Abstract
The cyclic diguanylate [bis-(3'–5')-cyclic dimeric guanosine monophosphate, c-di-GMP] riboswitch is the first known example of a gene-regulatory RNA that binds to a second messenger. C-di-GMP is widely employed by bacteria to regulate processes ranging from biofilm formation to the expression of virulence genes. The cocrystal structure of the c-di-GMP responsive GEMM riboswitch upstream of the tfoX gene of Vibrio cholerae reveals the second messenger binding the RNA at a three-helix junction. The 2-fold symmetric second messenger is recognized asymmetrically by the monomeric riboswitch employing canonical and non-canonical base pairing as well as intercalation. These interactions explain how the RNA discriminates against cyclic diadenylate (c-di-AMP), a putative bacterial second messenger. Small-angle X-ray scattering and biochemical analyses indicate that the RNA undergoes compaction and large-scale structural rearrangement in response to ligand binding, consistent with organization of the core three-helix junction of the riboswitch concomitant with binding of c-di-GMP.
C-di-GMP is a second messenger widespread in bacteria, where it regulates complex physiological processes associated with transition between motile-planktonic and sedentary-adhesive lifestyles, and impacts phenomena such as biofilm formation, the expression of virulence genes and the persistence of infections (reviewed in refs. 1–4). Diguanylate cyclases (DGCs), characterized by a GGDEF domain, synthesize c-di-GMP from two GTP molecules. C-di-GMP is degraded by phosphodiesterases (PDEs) with EAL and HD-GYP domains in response to various signals. Whole-genome sequencing demonstrated the ubiquity of proteins with these signatures in bacteria, where they are often found in large numbers in the genome of one organism. However, despite the ubiquity of DGCs and PDEs, only four types of effector proteins are known –the PilZ family of proteins, the FleQ transcription factor, PelD, and the I-site effectors. Thus, the mechanisms by which global changes in gene expression are modulated by c-di-GMP are a subject of intense research1–4.
Computational searches delineated a bacterial RNA motif widely associated with genes related to the environment, membranes and motility (GEMM)5. The diversity of genes associated with the GEMM motif is much broader than is typical of riboswitches, which generally only regulate a limited set of genes responsible for the biosynthesis, transport or degradation of their cognate metabolites. Therefore, it was hypothesized that the GEMM motif may respond to an intracellular signaling molecule5. Recently, it was discovered that the GEMM motif specifically binds to c-di-GMP and serves a direct role in the regulation of gene expression6, confirming predictions of the existence of such a riboswitch1,2. The c-di-GMP riboswitch is the first known example of a gene-regulatory RNA that recognizes a second messenger. It binds c-di-GMP with high specificity and Kd~1 nM. This affinity is ~1000 times tighter than that for the second messenger of an Escherichia coli PilZ c-di-GMP binding protein6, and is comparable to the tightest RNA-small molecule interactions known7. The broad distribution among bacteria of this RNA domain6, the physiological importance of c-di-GMP, and its involvement in controlling virulence of many pathogens make this riboswitch an important target for structure-function studies.
To provide a framework with which to understand how this RNA recognizes the bacterial second messenger, we have determined the cocrystal structure of a canonical c-di-GMP riboswitch from V. cholerae. The structure and previous biochemical characterization suggest that this riboswitch undergoes a conformational change upon ligand binding. We analyzed the solution behavior of the riboswitch through a combination of small-angle X-ray scattering (SAXS) and enzymatic probing. These studies indicate that c-di-GMP binding allosterically modulates the global architecture of the RNA, which switches between dramatically different free and c-di-GMP-bound conformations.
