Abstract
microRNAs (miRNAs) regulate numerous physiological processes such as cell division and differentiation in many tissue types including stem cells. To probe the role that miRNAs play in regulating processes relevant to embryonic stem cell biology, we used RNA interference to silence DICER and DROSHA, the two main miRNA processing enzymes. Consistent with a role for miRNAs in maintaining normal stem cell division and renewal, we found that perturbation of miRNA pathway function in human embryonic stem cells (hESCs) attenuates cell proliferation. Normal cell growth can be partially restored by introduction of the mature miRNAs miR-195 and miR-372. These miRNAs regulate two tumor suppressor genes, respectively: WEE1, which encodes a negative G2/M kinase modulator of the CycB/CDK complex and CDKN1A, which encodes p21, a CycE/CDK cyclin dependent kinase inhibitor that regulates the G1/S transition. We show that in wild-type hESCs, WEE1 levels control the rate of heSC division, whereas p21 levels must be maintained at a low level for hESC division to proceed. These data support a model for hESC cell cycle control in which miRNAs regulate negative cell cycle modulators at two phases of the cell cycle to ensure proper replenishment of the stem cell population.
Keywords: microRNAs, cell cycle, human embryonic stem cells, cell division, CDKN1A/p21, WEE1
Introduction
The formation of embryonic tissues as well as the renewal of adult tissues rely on stem cells. Because of their ability to assume almost any cell fate if given the appropriate cues, embryonic stem cells are considered to be pluripotent. Unlike other cell types, most stem cells maintain the capacity to divide throughout the life of the organism. Although stem cell research has intensified in recent years, the mechanisms that tightly regulate stem cell division are not fully understood.
Cell cycle progression is regulated by the sequential activation and inactivation of cyclin-dependent kinases (CDKs) by cyclins. Built-in cell cycle checkpoints control progression through the S and M phases of the cell cycle. One checkpoint mechanism that determines whether cells will continue proliferating or exit the cell cycle operates mainly in G1 and utilizes, among other regulators, the CDK inhibitor, CDKN1A/p21.1,77 Many other regulatory checkpoints have also been discovered,2 and it has been proposed that different cell types might be differentially sensitive to each checkpoint.
Whereas adult stem cells use the G1/S regulators p21 and p27 to control the timing of cell division, it is likely that embryonic stem cells (ESCs) do not actively respond to DNA damage-induced G1 checkpoints.3,4 ESCs exhibit an abbreviated cell cycle due to a short G1/S transition, and the cell cycle control mechanisms that normally function in G1 after DNA damage are reduced or absent.5 Accordingly, mouse ES cells (mESCs) do not show growth arrest after irradiation.6 Furthermore, p53 and several genes known to cooperate with p53 in the regulation of cell cycle, including p16, p19 and p21, are expressed at low levels, whereas DNA replication and repair related genes are expressed at high levels in mESCs and hESCs.7–9 The mechanism of dampening or regulation of the gap-phase checkpoints in hESCs is not well understood. However, recent work has shed light on some of the mechanisms that might be involved; the proliferation of mESCs is at least partly controlled during the DNA synthesis phase, independent of the DNA damage checkpoint9 and Nanog, a key stem cell transcription factor has been shown to have a positive role in the G1 to S transition by transcriptionally regulating CDK6 and CDC25A.11
miRNAs are especially attractive candidates for regulating stem cell proliferation and self-renewal as their ability to regulate many target genes simultaneously provides a means for coordinated control of cellular processes. miRNAs are endogenously transcribed in the nucleus and sequentially processed by the RNase III enzymes, DROSHA and DICER into small 20–22 nucleotide miRNAs.12,13 The mature miRNAs are loaded into the RNA-induced silencing complex (RISC), which facilitates interaction between miRNAs and their targets through imperfect base-pairing to transcript sequences that most often lie within the 3' untranslated region (3'UTR). This interaction leads to negative regulation of translation and/or destabilization of the mRNA.14
