTLPR association studies in depression have not held up on meta-analysis 47,48. The reasons for these discrepancies are presently unknown, much debated, and critical to resolve. One important possibility is that current diagnostic categories pool genetically heterogeneous phenocopies, as has specifically been proposed in depression 49. If so, the use of biological endophenotypes, such as in the present study, can potentially aid the dissection of these populations into genetically and pathophysiologically more homogenous categories.
Irrespectively of its possible role as a susceptibility factor, excessive reward-related DA-response to alcohol in carriers of OPRM1 118G might be of pharmacogenetic importance in treatment of excessive alcohol use. Indeed, two studies have found that the presence of the 118G allele was associated with a therapeutic response to the opioid antagonist naltrexone 50,51. Although one study failed to find such an effect 52, recent non-human primate data provide strong evidence for this notion under closely controlled experimental conditions 53. Our data are consistent with a selective role of endogenous opioids for alcohol reward in OPRM1 118G carriers, and provide a possible biological mechanism for the preferential naltrexone response in this population. However, the dissociation between objective measures of alcohol-induced DA-release and subjective reports of alcohol effects may argue against a simplistic view of alcohol-induced DA-release as an immediate mediator of drug reward. Instead, these data may be more compatible with a view of ventral striatal DA-activation as a signal predictive of alcohol reward, and perhaps involved in learning processes important for addiction 54. Of note, the level of alcohol-induce DA-activation in our study was overall lower than previously reported 10. A difference between the two studies is that alcohol administration was oral in the prior report, while we used intravenous administration. This may indicate that striatal DA response to alcohol is only in part driven by direct pharmacological actions of alcohol, while an additional component of the activation is evoked by smell or taste cues.
Our data demonstrate that the 118G allele, which encodes an amino acid substitution in the N-terminal extracellular loop of the receptor, confers a higher striatal DA-activation in response to alcohol regardless of whether it occurs in the context of other markers within its haplotype block or not. However, the molecular mechanism that mediates functional consequences of the 118G variant remains controversial. Initially described as a gain-of-function mutation due to increased affinity for β-End 11, OPRM1 118G has since been proposed to instead confer decreased expression in several systems, including human post-mortem brain tissue 55, and a mouse model where a mutation was introduced in a position corresponding to the human A118G marker 56. The latter model differs from that used in our study, in that it introduced the N → D substitution in the context of a murine N-terminal receptor protein sequence. In contrast, we generated mice expressing humanized receptors throughout this region by replacing all of exon 1, the main region of divergence between the two species (Figure 2A). Using this model, our present findings show that a key functional phenotype associated with the 118G variant, alcohol-induced striatal DA-release, is replicated across species in the absence of effects on receptor affinity, binding density, or signaling. A remaining possibility is altered oligomerization, a mechanism that is critical for opioid receptor desensitization, endocytosis, and re-insertion 57. OPRM1 A118G modifies a glycosylation site in the N-terminal extracellular loop of the receptor, and could thereby influence receptor dimerization and trafficking.
Functionally equivalent OPRM1 variants have evolved independently in two primate lineages, the rhesus macaque 13 and man 11. In both species, minor allele carriers are more sensitive to disruption of social attachment bonds 58,59. In rhesus, this is accompanied by bold - exploratory traits, while lower conscientiousness has been observed in human 118G carriers 13,58,60. These traits may have been advantageous in the evolutionary history of the respective species, and recent studies suggest that the 118G allele has been under positive selection 61. In contrast, the ability of these variants to modulate DA responses to alcohol is likely a coincidental evolutionary byproduct.
Identifying genetic sources of individual variation in complex behavioral phenotypes has proven challenging. Our findings support the notion that use of biological intermediate phenotypes and translational approaches can contribute important data to facilitate this difficult endeavor.
Supplementary Material
ACKNOWLEDGMENTS
Supported by the NIAAA Division of Intramural Clinical and Biological Research (Heilig, Hommer, George, Ramchandani), and grant AA014619 from NIAAA (Parsons) and PAI 433/06 from Consejeria de Innovacion y Ciencia (Pavon). None of the authors has any conflict of interest to declare. Data analysis by M. Kerich and R. Momenan, and biochemical analysis by E. Singley are gratefully acknowledged.