RESULTS
Crystal structure determination
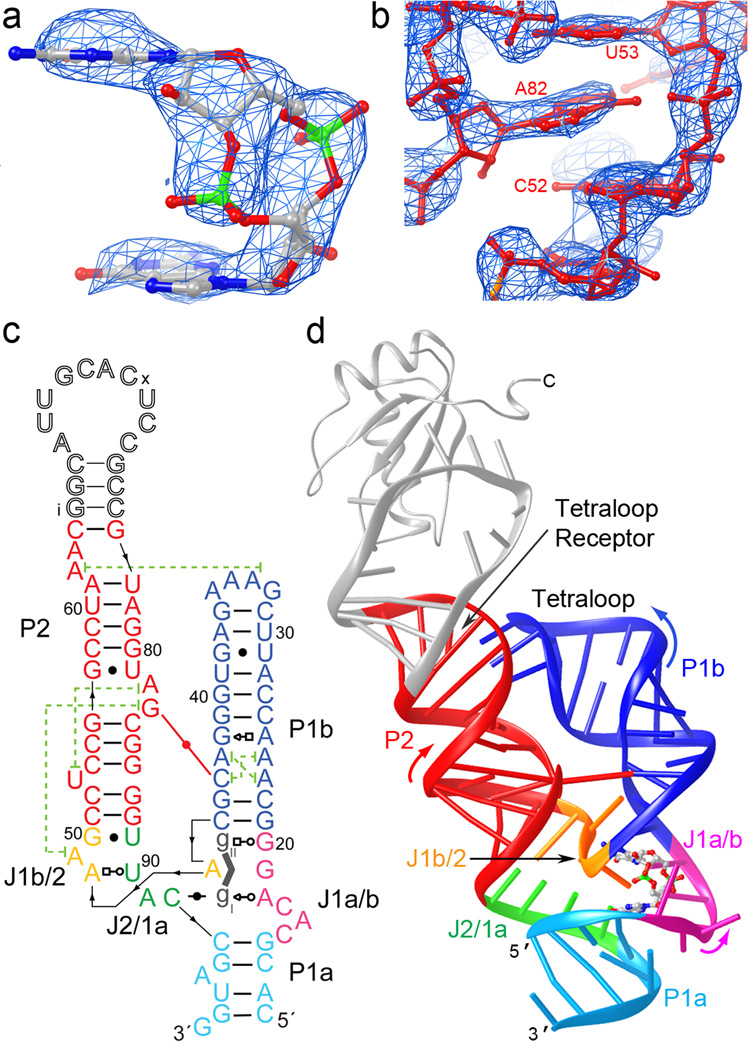
Previous analyses of the V. cholerae tfoX c-di-GMP riboswitch6 delineated the minimal 5'- and 3'- RNA boundaries needed for tight binding (Kd≤10 nM) to the second messenger (the 'aptamer domain'). A phylogenetically conserved helix (paired region P2) is closed by a loop (L2) that is highly variable5. Our crystallization construct encompasses these boundaries, and incorporates8 a binding site for the spliceosomal protein U1A in place of L2. Consistent with lack of conservation of L2, this construct binds both c-di-GMP and the U1A RNA-binding domain (RBD) (Supplementary Fig. 1). We solved the structure de novo by molecular replacement employing the U1A-RBD cognate complex9 and short A-form duplexes of arbitrary sequence as search models, analogous to what was reported for two ribozymes10,11 and an unrelated riboswitch12. This procedure13 unambiguously revealed the bound c-di-GMP in residual electron density maps phased with models that had never included the second messenger (Fig. 1a), and allowed complete tracing of both RNA molecules in the asymmetric unit (Fig. 1b). The crystallographic model (Rfree=29.2% at 3.2 Å resolution) has a mean coordinate precision of 0.5 Å (Methods and Table 1).
Figure 1.
Overall structure of the c-di-GMP riboswitch. (a) Unbiased |Fo|-|Fc| electron density corresponding to the bound c-di-GMP before it was included in the crystallographic model, superimposed on the refined model of the ligand (map contoured at 3.0 s.d.) (b) Portion of a composite simulated annealing-omit 2|Fo|-|Fc| Fourier synthesis42 calculated with the final crystallographic model, contoured around A82 at 1.5 s.d. (c) Schematic representation of the structure of the riboswitch bound to c-di-GMP. Thin black lines with arrowheads depict connectivity, dashed green lines, long-range stacking interactions, and the red line the inter-helical base pair. Leontis-Westhof symbols43 depict non-canonical base pairs. Except for the U1A binding site, the numbering scheme is that of ref. 6, which is used throughout. (d) Cartoon representation of the three-dimensional structure. Color coding as in (c). The U1A-RBD is shown as a gray ribbon.
Table 1.
Crystallographic intensity data and refinement statistics.
| Crystal I | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 31.8, 91.0, 280.1 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 30.0-3.2 (3.25- 3.2)* |
| Rmerge | 17.9 (43.8) |
| < I >/<σ(I)> | 7.9 (2.2) |
| Completeness (%) | 84.0 (75.4) |
| Redundancy | 4.4 (2.7) |
| Refinement | |
| Resolution (Å) | 30.0-3.2 (3.25-3.2) |
| No. reflections | 11257 (276) |
| Rwork / Rfree (%) | 22.2/29.2 |
| No. atoms | |
| RNA | 3984 |
| Protein | 1429 |
| c-di-GMP | 92 |
| B-factors | |
| RNA | 46.9 |
| Protein | 11.6 |
| c-di-GMP | 31.8 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.4 |
One crystal was used for the entire project.
Values in parentheses are for highest-resolution shell.
Overall ligand-bound structure
The c-di-GMP riboswitch is h-shaped (Fig. 1c,d). In addition to the two predicted helices (hereafter, P1b and P2), a third helix, P1a, is formed by the 5'- and 3'-most nucleotides of the aptamer domain, which were thought5,6 to be single-stranded. The c-di-GMP binding site is formed by nucleotides that join the three helical segments (J1a/b, J1b/2, J2/1a). One of the guanine bases of the bound c-di-GMP (hereafter, gI) stacks on the uppermost base pair of P1a, while the other (gII) stacks underneath the bottom base pair of P1b. The two nucleobases of the second messenger sandwich that of A47 from J1b/2, thus maintaining continuous base stacking between P1a and P1b. As predicted by phylogenetic analysis, P1b is capped by a GAAA tetraloop which makes canonical interactions (mediated by a ribose zipper and stacking of the base of A33 under that of A62) with a tetraloop receptor in P25,6.
