Summary
The Joint United Nations Programme on HIV/AIDS (UNAIDS) has decided to use Bayesian melding as the basis for its probabilistic projections of HIV prevalence in countries with generalized epidemics. This combines a mechanistic epidemiological model, prevalence data and expert opinion. Initially, the posterior distribution was approximated by sampling-importance-resampling, which is simple to implement, easy to interpret, transparent to users and gave acceptable results for most countries. For some countries, however, this is not computationally efficient because the posterior distribution tends to be concentrated around nonlinear ridges and can also be multimodal. We propose instead Incremental Mixture Importance Sampling (IMIS), which iteratively builds up a better importance sampling function. This retains the simplicity and transparency of sampling importance resampling, but is much more efficient computationally. It also leads to a simple estimator of the integrated likelihood that is the basis for Bayesian model comparison and model averaging. In simulation experiments and on real data it outperformed both sampling importance resampling and three publicly available generic Markov chain Monte Carlo algorithms for this kind of problem.
Keywords: Bayesian melding, Bayesian model selection, Bayesian model averaging, Epidemio-logical model, Integrated likelihood, Markov chain Monte Carlo, Prevalence, Sampling importance resampling, Susceptible infected removed model
1. Introduction
The Joint United Nations Programme on HIV/AIDS (UNAIDS) publishes updated estimates and projections of the number of people living with HIV/AIDS in the countries with generalized epidemics every two years. As part of this, statements of uncertainty are also provided. Generalized epidemics are defined by overall prevalence being above 1%, and the epidemic not being confined to particular subgroups; there are about 38 such countries (Ghys et al., 2004).
UNAIDS bases its estimates and projections on data from two main sources: prevalence estimates from antenatal clinics and estimates from Demographic and Health Surveys (DHS). The antenatal clinic data are sparser and less representative, but they are available in all the countries and are typically available for many time periods, allowing estimation of trends. The DHS data are more representative, but they are not available for all countries, and for most countries are available at only one time point. Thus the DHS data give better information about overall prevalence, when they are available, but the antenatal clinic data give better information about trends.
The quality of the data available varies widely from country to country. As a result, UNAIDS bases its estimates on a relatively simple method that can be supported by the data in all the countries, and so can give estimates that are comparable between countries. This is based on a simple standard epidemiological model with four adjustable parameters. The estimates and projections are produced using a Bayesian melding method that combines the epidemiological model with a hierarchical random effects model for the sampling variability of the data (Alkema et al., 2007, 2008). Out-of-sample predictive assessments by Alkema et al. (2007) indicated that the overall method performed well over the five-year time horizon of the projections.
This method was incorporated into the Estimation and Projection Package (EPP) produced by UNAIDS and available for download from http://www.epidem.org. This is used by the UNAIDS Secretariat and also by national o cials producing their own estimates and projections. It was used to produce the 2007 update of the UNAIDS estimates and projections (UNAIDS, 2007; Ghys et al., 2008). This update attracted a great deal of attention because it featured a large change, notably a revision of the estimate of the number of people with HIV/AIDS worldwide downwards from 39 million to 33 million.
The Bayesian melding posterior distribution is produced using the sampling-importance-resampling (SIR) algorithm, with the first sample drawn from the prior distribution (Rubin, 1987, 1988; Poole and Raftery, 2000; Alkema et al., 2007). This consists of simulating a large number of samples from the prior distribution of the model parameters, weighting these by their likelihoods, and then resampling them with replacement and with the computed weights. This method is simple to implement and explain, and works well for most of the countries involved. It is not computationally efficient, however, but for most countries this does not matter much: the model runs quickly and so it is possible to draw an initial sample of size 200,000, which is enough to give a reasonable estimate of the posterior in most cases.
For some countries, however, this was not enough to give a good picture of the uncertainty. Our goal here is to propose a generic method that is more efficient than the SIR algorithm while retaining its essential simplicity and transparency, and so can be used for all the countries involved. The problem is a difficult one because the posterior distribution tends to feature nonlinear ridges and to be multimodal, and so is unlike posterior distributions typical of standard statistical models. However, these features are common in posterior distributions derived from mechanistic models, because these are often underdetermined by the data, which are typically adequate only to estimate some nonlinear functions of the parameters well (Raftery et al., 1995; Poole and Raftery, 2000). This kind of posterior distribution is also challenging for generic Markov chain Monte Carlo (MCMC) algorithms.
We propose a new method, Incremental Mixture Importance Sampling (IMIS). The basic idea was first proposed in a different context by Steele et al. (2006). It is generic, relatively simple to implement and explain, and works well for countries where SIR does not. It also leads to a simple estimator of the integrated likelihood that is the basis for Bayesian model comparison and model averaging; this is challenging to obtain using MCMC (Han and Carlin, 2001; Raftery et al., 2007). In simulation experiments that mimic the ridge-like characteristics and multimodality of the posterior, it substantially outperformed SIR and three different generic Metropolis algorithms. It yields essentially independent samples from the posterior, avoiding the autocorrelation and burn-in problems that plague MCMC. It is a kind of adaptive importance sampling, a class of methods that also include sequential Monte Carlo and population Monte Carlo; we discuss other methods in Section 6.
In Section 2 we describe the EPP model and methdology used by UNAIDS in more detail. In Section 3 we describe our IMIS method for estimation, and also the simple estimator of the integrated likelihood to which it leads. In Section 4 we give results for some simulated examples, and in Section 5 we give results for estimation and projection for Zimbabwe, one of the countries for which SIR did not perform well.
2. The UNAIDS Estimation and Projection Package and Bayesian Melding
The UNAIDS Estimation and Projection Package is based on a simple susceptible-infected-removed epidemiological model that we will refer to as the “EPP model.” This involves four adjustable parameters or inputs, θ= (r, t0, f0, ϕ), where r is the force of infection, t0 is the start year of the HIV/AIDS epidemic, f0 is the initial fraction of the adult population at risk of infection, and ϕ is the behavior adjustment parameter. The output ρ is a sequence of yearly HIV prevalence rates.