Among other processes, miRNAs have been shown to function in stem cell division, maintenance and differentiation.15,78,79 Accordingly, some miRNAs are differentially expressed in stem cells, suggesting a specialized role in stem cell regulation.19–21 Recent work with mouse ESCs and Drosophila germ line stem cells (GSCs) mutant for components of the miRNA pathway lent support to the idea that miRNAs are important regulators of stem cell self-renewal and division.21–31 In mice, ES cells deficient in the miRNA pathway both fail to differentiate and proliferate more slowly compared with controls.29–32 These data suggest that the miRNA pathway is required for proper cell cycle progression in stem cells. To define cell cycle stages that are negatively affected by defects in the miRNA pathway, a detailed analysis in Drosophila GSCs was pursued. Tracking of cell cycle stage markers revealed that Dicer-1 GSCs were delayed in the CDK-inhibitor p21/p27/Dacapo-dependent G1/S transition, suggesting that miRNAs are required for GSCs to bypass the normal G1/S checkpoint.23 Similarly, work with mouse ESCs revealed that miRNAs function to maintain low levels of p21, which normally inhibits the G1/S transition.32
By reducing DICER and DROSHA, two enzymes needed for the processing of mature miRNA, we show that miRNAs are critical for hESC self-renewal. hESCs lacking the full complement of mature miRNAs due to gene knockdown of Dicer or Drosha are defective in stem cell proliferation. In parallel, our profiling of hESC miRNAs has revealed a candidate group of miRNAs that are highly enriched in hESCs compared with differentiated cells. We show that two miRNAs from that group, miR-195 and miR-372, can partially rescue the hESC cell division defect. Furthermore, in the wild-type hESC line H1, miR-195 overexpression is sufficient to reduce WEE1 levels and increase BrdU incorporation. This increase in BrdU incorporation is WEE1 dependent as reduction of WEE1 by RNAi increases BrdU incorporation two-fold. Therefore, WEE1 appears to be a critical component of hESC cell cycle regulation. miR-372 overexpression dramatically reduces p21 levels in Dicer-knockdown hESCs and HeLa cells. Overexpression of p21 reduces phospho-RB levels by 80% and BrdU incorporation by 40% showing that maintenance of a low level of p21 is also critical for hESC division. On the basis of our results, we propose that these two miRNAs cooperate in maintaining the proliferative capacity of hESCs.
Results
Dicer and Drosha can be knocked down by lentivirus-delivered shRNAs in hESCs
If miRNAs have an important function in hESC biology, then defects in the RNAi pathway should impact hESC self-renewal and/or proliferation. To examine the effects of global miRNA loss of function in hESCs we individually silenced the miRNA processing enzymes DROSHA and DICER by transducing cells with lentiviruses encoding short hairpin RNAs (shRNAs) targeting the associated gene transcripts. Along with regulating miRNA biogenesis, each enzyme has miRNA-independent functions. Furthermore, some miRNAs, for example mirtrons, bypass DROSHA but not DICER processing.33 Therefore, phenotypic overlaps between cells with either enzyme silenced would suggest a role for miRNAs in those cellular processes. To silence these genes, we first surveyed multiple commercially available lentiviral constructs designed to target the Dicer or Drosha transcripts for efficient gene suppression in HeLa cells by transient transfection of the corresponding plasmid DNA. We used the best performing constructs to generate viral stocks and stably transduced two separate hESC lines, H1 and H7. As a control, we stably transduced cells with the corresponding empty vectors (pLKO.1 and pCMV-GIN).
Figure 1 shows the performance of the different shRNA constructs in H1 cells as determined by real-time quantitative PCR (qPCR). We observed robust reduction of Dicer or Drosha mRNA levels in the hESC line H1 (6.3- and 5.3-fold for Dicer and 2.3- and 3.6-fold for Drosha). Similarly, we observed 4.5-fold reduction of Dicer in the hESC line H7 (Fig. 1C). We verified functional impacts of Dicer and Drosha silencing in these cells by showing that mature miRNAs were highly reduced (from ~3 to 54-fold for mir-10, miR-218 and miR-372; Fig. 1D). Thus it appears that the shRNAs we used silenced the two genes sufficiently to cause a phenotype relevant to miRNA processing.
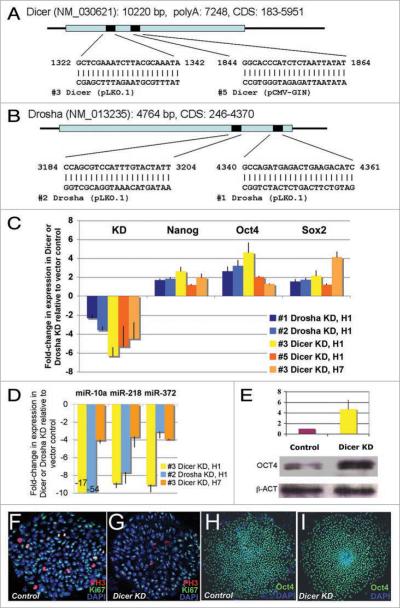
Figure 1.