A second inter-helical interaction mediating side-by-side packing of P1b and P2 is a cis-Watson-Crick pair between C44 (from P1b) and G83 (extruded from P2). Both nucleotides adopt the anti conformation, unlike in the hairpin ribozyme14,15 where the syn conformation of the guanosine making the inter-helical Watson-Crick pair enforces a closer approach of the two helices. The extrusion of G83 is accomplished by an S-shaped turn of the backbone, constructed by juxtaposition of the G57•U81 wobble pair and two base triples, that results in marked underwinding of P2 (Supplementary Fig. 2a). C44 projects into the minor groove of the overwound P1b. Overwinding results from cross-strand stacking of A24 on C44 and A43 on A23, and by formation of a sheared pair between A25 and G42 (Supplementary Fig. 2b). In addition to these non-standard conformations of P1b and P2, the inter-helical C44•G83 base pair appears to be stabilized by stacking of G83 on A49 (from J1b/2). Consistent with the importance of this inter-helical base pair, nucleotides corresponding to C44 and G83 are conserved in at least 97% of the 503 c-di-GMP riboswitches discovered by Sudarsan et al.6 in diverse bacterial genomes. Similarly, nucleotides whose interactions result in overwinding of P1b (corresponding to A23, A24 and A25) and underwinding of P2 (corresponding to U81 and A82) are 75–97% conserved6.
Second messenger recognition
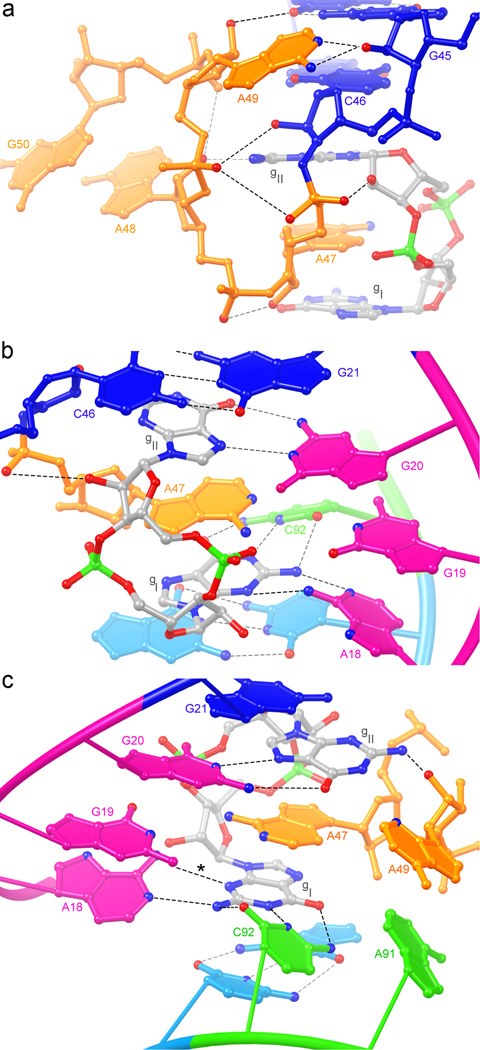
J1b/2 contributes to forming the c-di-GMP-binding site and to organizing the global architecture of the riboswitch. Nucleotides corresponding to A47, A48 and A49 are at least 97% conserved across all known c-di-GMP riboswitches, and are typically (>75%) followed by a guanine6. J1b/2 alternates between the P1a/b and the P2 stacks, thereby joining them (Fig. 1d,2a). While A47 together with the bound c-di-GMP forms part of the continuous stack between P1a and P1b, A48 forms part of the P2 stack by pairing with U90 through its Hoogsteen edge (Supplementary Fig. 3). The next residue of J1b/2, A49, flips back into P1b, stacking underneath the inter-helical C44•G83 pair. Finally, G50 pairs with U89 and forms part of the P2 stack.
Figure 2.
Specific binding of c-di-GMP. (a) The J1b/2 region stitches the three-helix junction and provides a nucleotide that intercalates between the c-di-GMP. View from the direction of Fig. 1d. Note alternation of J1b/2 residues between right (P1a/P1b) and left (P2). (b) View of the binding site from the major groove. (c) View from the minor groove. Additional interactions between the Watson-Crick faces of G19 and A47, and the sugar edge of A49 and gII are suggested by the location of hydrogen-bond donors and acceptors (colored blue or red denoting nitrogen or oxygen, respectively), but remain uncertain at the current resolution limit. Dashed lines denote putative hydrogen bonds (distance cutoff=3.8 Å; note that the mean precision of the atomic coordinates is 0.5 Å). In panel (c), the hydrogen bond marked (*) is between the N6 of A18 and the N3 of gI, not between G19 and the c-di-GMP.