The model divides the population at time t into three groups: a not-at-risk group X(t), an at-risk group Z(t) and an infected group Y(t). The model assumes a constant non-AIDS mortality rate μ and a constant fertility rate, and does not represent migration or age structure. The rate at which the sizes of the groups change are described by three differential equations:
| (1) |
where N(t) = X(t) + Z(t) + Y(t) the function g(τ) specifies the HIV death rate τ years after infection. The population being modeled is aged 15+. The number of new members at time t, E(t), depends on the population size 15 years ago, the birth rate and the survival rate from birth to age 15. When individuals survive to age 15, they are assigned to either the not-at-risk group X(t) or the at-risk group Z(t). The fraction of the new 15-year-old members entering the at-risk group Z(t) is given by .
The Bayesian melding approach (Raftery et al., 1995; Poole and Raftery, 2000) was applied to the EPP model by Alkema et al. (2007). It proceeds as follows. A prior distribution is specified for θ. The UNAIDS Reference Group on Estimates, Modelling and Projections agreed on a default prior distribution (see Section 5), but users can specify their own. The observed antenatal clinic and DHS data give the likelihood L(ρ) for the model output, using a hierarchical random effects model. A prior on the model output ρ can also be specified; this is currently restricted to being uniform between specified bounds for specific years.
In the initial UNAIDS implementation, the Bayesian melding procedure computed the posterior distributions using the SIR algorithm of Rubin (1987, 1988), with the prior as importance sampling distribution. It consists of the following stages:
Obtain N independent and identically distributed (i.i.d.) samples, {θ1, …, θN}, from the input prior distribution p(θ).
For each θi, determine the corresponding series of prevalence rates ρi by running the EPP model, and calculate the likelihood Li = L(ρi).
Set the importance weights for each ρi (and thus for each θi): .
Sample from the multinomial distribution of {θ1, …, θN} with probabilities {w1, …, wN} to approximate the posterior distribution of the inputs and outputs.
The default value of N is 200,000, and the EPP model runs fast enough for this to be feasible.
The SIR algorithm performs acceptably for most of the countries involved. For some, however, it does not work well. In these cases there is a small number of large importance weights, and so the resample is dominated by large numbers of repetitions of the same few points. Then there are few distinct values present in the resample and the target distribution is poorly sampled. This happens because the posterior distribution tends to be concentrated near thin curved manifolds, and in addition is often multimodal. In the most extreme case we have seen, for the urban parts of Ghana, SIR found only 8 unique points out of 3,000 resamples.
3. Incremental Mixture Importance Sampling
3.1 Basic IMIS Algorithm
To overcome this problem, we extend incremental mixture importance sampling (IMIS) to the Bayesian melding problem. IMIS is an iterative generalization of defensive mixture importance sampling (Hesterberg, 1995), and was introduced by Steele et al. (2006) in a very different context, that of a high-dimensional discrete distribution.
The basic idea of IMIS is that points with high importance weights are in areas where the target density is underrepresented by the importance sampling distribution. At each iteration, a multivariate normal distribution centered at the point with the highest importance weight is added to the current importance sampling distribution, which thus becomes a mixture of such functions and of the prior. In this way underrepresented parts of parameter space are successively identified and are given representation, ending up with an iteratively constructed importance sampling distribution that covers the target distribution well.
The algorithm ends when the importance sampling weights are reasonably uniform. Specifically, we end the algorithm when the expected fraction of unique points in the resample is at least 1 − 1/e = 0.632. This is the expected fraction when the importance sampling weights are all equal, which is the case when the importance sampling function is the same as the target distribution. The IMIS algorithm is as follows:
- Initial Stage:
- Sample N0 inputs θ1, θ2, …, θN0 from the prior distribution p(θ).
- For each θi, calculate the likelihood Li, and form the importance weights:
- Importance Sampling Stage: For k = 1, 2 …, repeat the following steps
- Choose the current maximum weight input as the center θ(k). Estimate Σ(k) as the weighted covariance of the B inputs with the smallest Mahalanobis distances to θ(k), where the distances are calculated with respect to the covariance of the prior distribution and the weights are taken to be proportional to the average of the importance weights and .
- Sample B new inputs from a multivariate Gaussian distribution Hk with covariance matrix Σ(k).
- Calculate the likelihood of the new inputs and combine the new inputs with the previous ones. Form the importance weights:
where c is chosen so that the weights add to 1, q(k) is the mixture sampling distribution , Hs is the s-th multivariate normal distribution, and Nk = N0 + Bk is the total number of inputs up to iteration k.
Resample Stage: Once the stopping criterion is satisfied, resample J inputs with replacement from θ1, …, θNk with weights w1, …, wNK, where K is the number of iterations at the importance sampling stage.
The algorithm has several control parameters to be set by the user: the number of initial samples N0, the sample size at each importance sampling iteration B, and the number of resamples J. The algorithm is unbiased for any choice of control parameters, because it is an importance sampling algorithm, but the control parameters can affect its efficiency. We have found good results with the choices N0 = 1000d, B = 100d and J = 3000, where d is the dimension of the integrand.
A great advantage of importance sampling is that it is effectively self-monitoring, in that poor coverage of the target distribution by the importance sampling is immediately seen by the presence of large importance weights. We use the following specific criteria to assess the performance of the various importance sampling algorithms considered here:
The maximum importance weight among the Nk inputs.
The variance of the rescaled importance weights, .
The entropy of the importance weights relative to uniformity,
The expected number of unique points after re-sampling, .
- The effective sample size, , where the coaffcient of variation CV is defined as (Kong et al., 1994). We can also write
Since IMIS will involve computing the likelihood that may be a product of several densities, numerical over- or under-flow is often encountered. Therefore, in practice it is often better to compute the weights as:
where , with C a suitable constant that will reduce computational problems, such as the maximum of the Lj’s.
3.2 IMIS with optimization
If the prior distribution disagrees strongly with the target distribution, particularly if the posterior is multimodal, the initial sample might miss a whole high probablity region. In this case, IMIS might not work, as it proceeds essentially by filling in the importance sampling distribution to make it cover the target. If there are no initial points at all in an important unexplored region, IMIS would find it difficult to extend the coverage to that region.