Footnotes
CONFLICT OF INTEREST
None of the authors has any competing interest, financial or otherwise.
Reference List
- 1.Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. The Lancet. 2009;373(9682):2223–2233. doi: 10.1016/S0140-6736(09)60746-7. [DOI] [PubMed] [Google Scholar]
- 2.Bouza C, Angeles M, Munoz A, Amate JM. Efficacy and safety of naltrexone and acamprosate in the treatment of alcohol dependence: a systematic review. Addiction. 2004;99(7):811–828. doi: 10.1111/j.1360-0443.2004.00763.x. [DOI] [PubMed] [Google Scholar]
- 3.Heilig M, Egli M. Pharmacological treatment of alcohol dependence: Target symptoms and target mechanisms. Pharm Therap. 2006;111(3):855–876. doi: 10.1016/j.pharmthera.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Spanagel R. Alcoholism: A Systems Approach From Molecular Physiology to Addictive Behavior. Physiol Rev. 2009;89(2):649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- 5.Schuckit MA. Vulnerability Factors for Alcoholism. In: Davis KL, Charney D, Coyle JT, Nemeroff C, editors. Neuropsychopharmacology: The fifth generation of progress. 41 ed. Philadelphia: Lippincott Williams & Wilkins; 2002. pp. 1399–1412. [Google Scholar]
- 6.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6(7):521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 7.Wise RA. Dopamine, learning and motivation. Nature Reviews Neuroscience. 2004;5(6):483–494. doi: 10.1038/nrn1406. [DOI] [PubMed] [Google Scholar]
- 8.Di Chiara G, Imperato A. Drugs abused by humans preferentially increase synaptic dopamine concentrations int he mesolimbic system of freely moving rats. Proc Natl Acad Sci U S A. 1988;85:5274–5278. doi: 10.1073/pnas.85.14.5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss F, Lorang MT, Bloom FE, Koob GF. Oral alcohol self-administration stimulates dopamine release in the rat nucleus accumbens: genetic and motivational determinants. J Pharmacol Exp Ther. 1993;267(1):250–258. [PubMed] [Google Scholar]
- 10.Boileau I, Assaad JM, Pihl RO, Benkelfat C, Leyton M, Diksic M, et al. Alcohol promotes dopamine release in the human nucleus accumbens. Synapse. 2003;49(4):226–231. doi: 10.1002/syn.10226. [DOI] [PubMed] [Google Scholar]
- 11.Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95(16):9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray LA, Hutchison KE. Effects of naltrexone on alcohol sensitivity and genetic moderators of medication response - A double-blind placebo-controlled study. Arch Gen Psychiatry. 2007;64(9):1069–1077. doi: 10.1001/archpsyc.64.9.1069. [DOI] [PubMed] [Google Scholar]
- 13.Miller GM, Bendor J, Tiefenbacher S, Yang H, Novak MA, Madras BK. A mu-opioid receptor single nucleotide polymorphism in rhesus monkey: association with stress response and aggression. Mol Psychiatry. 2004;9(1):99–108. doi: 10.1038/sj.mp.4001378. [DOI] [PubMed] [Google Scholar]
- 14.Barr CS, Schwandt M, Lindell SG, Chen SA, Goldman D, Suomi SJ, et al. Association of a functional polymorphism in the mu-opioid receptor gene with alcohol response and consumption in male rhesus macaques. Arch Gen Psychiatry. 2007;64(3):369–376. doi: 10.1001/archpsyc.64.3.369. [DOI] [PubMed] [Google Scholar]
- 15.Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: A meta-analysis. Drug Alcohol Depend. 2005 doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Endres CJ, Kolachana BS, Saunders RC, Su T, Weinberger D, Breier A, et al. Kinetic modeling of [11C]raclopride: combined PET-microdialysis studies. Journal of Cerebral Blood Flow & Metabolism. 1997;17(9):932–942. doi: 10.1097/00004647-199709000-00002. [DOI] [PubMed] [Google Scholar]