Binding to the riboswitch buries 69% of the solvent-accessible surface area of the c-di-GMP. The guanine bases of the second messenger face the same direction (Supplementary Fig. 4) and are deeply buried, while the 12-member ribose-phosphate ring of c-di-GMP is partially exposed in the broad major groove, and flanked on one side by nucleotides of J1a/b (Fig. 1d,2b). At the present resolution, cations and water molecules that may mediate additional contacts between the RNA and the ribose and phosphate moieties of c-di-GMP are unresolved. The gI nucleobase of the c-di-GMP makes a Watson-Crick base pair with the highly conserved6 C92 of J2/1a (Fig. 2c), and is also recognized through its sugar-edge face by A18. The latter nucleotide is not highly conserved, suggesting that this interaction varies among different c-di-GMP riboswitches. The Watson-Crick face of G20 recognizes the Hoogsteen edge of gII, which is upside-down relative to gI. Across all known c-di-GMP riboswitches, either a guanosine or an adenosine residue is present at the position corresponding to G20 of this riboswitch6, indicating that the specific interactions used to recognize gII are somewhat variable.
Ligand binding-induced global conformational change
In-line probing analysis of the V. cholerae tfoX riboswitch6 revealed reduction in self-scission of nucleotides 13–20, 47, and 93–96 upon c-di-GMP binding. Our structure shows that residues 13–20 and 93–96 assemble into P1a and J1a/b in the second-messenger-bound form of the RNA, and that residue 47 intercalates between the two nucleobases of c-di-GMP. Together, these results suggest that the riboswitch undergoes c-di-GMP-binding induced folding. We tested this hypothesis employing SAXS.
Guinier analysis16 reveals that the radius of gyration (Rg) of the c-di-GMP riboswitch aptamer compacts in response to both c-di-GMP and Mg2+ (Supplementary Table 1). At a Mg2+ concentration of 0.5 mM (below physiologic), the Rg (~32 Å) is insensitive to saturating c-di-GMP. However, in the presence of physiologic (2.5 mM) Mg2+, the free RNA compacts (Rg=28.5 Å), and undergoes a further c-di-GMP-dependent compaction (Rg=23.9 Å). This latter Rg is the same, within experimental precision, as that calculated with a model of the riboswitch based on our crystal structure (with a wild-type length L2 modeled in; Supplementary Table 2 and Supplementary Fig. 5). At highly stabilizing Mg2+ (10 mM), the RNA compacts even in the absence of c-di-GMP to a Rg that is comparable to that of its c-di-GMP-bound form at physiologic Mg2+ (Supplementary Table 1). However, Kratky analysis16 (Fig. 3a) shows that even at 10 mM Mg2+, the free RNA exhibits behavior at high q indicative of local disorder, suggesting that the c-di-GMP-binding pocket and/or P1a remain unfolded in the absence of ligand. P(r) analysis16 (Fig. 3b) reveals a marked ligand-induced compaction of the RNA at physiologic Mg2+, and also a modest c-di-GMP-induced conformational change at 10 mM Mg2+, likely indicative of folding of the second messenger-binding pocket.
Figure 3.
C-di-GMP and Mg2+ induced size and shape changes of the riboswitch, monitored by SAXS. Red indicates 2.5 mM Mg2+; black 10 mM Mg2+. Dots denote c-di-GMP-free conditions; solid lines, c-di-GMP-bound conditions ("a.u.", arbitrary units). (a) Kratky plot suggests local disorder of the ligand-free riboswitch even under high MgCl2 concentration (black dots). (b) Electron-pair probability analysis reveals compaction of the riboswitch upon binding the second messenger.
To gain further insight into the nature of the c-di-GMP-induced conformational change, we calculated low-resolution molecular envelopes16 employing SAXS data collected at physiologic Mg2+ concentration. Reconstructions based on the ligand-bound state SAXS data employing a range of starting maximum molecular dimension (Dmax) values robustly converged into an envelope with high shape complementarity (correlation coefficient = 0.94) to our cocrystal structure (with the U1A binding site replaced by a model of identical length to wild-type L2), and Dmax~90 Å (Fig. 4a and Supplementary Fig. 5, Supplementary Fig. 6a,b). Reconstructions based on the ligand-free state SAXS data converged on a more elongated molecular envelope (Dmax~100 Å) with two prominent arms (Fig. 4b and Supplementary Fig. 6c,d). The shape of this envelope is distinctly different from that of the c-di-GMP-bound state. In addition to obvious limitations due to its low resolution, interpretation of this envelope has the caveat that the ligand-free form of the RNA might explore multiple conformations in solution. However, we found that reconstructions based on ligand-free SAXS data do converge to molecular envelopes that are self-consistent at each of the three Mg2+ concentrations analyzed. Moreover, the variability of the reconstructions for any one of the three ligand-free conditions is comparable to that of the reconstructions from the ligand-bound SAXS data (Supplementary Fig. 7). We conclude that building molecular models to fit the SAXS-based molecular envelope reconstruction of the ligand-free riboswitch is justified, keeping in mind that the level of detail implied by such models is limited to the approximate location and orientation of helices.
Figure 4.