As a remedy for this, we suggest inserting an optimization stage after the initial stage. This yields a mixture of D multivariate normal distributions, each one centered around a local maximum of the target distribution. It works as follows:
- (1’) Optimization Stage:
- Set the maximum weight input from the initial stage as the first starting point, use the optimizer to get a local maximum of the posterior density , and store the inverse of the Hessian matrix as .
- For i = 2, …, D, exclude the starting points and the inputs that have the smallest Mahalanobis distances from the previous local maximum, . Among the remaining inputs, set the maximum weight input as the new starting point to get and as in step (a).
- Sample B inputs from each of the D multivariate normal distributions with centers and covariance matrices .
In our experiments, we used the L-BFGS-B method (Byrd et al., 1995) as implemented in the R function optim, and we set the maximum number of function evaluations to be 100. When the Hessian matrix was not positive definite, we used the first derivative to build a new information matrix, and added it to the inverse of the diagonal of the prior covariance matrix, diag(Σ(prior))−1. The inverse of the new information matrix is then defined as . We found good results with D = 10. We refer to this version of IMIS as IMIS-opt.
3.3 Estimating the Integrated Likelihood using IMIS
The standard Bayesian approach to model comparison is based on the Bayes factor between two models (Jeffreys, 1939; Kass and Raftery, 1995), and the standard Bayesian approach to accounting for model uncertainty is Bayesian model averaging (Leamer, 1978; Madigan and Raftery, 1994; Hoeting et al., 1999; Clyde and George, 2004). Both are based on the integrated likelihood of a model M, defined as
| (2) |
where D denotes the data, θ denotes the parameter vector (or set of inputs) of the model, p(Dǀθ) is the usual likelihood function, and π(θ) is the prior density of θ under the model. The Bayes factor for model M2 against model M1 is then
It has proven difficult to find a satisfactory general method to estimate the integrated likelihood using standard posterior simulation methods, including direct posterior simulation and MCMC. Many proposals have been made, however, and some work well for specific models; see Raftery et al. (2007) for a review of the literature. IMIS directly yields an estimate of the integrated likelihood that is simple to compute and has good theoretical properties.
We can rewrite (2) as
| (3) |
where q(θ) is the importance sampling distribution. The IMIS estimator of the integrated likelihood is then
| (4) |
where {θ1, …, θNK} is the sample from the final IMIS importance sampling distribution q(θ) = q(K)(θ). Then the IMIS estimator has several desirable theoretical properties, as shown by the following theorem, proved by Raftery and Bao (2009):
Theorem 1
If the posterior distribution of θ is proper, then, as an estimator of p(DǀM), is (i) unbiased, (ii) strongly consistent, and (iii) asymptotically normal, with
| (5) |
where q(K) is the mixture importance sampling function defined previously, consisting of a mixture of the prior distribution and K multivariate Gaussian distributions, , and the asymptotics refer to the amount of simulation increasing, so that NK tends to infinity at the K th stage.
Note that the finite variance of the IMIS estimator is not shared by other importance sampling estimators in general. It arises because the IMIS importance sampling distribution is a mixture of the prior and other distributions. The inclusion of the prior makes IMIS a defensive mixture method in the sense of Hesterberg (1995), and yields this desirable behavior. When the importance sampling distribution is the posterior itself, the importance sampling estimator of the integrated likelihood is the harmonic mean estimator of Newton and Raftery (1994), which is simulation-consistent but whose reciprocal has infinite variance except in special cases (Raftery et al., 2007). This is at the root of the difficulty in estimating the integrated likelihood directly from posterior simulation.
The Monte Carlo estimate of the variance of is given by
| (6) |
We can approximate the standard error of by
| (7) |
4. Simulated Examples
4.1 Methods for Comparison
We consider two simulated examples, to explore the behavior of IMIS in situations with features similar to those of the EPP problem. The EPP posterior is typically ridge-like and multimodal, so we show one example of simulating from a distribution concentrated around a thin curved manifold, and another of simulating from a bimodal distribution.
We compare IMIS and IMIS-opt with SIR, and also with generic random-walk Metropolis algorithms, which have become perhaps the most common way of simulating from posterior distributions. We include results for three different publicly available algorithms of this kind: a version that is close to theoretically optimal for simulating a multivariate normal distribution, a version that does not use optimization, and a version that was programmed in WinBUGS.
The first one is the random walk Metropolis algorithm implemented by the R function MCMCmetrop1R in the MCMCpack package (Martin and Quinn, 2007). This is close to the theoretically optimal algorithm for simulating from a multivariate normal distribution using a single-block random walk Metropolis algorithm with a multivariate normal proposal distribution (Gelman et al., 1996; Neal and Roberts, 2006). It invol ves numerically optimizing the posterior density to find the posterior mode and evaluating the inverse Hessian at that point. The variance of the proposal distribution is then 2.38/d times the estimated inverse Hessian at the posterior mode, where d is the dimension of the target distribution. The theoretical acceptance rate for a multivariate normal target ranges from 0.44 for d = 1 to 0.23 as d → ∞. We ran five chains, all starting from the mode found by the optimizer.
The second generic Metropolis algorithm uses the R function metrop in the mcmc package (Geyer, 2005), with a multivariate normal proposal distribution with variance proportional to the identity matrix. We simulated five starting values from the prior, and ran five chains, tuning the proposal variance so that the acceptance rate was close to 0.23. Finally, we also ran the WinBUGS program (Lunn et al., 2000) using the “zero trick” to specify a new distribution (Cowles, 2004), again running five chains from starting values simulated from the prior. This random walk Metropolis algorithm is used in WinBUGS for nonconjugate continuous full conditionals with an unrestricted range.