- 17.Gunning-Dixon FM, Head D, McQuain J, Acker JD, Raz N. Differential aging of the human striatum: A prospective MR imaging study. American Journal of Neuroradiology. 1998;19(8):1501–1507. [PMC free article] [PubMed] [Google Scholar]
- 18.Urban N, Slifstein M, Kegeles LS, Martinez D, Xu X, Sakr E, et al. In vivo evidence for gender differences in alcohol induced striatal dopamine release: a PET imaging study with [11C]; Annual Meeting of the American College of Neuropsychopharmacology; 2009. [Google Scholar]
- 19.Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: A meta-analysis. Drug Alcohol Depend. 2005 doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 20.Ramchandani VA, Plawecki M, Li TK, O'Connor S. Intravenous Ethanol Infusions Can Mimic the Time Course of Breath Alcohol Concentrations Following Oral Alcohol Administration in Healthy Volunteers. Alcoholism-Clinical and Experimental Research. 2009;33(5):938–944. doi: 10.1111/j.1530-0277.2009.00906.x. [DOI] [PubMed] [Google Scholar]
- 21.Ramchandani VA, Bolane J, Li TK, O'Connor S. A physiologically-based pharmacokinetic (PBPK) model for alcohol facilitates rapid BrAC clamping. Alcoholism: Clinical and Experimental Research. 1999;23(4):617–623. [PubMed] [Google Scholar]
- 22.Blekher T, Ramchandani VA, Flury L, Foroud T, Kareken D, Yee RD, et al. Saccadic eye movements are associated with a family history of alcoholism at baseline and after exposure to alcohol. Alcoholism: Clinical and Experimental Research. 2002;26(10):1568. doi: 10.1097/01.ALC.0000033121.05006.EF. [DOI] [PubMed] [Google Scholar]
- 23.Kwo PY, Ramchandani VA, O'Connor S, Amann D, Carr LG, Sandrasegaran K, et al. Gender differences in alcohol metabolism: relationship to liver volume and effect of adjusting for body mass. Gastroenterology(New York, NY 1943) 1998;115(6):1552–1557. doi: 10.1016/s0016-5085(98)70035-6. [DOI] [PubMed] [Google Scholar]
- 24.Morzorati SL, Ramchandani VA, Flury L, Li TK, O'Connor S. Self-reported subjective perception of intoxication reflects family history of alcoholism when breath alcohol levels are constant. Alcoholism-Clinical and Experimental Research. 2002;26(8):1299–1306. doi: 10.1097/01.ALC.0000025886.41927.83. [DOI] [PubMed] [Google Scholar]
- 25.Ramchandani VA, Flurry L, Morzorati SL, Kareken D, Blekher T, Foroud T, et al. Recent Drinking History: Association with Family History of Alcoholism and the Acute Response to Alcohol during a 60 Mg% Clamp*. J Stud Alcohol. 2002;63(6):734–745. doi: 10.15288/jsa.2002.63.734. [DOI] [PubMed] [Google Scholar]
- 26.Neumark YD, Friedlander Y, Durst R, Leitersdorf E, Jaffe D, Ramchandani VA, et al. Alcohol dehydrogenase polymorphisms influence alcohol-elimination rates in a male Jewish population. Alcohol Clin Exp Res. 2004;28(1):10–14. doi: 10.1097/01.ALC.0000108667.79219.4D. [DOI] [PubMed] [Google Scholar]
- 27.Gilman JM, Ramchandani VA, Davis MB, Bjork JM, Hommer DW. Why we like to drink: a functional magnetic resonance imaging study of the rewarding and anxiolytic effects of alcohol. J Neurosci. 2008;28(18):4583–4591. doi: 10.1523/JNEUROSCI.0086-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Wit H, McCracken SG. Ethanol self-administration in males with and without an alcoholic first-degree relative. Alcoholism: Clinical and Experimental Research. 1990;14(1):63–70. doi: 10.1111/j.1530-0277.1990.tb00448.x. [DOI] [PubMed] [Google Scholar]
- 29.Lammertsma AA, Hume SP. Simplified reference tissue model for PET receptor studies. Neuroimage. 1996;4(3):153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- 30.Skinner HA, Sheu WJ. Reliability of alcohol use indices. The Lifetime Drinking History and the MAST. J Stud Alcohol. 1982;43(11):1157. doi: 10.15288/jsa.1982.43.1157. [DOI] [PubMed] [Google Scholar]