Global rearrangement of the riboswitch induced by c-di-GMP binding. (a) Low-resolution molecular envelope reconstruction based on SAXS data of the riboswitch in the presence of c-di-GMP (at 2.5 mM Mg2+). The maximum linear dimension of the reconstruction is ~90 Å. (b) Molecular envelope based on SAXS data of the riboswitch in the absence of c-di-GMP (at 2.5 mM Mg2+). The maximum linear dimension of the reconstruction is ~100 Å. The two arms of this reconstruction have dimensions approximately corresponding to those of P2 and P1b. (c) Results of nuclease probing summarized on the bound-state secondary structure. Stars denote RNase T1 protection upon c-di-GMP binding; filled ovals, RNase V1 protection upon c-di-GMP binding; open ovals, increased RNase V1 cleavage upon c-di-GMP binding. (d) Section of the autoradiogram highlighting protections observed due to their presence in the inter-helical interface, or indirectly, from stabilization of the interface between P1b and P2. (See Supplementary Fig. 10 for complete autoradiogram; this panel shows the lanes corresponding to 3.0 mM Mg2+). "+" and "−" denote conditions with 70 µM and 0 µM c-di-GMP, respectively. "NR" and "BH" denote "no reaction" and "base hydrolysis".
In-line probing of the ligand-free riboswitch suggests that P1a is destabilized in the absence of c-di-GMP6. A molecular model of the free state based on our cocrystal structure in which the two strands of P1a have simply been separated, however, fails to recapitulate the experimentally determined change in Rg between the free and c-di-GMP-bound states (ΔRg of melted P1a model = 2.2 Å; experimental ΔRg=4.6 Å) and fits the SAXS envelope poorly (Supplementary Table 2 and Supplementary Fig. 8). We noted that the dimensions of P2 fit the long arm of the free state SAXS-based envelope, and the dimensions of P1b are a good fit for the short arm of this envelope (Fig. 4b, and Supplementary Fig. 6c,d). A model of the c-di-GMP-free state constructed by manually rotating P1b by ~180° with the center of rotation at A47 (Fig. 4b) has a calculated ΔRg=4.7 Å (Supplementary Table 2) and a correlation coefficient with the envelope of 0.91. Moreover, the scattering profile calculated from this model closely approximates the experimental scattering data, except at high q (Supplementary Fig. 9). Therefore, we assign the two arms of the ligand-free SAXS reconstruction to helices P1b and P2, while emphasizing that this model cannot capture molecular details (as underscored by the poor high-q fit).
Our tentative model of the ligand-free form of the RNA implies that in addition to unfolding of the c-di-GMP binding pocket (and possibly P1a), the inter-helical interactions mediating side-by-side packing of P1b and P2 in the c-di-GMP-bound state are broken in the ligand-free state. To test this, we subjected the riboswitch to nuclease probing. Protection from nucleases T1 and V1 is indicative of structure formation17,18. Steric clash between nucleases and a structured substrate RNA can also protect RNA segments that would otherwise be readily cleaved (refs. 19–23). For instance, in two separate studies of RNase P, it was found that relative protection from nuclease V1 was consistent with occlusion of nucleotides due to tertiary structure17 or protein binding24. RNase V1 probing of the c-di-GMP riboswitch reveals protection patterns that suggest occlusion due to structure formation. Specifically, nucleotides in the c-di-GMP binding pocket and three nucleotides found at the inter-helical interface (A37, A48, C59) become protected from cleavage by nuclease V1 upon addition of c-di-GMP (Fig. 4c,d and Supplementary Fig. 10). In addition, we find protection of guanosine residues of the riboswitch from RNase T1 in the presence of c-di-GMP, consistent with stabilization of P1a, J1a/b and inter-helical regions of the RNA. Noteworthy are c-di-GMP-induced RNase T1 protections of G32 (of the tetraloop) and G83 (of the inter-helical G83•C44 base-pair). These nucleotides are likely protected by their participation in P1b–P2 interactions. Our nuclease probing results are therefore consistent with compaction of the riboswitch induced by c-di-GMP binding, and with a free-state structure that lacks the side-by-side packing of P1b and P2 of the bound state.
DISCUSSION
The ligand-bound structures of the purine25,26 and c-di-GMP riboswitches exhibit some general resemblance to each other: both RNAs are comprised of two helical stacks that pack side-by-side, their ligand-binding pockets are both at a three-helix junction, and in both cases, the bound small molecule continues the helical stack between two of the three helices. Consistent with the dissimilarity between a free purine and c-di-GMP, the nature of the binding pockets of the two corresponding riboswitches is completely different. The long-range interactions that hold their respective two helical stacks in side-by-side arrangements are distinctly different as well, suggesting independent evolutionary origins for these two riboswitch classes.