As our primary criterion for comparing all six methods, we use Efficiency, defined as the Effective Sample Size (ESS) divided by the total number of function evaluations. For the MCMC methods, we used the estimated effective sample size using an autoregressive estimate of the spectrum at zero, as implemented in the R function effectiveSize of the coda package (Plummer et al., 2006). To estimate the ESS for the MCMC methods, we used the diagnostic of Gelman and Rubin (1992) to assess convergence using the five chains, and then ran one of the chains for as many iterations as used by IMIS, discarding the iterations before the five chains had converged as burn-in.
4.2 Ridge-like Simulated Example
The posterior distributions from mechanistic deterministic models are often ridge-like, that is, concentrated around thin curved manifolds. This is because they are often underdetermined by the data, with only functions of the parameters being well estimated. We simulate a situation of this kind, with a setup first used by Bates (2001).
The deterministic model which maps the inputs θ = (θ1, …, θ6) to the outputs ϕ = (ϕ1, …, ϕ4) is given by
| (8) |
The prior distributions on the inputs are independent and normally distributed with means {6.0, 0.5, 5.5, 0.15, 3.0, 0.6} and standard deviations {1.3, 0.14, 0.289, 0.029, 0.04, 0.1}. In addition, there are independent normal likelihoods on the outputs, with means {7.0, 0.0525, 2.0, 4.0} and standard deviations {0.5, 0.00144, 0.01, 0.01} respectively.
IMIS converged after 35,400 function evaluations, and IMIS-opt converged after 16,800 evaluations. The e ciencies of the different methods are shown in Table 1. SIR was clearly much less efficient than the other methods. IMIS was far more efficient, and IMIS-opt was about 50% more efficient again, although there were not multiple modes in this case. The MCMC methods were more efficient than SIR, and the MCMCMetrop1R method, with its optimality propertes, greatly outperformed the metrop and WinBUGS methods, by a factor of at least 7. However, IMIS and IMIS-opt were much more efficient than the best of the generic MCMC methods, by a factor of at least 15.
Table 1.
Efficiencies of Different Methods for the Simulated Examples and EPP for Zimbabwe. The value 1.0 corresponds to direct independent sampling from the posterior distribution.
| Method | Ridge-like | Bimodal d = 4 | Bimodal d = 20 | EPP |
|---|---|---|---|---|
| SIR | 2 × 10−5 | .0002 | 10−6 | 8 × 10−6 |
| IMIS | .0675 | .2063 | .0073 | .1361 |
| IMIS-opt | .1040 | .4047 | .1758 | .1553 |
| MCMCMetrop1R | .0045 | .0437 | .0143 | .0031 |
| metrop | .0006 | .0047 | .0004 | NA |
| WinBUGS | .0005 | .0223 | .0049 | NA |
4.3 Bimodal Example
Our second example was bimodal, and the target distribution was the product of a likelihood and a prior. The likelihood was a mixture of two multivariate normal distributions:
where AR(ρ) represents a first-order autoregressive covariance matrix with correlation parameter ρ and marginal variance 1. The prior was .
We considered two cases: d = 4, the same dimension as the EPP model, and d = 20, corresponding to larger deterministic models that have been analyzed using Bayesian melding, for example in forestry (Radtke et al., 2002) and hydrology (Hong et al., 2005). In the four-dimensional case, the high density region for each mode (defined as the sphere centered at the mode with radius three standard deviations) is 4 × 10−6 times the volume of the prior distribution’s support, while in the 20-dimensional case the ratio is about 10−35. This experiment was modeled on an experiment of Warnes (2001).
When d = 4, IMIS converged after 13,600 function evaluations and IMIS-opt after 8,600 evaluations. When d = 20, the numbers were much larger: 150,000 for IMIS and 41,000 for IMIS-opt. However, when d = 20, IMIS had not sampled the second mode, while IMIS-opt was the only one of the methods we consider here that sampled both modes in this challenging example.
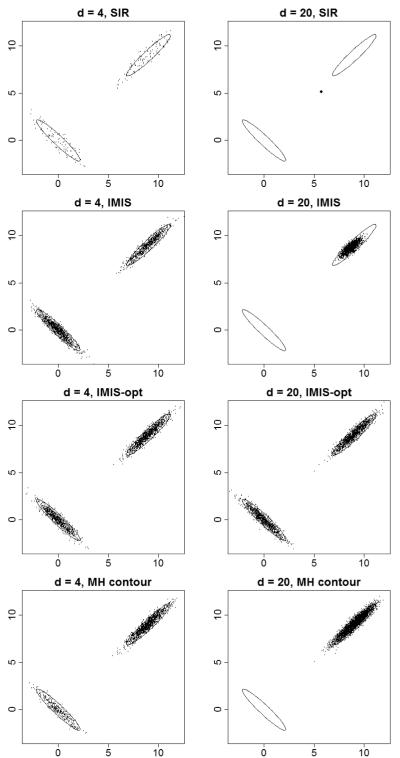
Contour plots of the simulated values are shown in Figure 1, for the same number of function evaluations for all methods (13,600 for d = 4 and 150,000 for d = 20). SIR did not perform well: it sampled both modes for d = 4, but only sparsely, and for d = 20 it missed both modes. IMIS did an excellent job for d = 4, but for d = 20 only partly explored one mode and missed the other. IMIS-opt did a very good job in both cases.
Figure 1.
Results for the Bimodal Simulated Example, for SIR, IMIS, IMIS-opt, and the best performing of the three generic Metropolis algorithms considered. The results are for dimensions 4 and 20, and are projected onto the first two dimensions. The true elliptical contours of the densities are shown in black.
For d = 4, Metrop1R performed best of the three MCMC methods we used. For d = 20, however, Metrop1R did not work, but we implemented a similar method using the metrop function, and that is shown in Figure 1. For d = 4, the MCMC method found both modes, but did not weight them correctly. For d = 20, the MCMC method found only one of the modes.
These impressions are confirmed by the e ciencies in Table 1. For both d = 4 and d = 20, IMIS-opt performed best by far; IMIS was a respectable second for d = 4, but far behind for d = 20. IMIS-opt was about 10 times more efficient than the best of the three generic MCMC methods in both cases.