- 31.Zhang HP, Luo XG, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, et al. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15(6):807–819. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 1993;112(4):503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- 33.Crawley JN, Paylor R. A proposed test battery and constellations of specific behavioral paradigms to investigate the behavioral phenotypes of transgenic and knockout mice. Horm Behav. 1997;31(3):197–211. doi: 10.1006/hbeh.1997.1382. [DOI] [PubMed] [Google Scholar]
- 34.Morris BJ, Millan MJ, Herz A. Antagonist-induced opioid receptor up-regulation. II. Regionally specific modulation of mu, delta and kappa binding sites in rat brain revealed by quantitative autoradiography. J Pharmacol Exp Ther. 1988;247(2):729–736. [PubMed] [Google Scholar]
- 35.Sim LJ, Selley DE, Childers SR. In vitro autoradiography of receptor-activated G proteins in rat brain by agonist-stimulated guanylyl 5'-[gamma-[35S]thio]-triphosphate binding. Proc Natl Acad Sci U S A. 1995;92(16):7242–7246. doi: 10.1073/pnas.92.16.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. 2nd ed. London: Academic Press; 2001. [Google Scholar]
- 37.Margas W, Mahmoud S, Ruiz-Velasco V. Muscarinic Acetylcholine Receptor Modulation of Mu ({mu}) Opioid Receptors in Adult Rat Sphenopalatine Ganglion Neurons. J Neurophysiol. 2009 doi: 10.1152/jn.00295.2009. 00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frantz KJ, Hansson KJ, Stouffer DG, Parsons LH. 5-HT6 receptor antagonism potentiates the behavioral and neurochemical effects of amphetamine but not cocaine. Neuropharmacology. 2002;42(2):170–180. doi: 10.1016/s0028-3908(01)00165-4. [DOI] [PubMed] [Google Scholar]
- 39.Munafo MR, Brown SM, Hariri AR. Serotonin Transporter (5-HTTLPR) Genotype and Amygdala Activation: A Meta-Analysis. Biol Psychiatry. 2008;63(9):852–857. doi: 10.1016/j.biopsych.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furmark T, Appel L, Michelgard A, Wahlstedt K, Ahs F, Zancan S, et al. Cerebral blood flow changes after treatment of social phobia with the neurokinin-1 antagonist GR205171, citalopram, or placebo. Biol Psychiatry. 2005;58(2):132–142. doi: 10.1016/j.biopsych.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 41.Newlin DB, Thomson JB. Alcohol Challenge with Sons of Alcoholics - A Critical-Review and Analysis. Psychol Bull. 1990;108(3):383–402. doi: 10.1037/0033-2909.108.3.383. [DOI] [PubMed] [Google Scholar]
- 42.Bart G, Kreek MJ, Ott J, LaForge KS, Proudnikov D, Pollak L, et al. Increased attributable risk related to a functional mu-opioid receptor gene polymorphism in association with alcohol dependence in central Sweden. Neuropsychopharmacology. 2005;30(2):417–422. doi: 10.1038/sj.npp.1300598. [DOI] [PubMed] [Google Scholar]
- 43.Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the [mu]-opioid receptor gene with substance dependence: A meta-analysis. Drug Alcohol Depend. 2006;83(3):262–268. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Hariri AR, Mattay VS, Tessitore A, Kolachana B, Fera F, Goldman D, et al. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297(5580):400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- 45.Bennett AJ, Lesch KP, Heils A, Long JC, Lorenz JG, Shoaf SE, et al. Early experience and serotonin transporter gene variation interact to influence primate CNS function. Mol Psychiatry. 2002;7(1):118–122. doi: 10.1038/sj.mp.4000949. [DOI] [PubMed] [Google Scholar]