Although the c-di-GMP riboswitch employs separate binding pockets to recognize the two nucleobases of the second messenger, the presence of both, in cyclically linked form, is required for high affinity binding (ref. 6 and Supplementary Fig. 11a). Our structure predicts that this riboswitch should discriminate against c-di-AMP, a recently discovered putative second messenger of archaea and bacteria27. If bound in an equivalent manner, the nucleobase of c-di-AMP corresponding to gI would be forced to make unfavorable hydrogen bonds with the highly conserved C92, and the other would not be able to make the interactions with G20 and A49 in which gII engages (Fig. 3c). This is borne out by electrophoretic mobility-shift analysis, in which, the riboswitch does not bind c-di-AMP even at a ligand concentration of 70 µM (Supplementary Fig. 11b). Since bacteria employ both c-di-GMP and c-di-AMP, it is possible that paralogs of the V. cholerae tfoX riboswitch might exist that recognize c-di-AMP, or that do not discriminate between the two second messengers, allowing for crosstalk between pathways. The purine riboswitches are selective for either adenine or guanine, and their selectivity derives almost exclusively from a single pyrimidine residue that Watson-Crick base pairs with their respective ligand25,26. Because the c-di-GMP riboswitch employs Watson-Crick pairing as only one of multiple strategies to recognize gI of its cognate ligand, and does not employ Watson-Crick pairing to recognize gII, it is unlikely that two point mutations would result in high-affinity (Kd~1 nM) binding of c-di-AMP. Sequence alignments6 suggest that some GEMM riboswitches have a uridine at the position that corresponds to C92, raising the possibility that they bind c-di-AMP. Although the ligand specificity of such RNAs will have to be determined experimentally, riboswitches have been shown to exhibit considerable plasticity in their binding sites28,29, and it is possible that a few mutations may allow evolution of a high-affinity c-di-AMP riboswitch from a c-di-GMP riboswitch.
Except for the glmS ribozyme-riboswitch30,31, most riboswitches function by a mechanism in which stabilization of the aptamer domain (through the free energy of binding to their cognate ligand) results in sequestration of an RNA segment that would otherwise form part of an alternative structure, for instance, a rho-independent transcriptional terminator, or a ribosome binding site32–34. Typically, this sequestered segment is part of P1 in the ligand-bound state. Analyses of the genomic contexts of c-di-GMP riboswitches5 and in vivo reporter assays6 suggest that such a mechanism is operative for at least some of these riboswitches. Our combined crystallographic, SAXS and biochemical analysis of the c-di-GMP riboswitch shows that it undergoes a pronounced global structural rearrangement in response to ligand binding (Fig. 4). This rearrangement probably involves folding of the J1a/b, J1b/2 and J2/1b regions (Fig. 1c) to form the second messenger-binding site, resulting in stabilization of helix P1a, and concomitantly, in stabilization of the side-by-side orientation of P1b and P2 through inter-helical contacts and the ligand-induced structure of the three-helix junction itself. Comparison with other studies shows that such a conformational switch is not a general property of riboswitches. SAXS analysis of the glycine riboswitch35 demonstrated that that RNA exhibits pronounced Mg2+ and ligand-dependent conformational changes. In contrast, the lysine riboswitch36,37 has been shown to adopt essentially indistinguishable three-dimensional structures in the presence or absence of the amino acid, and even in the absence of free Mg2+. Thus, it is possible that the large conformational switch we have uncovered is solely a consequence of the stabilization of the riboswitch aptamer domain structure by ligand binding. Alternatively, the conformational switch may have evolved to modulate the affinity38,39 of the riboswitch or its rate of folding40,41 so as to optimize its switching behavior in its physiological context.
METHODS
Reagent preparation and biochemical assays
We carried out transcription and purification of the 93 nucleotide (nt) crystallization RNA construct, preceded by a hammerhead ribozyme and followed by a VS ribozyme substrate stem-loop, essentially as described44,45. We prepared the 88 nt wild-type V. cholerae c-di-GMP riboswitch aptamer domain6 similarly. Ref. 8 describes U1A-RBD (Y31H, Q36R) expression and purification. We purchased c-di-GMP and c-di-AMP from Axxora, LLC (San Diego, CA), and used both without further purification.
For electrophoretic mobility shift assays (EMSA), we diluted RNA to 35 µM in a buffer comprising 5 mM Tris-HCl (pH 8.0), 3 mM MgCl2, 10 mM NaCl, and 100 mM KCl and small molecule (0 or 70 µM). Following 30' incubation at 310 K, we resolved the samples on 10% polyacrylamide gels with 0.5x THE supplemented with 1mM MgCl2 as the running buffer. We stained the gels with toluidine blue. For nuclease probing, we prepared 32P-labeled RNA in 50 mM Tris-HCl (pH 8.0), 30 mM NaCl, 30 mM KCl and appropriate final MgCl2 concentration (1.8 mM, 2.4 mM, 3.0 mM) and either 0 or 70 µM c-di-GMP. We added 1 µL of nuclease (T1, 0.01 U µl−1; V1, 0.0025 U µL−1) to initiate structural probing. We performed reactions at 310 K for 5 minutes in a final volume of 5 µL, terminated them by addition of 5 µL gel loading buffer (Ambion), and loaded them immediately onto 10% polyacrylamide-8M urea gels. We performed autoradiography by phosphorimaging. We performed nuclease mapping experiments in triplicate. We quantified cleavage bands that could be resolved as singular using ImageQuantTL software (GE Healthcare) and normalized them to a uniform band (nucleotide 17) across all lanes. We calculated protection ratios by dividing the intensity of the band in the absence of c-di-GMP by the intensity of the band in the presence of c-di-GMP. We report nucleotides with a protection ratio greater than 1.75 (T1 nuclease) and 1.25 (V1 nuclease) in Fig. 4b,4c. Similarly, we report increased cleavage by nuclease V1 for ratios less than 0.8.