5. Application to EPP for Zimbabwe
5.1 Estimation and Projection Results
We now apply IMIS to Bayesian melding for the EPP model for the urban parts of Zimbabwe. We use prevalence data from 18 antenatal clinics from 1989 to 2004. The observed prevalences are shown by the dots in Figure 2, with different colors corresponding to different clinics.
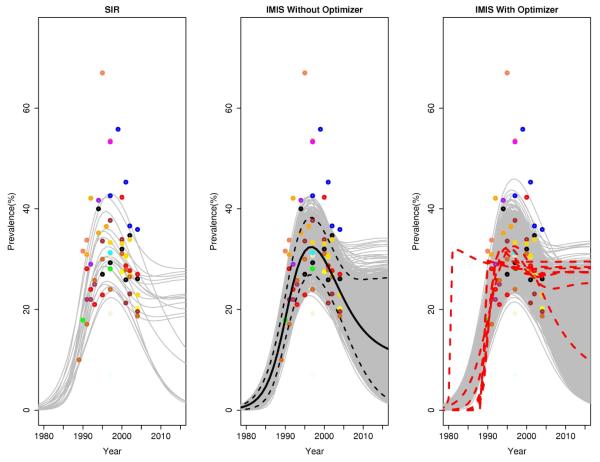
Figure 2.
EPP model for 2004 Zimbabwe Urban Data: Uncertainty assessment using SIR (left), IMIS (middle), and IMIS-opt (right). The dots show the observed prevalences, with different colors corresponding to different antenatal clinics. The gray lines are 3,000 resampled trajectories. In the middle figure, the solid black line is the posterior mean and the dashed black lines are 2.5% and 97.5% posterior quantiles. In the right figure, the red lines are 10 trajectories corresponding to the local optima.
We implemented Bayesian melding using the random effects likelihood function described by Alkema et al. (2007) and the default prior distributions agreed by the UNAIDS Reference Group, namely log(r) ~ U[log(0.5), log(150)], f0 ~ U[0,1], t0 ~ Discrete Uniform[1970, 1971, …, 1990], and ϕ ~ Logistic(100, 50). The starting time of the epidemic, t0, is treated as an integer by the EPP model because the data typically come in yearly increments, but our implementation of IMIS is designed for continuous parameters. We deal with this by treating t0 as continuous in the importance sampling distribution, with a piecewise constant likelihood.
Figure 2(a) shows the results of using SIR. The J = 3, 000 resamples yielded only 19 unique trajectories, each one corresponding to a different set of parameter values. The largest weight was 0.81, so over 2,400 of the 3,000 resampled trajectories were actually the same.
Figures 2(b) and (c) show the IMIS and IMIS-opt results. Clearly they cover the posterior distribution much more fully and smoothly. In IMIS-opt, the red dashed lines correspond to the local optima found; most of these had little support in the posterior.
Table 2 shows performance measures for the various methods used. Among the three MCMC methods we show results only for MCMCMetrop1R, as it performed better than theother two in this case. With the IMIS importance sampling distribution, the 3,000 resamples generated 1,929 unique points, which is close to the expected number of unique points when the importance sampling distribution is equal to the target (1,896). IMIS-opt was slightly more efficient than IMIS in this case, but not much more so, reflecting the fact that the secondary modes were much smaller than the largest one. The best of the MCMC methods was far less eiffcient than IMIS, by a factor of more than 40. In fact, when we ran the five chains from different starting points, they had not converged within 20,400/5 = 4,088 iterations, according to the Gelman-Rubin diagnostic.
Table 2.
Efficiencies of Different Methods for the EPP applied to the Zimbabwe Urban urban
| Method | total inputs | variance | entropy | max weight | unique points | ESS |
|---|---|---|---|---|---|---|
| SIR | 200,000 | 13,317 | 0.81 | 0.06 | 24 | 1.5 |
| IMIS | 20,400 | 6.5 | 0.0010 | 0.82 | 1929 | 2776 |
| IMIS-opt | 17,447 | 5.5 | 0.0012 | 0.84 | 1915 | 2709 |
| MCMCMetrop1R() | 20,447 | 62.7 |
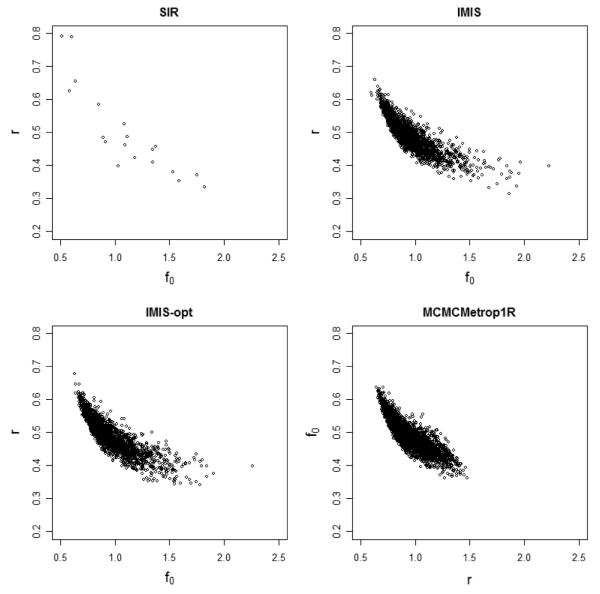
Figure 3 shows plots of the samples from the joint posterior distribution of f0 and r found by the four methods. SIR covered the posterior distribution completely but sparsely, while generic MCMC (in this case a single chain starting from the SIR mean and run for over 168,000 iterations) missed substantial parts of the posterior distribution.
Figure 3.
EPP model for Zimbabwe urban data: Comparison of the marginal posterior distribution of (f0, r) obtained using SIR with 200,000 inputs (upper left), IMIS without optimizer 20,400 inputs (upper right), IMIS with 10 optimizers 17,447 inputs (lower left) and generic Metropolis random walk starting from the SIR mean and with 168,074 iterations (lower right).