- 46.Barr CS, Newman TK, Schwandt M, Shannon C, Dvoskin RL, Lindell SG, et al. Sexual dichotomy of an interaction between early adversity and the serotonin transporter gene promoter variant in rhesus macaques. Proc Natl Acad Sci U S A. 2004;101(33):12358–12363. doi: 10.1073/pnas.0403763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 48.Risch N, Herrell R, Lehner T, Liang KY, Eaves L, Hoh J, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. JAMA. 2009;301(23):2462. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kendler KS, Thornton LM, Gardner CO. Genetic risk, number of previous depressive episodes, and stressful life events in predicting onset of major depression. Am J Psychiatry. 2001;158(4):582–586. doi: 10.1176/appi.ajp.158.4.582. [DOI] [PubMed] [Google Scholar]
- 50.Oslin DW, Berrettini W, Kranzler HR, Pettinati H, Gelernter J, Volpicelli JR, et al. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacology. 2003;28(8):1546–1552. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- 51.Anton RF, Oroszi G, O'Malley S, Couper D, Swift R, Pettinati H, et al. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry. 2008;65(2):135–144. doi: 10.1001/archpsyc.65.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gelernter J, Gueorguieva R, Kranzler HR, Zhang HP, Cramer J, Rosenheck R, et al. Opioid receptor gene (OPRM1, OPRK1, and OPRD1) variants and response to naltrexone treatment for alcohol dependence: Results from the VA cooperative study. Alcoholism-Clinical and Experimental Research. 2007;31(4):555–563. doi: 10.1111/j.1530-0277.2007.00339.x. [DOI] [PubMed] [Google Scholar]
- 53.Barr CS, Chen SA, Schwandt ML, Lindell SG, Sun H, Suomi SJ, et al. Suppression of alcohol preference by naltrexone in the rhesus macaque: A critical role of genetic variation at the µ-opioid receptor gene (OPRM1) locus. Biol Psychiatry. 2009 doi: 10.1016/j.biopsych.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schultz W. Behavioral theories and the neurophysiology of reward. Annu Rev Psychol. 2006;57:87–115. doi: 10.1146/annurev.psych.56.091103.070229. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Wang DX, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280(38):32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 56.Mague SD, Isiegas C, Huang P, Liu-Chen LY, Lerman C, Blendy JA. Mouse model of OPRM1 (A118G) polymorphism has sex-specific effects on drug-mediated behavior. Proc Natl Acad Sci U S A. 2009;106(26):10847–10852. doi: 10.1073/pnas.0901800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He L, Fong J, von Zastrow M, Whistler JL. Regulation of opioid receptor trafficking and morphine tolerance by receptor oligornerization. Cell. 2002;108(2):271–282. doi: 10.1016/s0092-8674(02)00613-x. [DOI] [PubMed] [Google Scholar]
- 58.Barr CS, Schwandt ML, Lindell SG, Higley JD, Maestripieri D, Goldman D, et al. Variation at the mu-opioid receptor gene (OPRM1) influences attachment behavior in infant primates. Proc Natl Acad Sci U S A. 2008;105(13):5277–5281. doi: 10.1073/pnas.0710225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Way BM, Taylor SE, Eisenberger NI. Variation in the µ-opioid receptor gene (OPRM1) is associated with dispositional and neural sensitivity to social rejection. Proceedings of the National Academy of Sciences. 2009;106(35):15079–15084. doi: 10.1073/pnas.0812612106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wand GS, McCaul M, Yang X, Reynolds J, Gotjen D, Lee S, et al. The mu-opioid receptor gene polymorphism (A118G) alters HPA axis activation induced by opioid receptor blockade. Neuropsychopharmacology. 2002;26(1):106–114. doi: 10.1016/S0893-133X(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 61.Pang GSY, Wang JB, Wang ZH, Goh C, Lee CGL. The G allele of SNP E1/A118G at the mu-opioid receptor gene locus shows genomic evidence of recent positive selection. Pharmacogenomics. 2009;10(7):1101–1109. doi: 10.2217/pgs.09.63. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.