Crystallization and structure determination
We mixed RNA incubated as for EMSA, but at a final concentration of 250 µM (with one molar equivalent each of c-di-GMP and U1A-RBD, and 1 mM spermine) with an equal volume of a reservoir solution comprising 30% (w/v) PEG 3350, 300 mM NH4OAc (pH 7.0), 100 mM Tris-HCl (pH 8.5). We obtained thin, plate-shaped crystals by vapor diffusion at 303 K over the course of a day. We flash froze a crystal directly from mother liquor by plunging it into liquid nitrogen. We thawed and then annealed46 this crystal in a solution comprising 35% (w/v) PEG 3350, 320 mM NH4OAc (pH 7.0), 100 mM Tris-HCl (pH 8.5), 3 mM MgCl2, and flash froze it in the same manner. We collected diffraction data by the oscillation method with 1 Å X-radiation at 100 K from one crystal at beamline 8.2.2 of the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory, and reduced the data with the HKL package47. High crystal mosaicity (~0.9°) and the relatively long c unit cell dimension limited the completeness and precision of the data.
We initiated structure determination by molecular replacement with PHASER48 with four independent search models (two U1A-RBDs and their binding sites, and two 5 bp duplexes). In the first round, PHASER successfully placed two U1A-RBDs and their binding sites and one of the duplexes (translation function Z-scores of 10.0, 16.8, and 10.1, respectively, and an overall log-likelihood gain of 446). We carried out iterative rounds of simulated annealing, energy minimization, and tightly restrained individual B-factor refinement42 (with a random 10% of the data reserved for cross-validation), interspersed with further rounds of molecular replacement to achieve complete tracing49 of both RNA-protein complexes in the asymmetric unit. This model had a free-R factor of 31.4%, and a residual Fourier synthesis calculated with phases derived from it revealed clear electron density for the two bound c-di-GMP molecules (Fig. 1a). We manually placed the second messengers into the residual electron density, and carried out further refinement, which smoothly reduced the free-R factor to 29.2%. We employed an anisotropic B-factor correction and an automatic solvent mask throughout. We did not employ non-crystallographic symmetry restraints. We chose B-factor restraints to minimize the free-R factor (the r.m.s.d. of B-factors of covalently bonded atoms is 0.49 Å2). The current model has a cross-validated σA coordinate precision of 0.54 Å. 95.8% of residues of the U1A-RBDs lie in the most favored and additionally allowed regions of the Ramachandran plot50. There are no residues with disallowed backbone conformations. We prepared crystal structure figures with RIBBONS51 using the coordinates of RNA chain A.
Small-angle X-ray scattering
We prepared SAXS samples similarly to the nuclease probing samples, with slight modifications (20 µM RNA, 100 µL final volume). We also prepared identical buffers (Supplementary Methods). We collected data under continuous flow at 310 K at the BioCAT beamline of the Advanced Photon Source (APS), Argonne National Laboratory. We analyzed buffer-subtracted data using IGOR (Wavemetrics, Inc.). We used Guinier analysis in the range q*Rg~1 to calculate the radii of gyration. We averaged multiple DAMMIN52 reconstructions using DAMAVER53. We converted the filtered, averaged reconstructions to electron density map using SITUS54. We display these molecular envelope reconstructions with CHIMERA55. We computed the scattering profile of molecular models using CRYSOL56.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the staff of ALS beamline 8.2.2 and J. Bolduc for assistance with synchrotron and home laboratory single-crystal diffraction data collection, respectively, L. Guo from BioCAT at APS for assistance with SAXS data collection, and T. Hamma, J. Pitt, J. Posakony, A. Roll-Mecak, and H. Suga for discussions. Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38. BioCAT is a NIH-supported Research Center (RR-08630). This work was supported by the Howard Hughes Medical Institute (HHMI) and the W.M. Keck Foundation. A.R.F. is an Investigator of the HHMI.
Footnotes
Accession codes
Protein Data Bank: Coordinates and structure factor amplitudes for the V. cholerae tfoX c-di-GMP riboswitch in complex with c-di-GMP have been deposited with accession code 3IWN.
AUTHOR CONTRIBUTIONS N.K. designed and prepared RNA constructs, analyzed ligand binding, obtained crystals, carried out diffraction data collection, and participated in structure determination and in SAXS data collection. N.B. participated in SAXS data collection, analyzed the SAXS data, and designed, performed and analyzed the nuclease probing experiments. A.F. participated in diffraction data collection, structure determination and refinement. All authors contributed to manuscript preparation.