5.2 Model Comparison and Model Averaging
One question of great interest in the context of HIV/AIDS is whether the pattern of the epidemic has changed, for example due to a change in behavior. One way of representing this is the so-called “R-jump model” proposed by Brown et al. (2008), which allows for a onetime change in the force of infection, r, at time τ. The R-jump model specifies that r depends on time t, denoted by r(t), according to r(t) = r1 if t0 ≤ t ≤ τ, and r(t) = r2 if t > τ. We use a joint prior distribution for (t0, τ) specified by t0 ~ U[1970, 1990], and τǀt0 ~ U[t0, 2004]. The priors for r1 and r2 are taken to be independent, with log(rj) ~ U[0.5, 150].
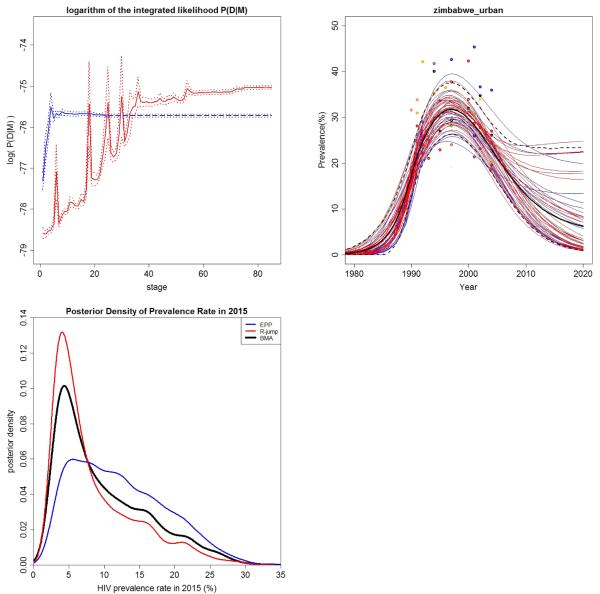
This question can be addressed by computing the Bayes factor for the R-jump model, M2, against the standard EPP model, M1. The estimated values and 95% Bayesian confidence intervals of the log integrated likelihoods after each stage of IMIS are shown in Figure 4. The values increase and then stablize, which is a result of the “pseudo-bias” that can a ict importance sampling (Ventura, 2002). This underlines the importance of running IMIS to convergence.
Figure 4.
Model selection and model averaging for Zimbabwe urban data: (a) Estimated log integrated likelihood (solid lines) plus or minus twice its approximate standard error (dashed lines). The standard EPP model is in blue and the R-jump model in red. IMIS required 26 stages for the standard EPP model and 86 for the R-jump model. The lines for the standard EPP model are extended horizontally to allow easier visual comparison with the R-jump model. (b) A sample of 50 trajectories from the combined BMA posterior distribution for 2006 Zimbabwe urban data. The trajectories from the standard EPP model are in blue and those from the R-jump model are in red. (c) Kernel density estimate of the posterior distribution of HIV prevalence in 2015. The standard EPP model has median 11.3% and 95% confidence interval (3.0%, 26.0%), the R-jump model has median 6.2% and 95% interval (2.6%, 23.2%), and the BMA combined posterior has median 7.6% and 95% interval (2.7%, 24.9%).
The estimated log Bayes factor was log(B21) = 75.73 − 75.04 = 0.69. The standard error of the log integrated likelihood was 0.018 for the standard EPP model and 0.020 for the R-jump model, and so the standard error of the log Bayes factor was . Thus the estimated Bayes factor was just 2.0, “evidence worth no more than a bare mention,” in the words of Jeffreys (1939). Thus there is slight evidence for the R-jump model against the standard EPP model for Zimbabwe, but nothing conclusive.
When multiple competing models are considered plausible, Bayesian model averaging can be used to obtain combined inference about the trajectory of the disease prevalence, taking account of the model uncertainty. We illustrate this here by combining the results of the standard EPP and R-jump models. We took trajectories from the standard EPP posterior distribution; and trajectories from the R-jump model. Figure 4(b) shows 50 trajectories randomly selected from this combined sample. In this case the projections for 2015 did not differ greatly between the models, so model uncertainty was not a major component of uncertainty about this quantity of interest.
6. Discussion
We have proposed a new computational method for implementing Bayesian melding for the EPP model of the HIV/AIDS epidemic used by UNAIDS. This is incremental mixture importance sampling (IMIS), a form of adaptive importance sampling. IMIS outperformed both simple SIR and three publicly available variants of generic MCMC in simulations and real data relevant to the problem at hand. IMIS also yields a simple estimate of the integrated likelihood required for Bayesian model comparison and model averaging; this is hard to obtain in general using MCMC. Moreover, the importance sampling framework allows easy assessment of the Monte Carlo error and facilitates parallel implementation.
These results are relevant to problems like the present one, with posterior distributions of moderate dimension (up to at least 30) that tend to be concentrated close to ridges, or thin curved manifolds, and also can be multimodal. Many problems involving deterministic or mechanistic models have these characteristics, so this method may be more widely useful. It is unlikely to be as useful, however, for some classes of models that are routinely and successfully handled by MCMC, such as Bayesian hierarchical models with hundreds or thousands of parameters.
IMIS is an adaptive important sampling method, and other adaptive importance methods have been proposed, including global adaptive importance sampling (Oh and Berger, 1993; West, 1992, 1993; Zhang, 1996), local adaptive importance sampling (Givens and Raftery, 1996), defensive mixture importance sampling (Hesterberg, 1995; Raghavan and Cox, 1998; Owen and Zhou, 2000), population Monte Carlo (PMC) (Cappé et al 2008 and references therein), and stochastic approximation Monte Carlo (Liang et al., 2007). IMIS more directly attacks the fundamental problem of importance sampling: important regions of parameter space that are unrepresented or sparsely represented in the original sample from the important sampling function. The presence of these is identified by large importance weights, and IMIS directly adds mass to the importance sampling function where there are large weights. In this way it learns an effective importance sampling function. Also, the proposed optimization step of IMIS-opt makes it easier to reach modes not initially sampled.