Dicer and Drosha knockdown H1 hESCs. (A and B) two different lentiviral short hairpin RNA (shRNA) constructs per gene were used in hESCs to knock down Dicer (A) or Drosha (B). (C) the silencing levels for Drosha and Dicer in the H1 lines and Dicer in the H7 line are shown as fold changes in expression compared with cells treated with the empty vector control lines. The mean (±SD) was calculated from at least three independent experiments. Stem cell regulators, Nanog, Oct4 and Sox2 were upregulated in these knockdown lines. Standard deviations were calculated from 6 independent experiments for Nanog and Oct4 and two experiments for Sox2 in H1 and H7 cells. (D) the levels of mature miRNAs 10a, 218 and 372 were reduced in the Drosha- or Dicer-knockdown hESCs. The mean (±SD) was calculated from three independent experiments. (E) OCt4 protein expression was increased four-fold in the Dicer knockdown H1 compared with the control line. The mean (±range) was calculated from two independent experiments. (F and G) Morphologically the Dicer knockdown H1 hESC colonies are similar to control H1 hESC colonies. Similar colony structures were observed for Drosha knockdown lines #1 and #2, Dicer knockdown lines #3 and #5 and control lines #4 and #7. Immunofluorescence staining for DAPI, Ki67 and phospho-Histone 3 were examined by confocal microscopy and are represented in blue, green and red respectively. (H and I) Dicer-knockdown H1 hESCs and the control cells both express the stem cell marker OCT4. OCT4 is green and DAPI is blue.
Dicer- or Drosha-knockdown hESC lines retain expression of stem cell-specific markers
To identify the processes that are most sensitive to perturbations in mature miRNA levels in hESCs, we first tested whether the Dicer- and Drosha-knockdown lines showed abnormal colony morphology and whether they continued to express stem cell markers such as Nanog, Oct4 and Sox2. Nanog, Oct4 and Sox2 are highly expressed in mouse and human embryonic stem cell lines34,35 and are required for stem cell pluripotency.36–40 In hESCs, downregulation of either Nanog, Oct4 or Sox2 leads to differentiation.41–43 Moreover, experiments in which primary cells are reprogrammed into induced-pluripotent stem cells (iPSCs) revealed that OCT4 and SOX2 were essential for this reprogramming,25,44 again supporting a critical role for these genes in maintaining stem cell properties.
Interestingly, our Dicer- and Drosha-knockdown hESCs still appeared hESC-like with characteristic hESC colony morphology and Oct4 expression (Fig. 1F–I). Further, the levels of Nanog, Oct4 and Sox2 were upregulated in the H1 Drosha- and Dicer-knockdown lines and the H7 Dicer-knockdown line (Fig. 1C). We confirmed that the OCT4 protein was also upregulated in the H1 Dicer-knockdown line (4.7-fold upregulation; Fig. 1E) and that most early differentiation markers were not upregulated compared with levels observed in the control line (Table S1).
Because stem cell markers are upregulated in Drosha- and Dicer-knockdown cells, it is possible that these hESC markers may be regulated by miRNAs. Indeed, recent reports have shown that critical stem cell markers in human and mouse ESCs are regulated by miRNAs.45,46 In support of this hypothesis, the miRNA that was shown to act as a rheostat for stem cell markers,45 miR-145, is significantly downregulated in these lines (3.7-fold and 8.5-fold downregulation in Dicer and Drosha-knockdown hESCs, respectively).
Previous work has shown that overexpression of key stem cell markers in mESCs or hESCs causes defects in stemness.11,38,47,80 NANOG overexpression in hESCs has been shown to enable feeder-free growth while inducing aspects of primitive ectoderm,47 and to hasten S-phase entry.11 However, our Drosha- and Dicer-knockdown hESC lines appear morphologically similar to wild-type hESCs, the primitive ectoderm marker FGF5 is not upregulated, the inner cell mass (ICM) markers, REX1 and GBX2 are not downregulated (Table S2) and the cell cycle rate is attenuated, not increased (Fig. 2A).