REFERENCES
- 1.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 2.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfe A, Visick K. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr Opin Microbiol. 2009;12:170–176. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg Z, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudarsan, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao H, Edwards TE, Ferré-D'Amaré AR. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem Biol. 2008;15:1125–1137. doi: 10.1016/j.chembiol.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferré-D'Amaré AR, Doudna JA. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J Mol Biol. 2000;295:541–556. doi: 10.1006/jmbi.1999.3398. [DOI] [PubMed] [Google Scholar]
- 9.Oubridge C, Ito N, Evans PR, Teo C-H, Nagai K. Crystal structure at 1.92Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 10.Robertson MP, Scott WG. The structural basis of ribozyme-catalyzed RNA assembly. Science. 2007;315:1549–1553. doi: 10.1126/science.1136231. [DOI] [PubMed] [Google Scholar]
- 11.Xiao H, Murakami H, Suga H, Ferré-D'Amaré AR. Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme. Nature. 2008;454:358–361. doi: 10.1038/nature07033. [DOI] [PubMed] [Google Scholar]
- 12.Klein D, Edwards T, Ferré-D'Amaré A. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson MP, Scott WG. A general method for phasing novel complex RNA crystal structures without heavy-atom derivatives. Acta Crystallogr D. 2008;64:738–744. doi: 10.1107/S0907444908011578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rupert PB, Ferré-D'Amaré AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 15.Ferré-D'Amaré AR. The hairpin ribozyme. Biopolymers. 2004;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- 16.Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annual Rev Biophys Biomolec Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 17.Baird N, Westhof E, Qin H, Pan T, Sosnick T. Structure of a folding intermediate reveals the interplay between core and peripheral elements in RNA Folding. J Mol Biol. 2005;352:712–722. doi: 10.1016/j.jmb.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Ehresmann C, et al. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dock-Bregeon AC, Garcia A, Giegé R, Moras D. The contacts of yeast tRNA(Ser) with seryl-tRNA synthetase studied by footprinting experiments. Eur J Biochem. 1990;188:283–290. doi: 10.1111/j.1432-1033.1990.tb15401.x. [DOI] [PubMed] [Google Scholar]
- 20.Mei HY, et al. Inhibitors of protein-RNA complexation that target the RNA: specific recognition of human immunodeficiency virus type 1 TAR RNA by small organic molecules. Biochemistry. 1998;37:14204–14212. doi: 10.1021/bi981308u. [DOI] [PubMed] [Google Scholar]
- 21.Hallegger M, Taschner A, Jantsch MF. RNA aptamers binding the double-stranded RNA-binding domain. RNA. 2006;12:1993–2004. doi: 10.1261/rna.125506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pouch-Pélissier MN, et al. SINE RNA induces severe developmental defects in Arabidopsis thaliana and interacts with HYL1 (DRB1), a key member of the DCL1 complex. PLoS Genet. 2008;4:e1000096. doi: 10.1371/journal.pgen.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savochkina L, Alekseenkova V, Belyanko T, Dobrynina N, Beabealashvilli R. RNA protections from RNase V1 due to RNA structure alone. BMC Res Notes. 2008;1:15. doi: 10.1186/1756-0500-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esakova O, Perederina A, Quan C, Schmitt ME, Krasilnikov AS. Footprinting analysis demonstrates extensive similarity between eukaryotic RNase P and RNase MRP holoenzymes. RNA. 2008;14:1558–1567. doi: 10.1261/rna.1106408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 26.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–17241. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witte G, Hartung S, Büttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 28.Edwards TE, Ferré-D'Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein DJ, Been MD, Ferré-D'Amaré AR. Essential role of an active-site guanine in glmS ribozyme catalysis. J. Am. Chem. Soc. 2007;129:14858–14859. doi: 10.1021/ja0768441. [DOI] [PubMed] [Google Scholar]
- 32.Edwards TE, Klein DJ, Ferré-D'Amaré AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Op Struct Biol. 2007;17:273–279. doi: 10.1016/j.sbi.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Henkin T. Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 2008;22:3383–3390. doi: 10.1101/gad.1747308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serganov A. The long and the short of riboswitches. Curr Op Struct Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipfert J, et al. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serganov A, Huang L, Patel D. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garst AD, Héroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulhbacher J, Lafontaine DA. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomsic J, Mcdaniel B, Grundy F, Henkin T. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM in vivo and in vitro. J Bacteriol. 2007;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 41.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 42.Brünger AT, et al. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferré-D'Amaré AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5' and 3' heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rupert PB, Ferré-D'Amaré AR. Crystallization of the hairpin ribozyme: illustrative protocols. Meth Mol Biol. 2004;252:303–311. doi: 10.1385/1-59259-746-7:303. [DOI] [PubMed] [Google Scholar]
- 46.Heras B, Martin JL. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr D. 2005;61:1173–1180. doi: 10.1107/S0907444905019451. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 50.Laskowski RJ, Macarthur MW, Moss DS, Thornton JM. PROCHECK: a program to check stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–290. [Google Scholar]
- 51.Carson M. Ribbons. Meth Enzymol. 1997;277:493–505. [PubMed] [Google Scholar]
- 52.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophysical J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 55.Pettersen E, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56.Svergun DI, Bargerato C, Koch MHJ. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.