There are also similarities between IMIS and sequential Monte Carlo (SMC), since they are both iterated importance sampling schemes. SMC methods were originally introduced to deal with sequential inference problems in statistics; see Liu (2002) and Moral et al. (2007) for a review. It was soon pointed out that SMC can also be used for static inference, and several methods for doing this have been proposed, including annealed importance sampling (Neal, 2001), resample-move (Gilks and Berzuini, 2001), sequential particle filtering (Chopin, 2002), and the dynamically weighted importance sampling method (Liang, 2002); see Jasra et al. (2007) for a review. A problem for SMC samplers is that the particles may only represent a single mode of the target (Jasra et al., 2008).
The main differences between IMIS and the other adaptive importance sampling methods mentioned (including PMC and SMC methods) are that IMIS updates the importance sampling function in a different and more direct way, IMIS uses all the values simulated and not just those simulated at the last stage, and IMIS (in the IMIS-opt version) uses optimization to reach local modes not reached by the sample from the initial importance sampling function. The IMIS idea was introduced in a different context by Steele et al. (2006), and the present work extends this to the problem of simulating from a continuous posterior distribution. Cappé et al. (2008) subsequently proposed a PMC method that is an adaptive importance sampling method with an adaptively estimated mixture of multivariate normals and the prior as importance sampling function. IMIS differs from their approach in the way the importance sampling function is updated, its use of all the samples simulated and the optimization step.
An alternative to our approach to inference about deterministic models is the approach based on interpolation using model emulators (see Liu and West 2009 and references therein). This seems more complex than our approach, especially when the dimension of the input and output are not very small.
The application of IMIS to the EPP model is implemented as part of the UNAIDS EPP software, publicly and freely available from http://www.epidem.org.
Acknowledgements
This research was supported by NICHD grant R01 HD054511. The authors are grateful to the Editor, the Associate Editor and two anonymous referees for helpful comments that improved the manuscript, and to Leontine Alkema, Tim Brown, Peter Ghys and Eleanor Gouws for helpful discussions and for sharing data.
References
- Alkema L, Raftery AE, Brown T. Bayesian melding for estimating uncertainty in national HIV prevalence estimates. Sexually Transmitted Infections. 2008;84:i11–i16. doi: 10.1136/sti.2008.029991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkema L, Raftery AE, Clark SJ. Probabilistic projections of HIV prevalence using Bayesian melding. Annals of Applied Statistics. 2007;1:229–248. [Google Scholar]
- Bates S. PhD thesis. University of Washington; 2001. Bayesian inference for deterministic simulation models for environmental assessment. [Google Scholar]
- Brown T, Salomon JA, Alkema L, Raftery AE, Gouws E. Progress and challenges in modelling country-level HIV/AIDS epidemics: the UNAIDS Estimation and Projection Package 2007. Sexually Transmitted Infections. 2008;84:i5–i10. doi: 10.1136/sti.2008.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd RH, Lu P, Nocedal J, Zhu CY. A limited memory algorithm for bound constrained optimization. SIAM Journal on Scientific Computing. 1995;16:1190–1208. [Google Scholar]
- Cappé O, Douc R, Guillin A, Marin J-M, Robert C. Adaptive importance sampling in general mixture classes. Statistics and Computing. 2008;18:447–459. [Google Scholar]
- Chopin N. A sequential particle filter method for static models. Biometrika. 2002;89:539–551. [Google Scholar]
- Clyde M, George EI. Model uncertainty. Statistical Science. 2004;19:81–94. [Google Scholar]
- Cowles MK. Review of winbugs 1.4. The American Statistician. 2004;58:330–336. [Google Scholar]
- Gelman A, Roberts G, Gilks W. E cient Metropolis jumping rules. Bayesian Statistics. 1996;5:599–607. [Google Scholar]
- Gelman A, Rubin D. Inference from iterative simulation using multiple sequences. Statistical Science. 1992;7:457–511. [Google Scholar]
- Geyer C. MCMC R Package. 2005 At http://www.stat.umn.edu/geyer/mcmc/
- Ghys PD, Brown T, Grassly NC, Garnett G, Stanecki KA, Stover J, Walker N. The UNAIDS estimation and projection package: A software package to estimate and project national HIV epidemics. Sexual Transmitted Infections. 2004;80:i5–i9. doi: 10.1136/sti.2004.010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghys PD, Walker N, McFarland W, Miller R, Garnett GP. Improved data, methods and tools for the 2007 HIV and AIDS estimates and projections. Sexually Transmitted Infections. 2008;84:i1–i4. doi: 10.1136/sti.2008.032573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks WR, Berzuini C. Following a moving target Monte Carlo inference for dynamic Bayesian models. Journal of The Royal Statistical Society Series B. 2001;63:127–146. [Google Scholar]
- Givens GH, Raftery AE. Local adaptive importance sampling for multivariate densities with strong nonlinear relationships. Journal of the American Statistical Association. 1996;91:132–141. [Google Scholar]
- Han C, Carlin BP. Markov chain Monte Carlo methods for computing Bayes factors. Journal of the American Statistical Association. 2001;96:1122–1132. [Google Scholar]
- Hesterberg T. Weighted average importance sampling and defensive mixture distributions. Technometrics. 1995;37:185–194. [Google Scholar]
- Hoeting JA, Madigan D, Raftery AE, Volinsky CT. Bayesian model averaging: A tutorial (with discussion) Statistical Science. 1999;14:382–417. [Google Scholar]
- Hong B, Strawderman R, Swaney DP, Weinstein D. Bayesian estimation of input parameters of a nitrogen cycle model applied to a forested reference watershed, Hubbard Brook Watershed Six. Water Resources Research. 2005;41:W03007. [Google Scholar]
- Jasra A, Doucet A, Stephens DA, Holmes CC. Interacting sequential Monte Carlo samplers for trans-dimensional simulation. Computational Statistics and Data Analysis. 2008;52:1765–1791. [Google Scholar]
- Jasra A, Stephens DA, Holmes CC. On population-based simulation for static inference. Statistics and Computing. 2007;17:263–279. [Google Scholar]
- Jeffreys H. Theory of Probability. Oxford University Press; 1939. [Google Scholar]
- Kass RE, Raftery AE. Bayes factors. Journal of the American Statistical Association. 1995;90:773–795. [Google Scholar]
- Kong A, Liu J, Wong W. Sequential imputations and Bayesian missing data problems. Journal of the American Statistical Association. 1994;89:278–288. [Google Scholar]
- Leamer E. Specification Searches: Ad Hoc Inference with Nonexperimental Data. John Wiley; New York: 1978. [Google Scholar]
- Liang F. Dynamically weighted importance sampling in Monte Carlo computation. Journal of the American Statistical Association. 2002;97:807–821. [Google Scholar]
- Liang FM, Liu CH, Carroll RJ. Stochastic approximation in monte carlo computation. Journal of the American Statistical Association. 2007;102:305–320. [Google Scholar]
- Liu F, West M. A dynamic modelling strategy for Bayesian computer model emulation. Bayesian Analysis. 2009;4:393–412. [Google Scholar]
- Liu JS. Monte Carlo Strategies in Scientific Computing. Springer; New York: 2002. [Google Scholar]
- Lunn DJ, Thomas A, Best N, Spiegelhalter D. WinBUGS – a Bayesian modelling framework: Concepts, structure, and extensibility. Statistics and Computing. 2000;10:325–337. [Google Scholar]
- Madigan D, Raftery AE. Model selection and accounting for model uncertainty in graphical models using Occam’s window. Journal of the American Statistical Association. 1994;89:1335–1346. [Google Scholar]
- Martin A, Quinn K. MCMCpack: Markov chain Monte Carlo (MCMC) Package. 2007 At http://mcmcpack.wustl.edu. [Google Scholar]
- Moral PD, Doucet A, Jasra A. Sequential Monte Carlo for Bayesian computation. In: Bernardo JM, Bayarri MJ, Berger JO, Dawid AP, Heckerman D, Smith AFM, West M, editors. Bayesian Statistics 8. Oxford University Press; 2007. pp. 115–148. [Google Scholar]
- Neal P, Roberts G. Optimal scaling for partially updating mcmc algorithms. Annals of Applied Probability. 2006;16:475–515. [Google Scholar]
- Neal RM. Annealed importance sampling. Statistics and Computing. 2001;11:125–139. [Google Scholar]
- Newton MA, Raftery AE. Approximate Bayesian inference by the weighted likelihood bootstrap (with discussion) Journal of the Royal Statistical Society, Series B. 1994;56:3–48. [Google Scholar]
- Oh M. Integration of multimodal functions by Monte Carlo importance sampling. Journal of the American Statistical Association. 1993;88:450–456. [Google Scholar]
- Owen A, Zhou Y. Safe and effective importance sampling. Journal of the American Statistical Association. 2000;95:135–143. [Google Scholar]
- Plummer M, Best N, Cowles K, Vines K. CODA: Convergence diagnosis and output analysis for MCMC. R News. 2006;6:7–11. [Google Scholar]
- Poole D, Raftery AE. Inference for deterministic simulation models: The Bayesian melding approach. Journal of the American Statistical Association. 2000;95:1244–1255. [Google Scholar]
- Radtke PJ, Burk TE, Bolstad PV. Bayesian melding of a forest ecosystem model with correlated inputs. Forest Science. 2002;48:701–711. [Google Scholar]
- Raftery AE, Bao L. Department of Statistics. University of Washington; Seattle, Wash: 2009. Estimating and projecting trends in HIV/AIDS generalized epidemics using incremental mixture importance sampling. Technical Report 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery AE, Givens GH, Zeh JE. Inference from a deterministic population dynamics model for bowhead whales. Journal of the American Statistical Association. 1995;90:402–416. [Google Scholar]
- Raftery AE, Newton MA, Satagopan JM, Krivitsky P. Estimating the integrated likelihood via posterior simulation using the harmonic mean identity (with discussion) In: Bernardo JM, Bayarri MJ, Berger JO, Dawid AP, Heckerman D, Smith AFM, West M, editors. Bayesian Statistics 8. Oxford University Press; 2007. pp. 1–45. [Google Scholar]
- Raghavan N, Cox D. Adaptive mixture importance sampling. Journal of Statistical Computation and Simulation. 1998;60:237–259. [Google Scholar]
- Rubin DB. The calculation of posterior distributions by data augmentation: Comment: A noniterative sampling/importance resampling alternative to the data augmentation algorithm for creating a few imputations when fractions of missing information are modest: The SIR algorithm. Journal of the American Statistical Association. 1987;82:543–546. [Google Scholar]
- Rubin DB. Using the SIR algorithm to simulate posterior distributions. In: Bernardo JM, Berger JO, Dawid AP, Smith AFM, editors. Bayesian Statistics 3. Oxford University Press; 1988. pp. 395–402. [Google Scholar]
- Steele RJ, Raftery AE, Emond MJ. Computing normalizing constants for finite mixture models via incremental mixture importance sampling (IMIS) Journal of Computational and Graphical Statistics. 2006;15(23):712–734. [Google Scholar]
- UNAIDS . 2007 AIDS Epidemic Update. UNAIDS; Geneva: 2007. Available from http://www.unaids.org/en/Publications. [Google Scholar]
- Ventura V. Non-parametric bootstrap recycling. Statistics and Computing. 2002;12:261–271. [Google Scholar]
- Warnes G. Technical Report 395. Department of Statistics, University of Washington; Seattle, Wash: 2001. The normal kernel coupler: An adaptive Markov chain Monte Carlo method for efficiently sampling from multi-modal distributions. [Google Scholar]
- West M. Modelling with mixtures. In: Bernardo J, Berger J, Dawid A, Smith A, editors. Bayesian Statistics 4. Oxford University Press; 1992. pp. 503–524. [Google Scholar]
- West M. Approximating posterior distributions by mixtures. Journal of the Royal Statistical Society. Series B. 1993;55:409–422. [Google Scholar]
- Zhang P. Nonparametric importance sampling. Journal of the American Statistical Association. 1996;91:1245–1253. [Google Scholar]