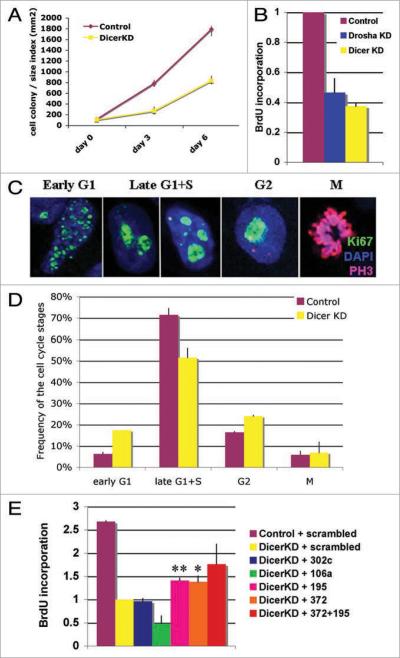
Figure 2.
Dicer and Drosha knockdown in H1 hESCs results in a cell division defect. (A) The Dicer-knockdown hESC line H1(#3) grows significantly slower than the control line (#4) based on a colony count assay. (B) Both Drosha- (#1) and Dicer- (#3) knockdown lines show significant reduction in BrdU incorporation compared with the control line (#4). (C and D) Based on Ki67 staining patterns (C), cells were categorized into different cell cycle phases. (D) Dicer-knockdown H1 hESCs (yellow bars) show an increase in frequency of G1 and G2 stages and a decrease in S-phase frequency compared to the control lines (brown bars). (E) miR-195 and miR-372 partially rescue the cell cycle defect in Dicer-knockdown hESCs as determined by BrdU incorporation assay. The standard deviations or ranges are calculated from two independent experiments for miR-302c and 372 + 195, three experiments for miR-106a and four for miR-195 and 372. The miRNA overexpression was verified by qPCR analysis.
Because OCT4, NANOG and SOX2 proteins form a transcriptional complex,48 we propose that simultaneous changes of Oct4, Nanog and/or Sox2 levels overcome the defects observed when each individual component is independently overexpressed. Because it has been proposed that reciprocal inhibition between lineage-specific transcription factors might be involved in the first events to occur during differentiation,35,49,50 the outcome of upregulation of all three stem cell markers might be very different from Nanog or Oct4 upregulation alone.
Dicer and Drosha are required for hESC cell cycle control
Cells lacking Dicer or Drosha grew significantly slower than cells transduced with control lentiviral vectors (Fig. 2A and B). To monitor cell growth, we counted the mean cell colony number multiplied by the mean colony area and found that Dicer-knockdown cells grow 2.6-fold slower than control cells (Fig. 2A). Similarly, BrdU incorporation assays revealed that Dicer- and Drosha-knockdown cells incorporate 2.6- and 2.5-fold less BrdU than control cells, respectively (p < 0.009 or 0.0075; Fig. 2B). A similar, but less severe, defect was observed in the H7 Dicer-knockdown line (1.5-fold reduction, p < 0.007). These data suggest that both Dicer and Drosha knockdown affect hESC cell growth. To investigate this further, we have focused on analyzing the Dicer-knockdown line.
To understand the cause of slow growth and in particular, to uncover the cell cycle stages that might be delayed, we assessed Ki67 patterns in Dicer-knockdown and control hESC lines. Ki67 antibody staining has been used to track different cell cycle stages; Ki67 shows no staining in G0 and distinct staining patterns in early G1, late G1 + S, G2 and M phase.50 hESCs exhibit atypical cell cycle profiles compared with other cell lines showing a longer S phase and shorter G1 phase (Fig. 2D and reference 5). As shown in Figure 2D, Ki67 analysis revealed an increase in early G1 and G2 and a decrease in late G1/S in Dicer-knockdown cells compared with control lines. These data suggest that reduction of miRNAs results in G1/S and G2/M transition delays in hESCs. We hypothesized that critical checkpoint regulators normally controlled by miRNAs in G1 and G2 were upregulated in the Dicer-knockdown lines and performed genome wide miRNA qPCR and mRNA microarray profiling to identify such genes.
qPCR-based miRNA profiling identifies differentially expressed miRNAs in hESC lines H1, H7 and HSF6
To identify the miRNAs responsible for the defects observed in Dicer- and Drosha-knockdown lines, we first looked for miRNAs that are enriched in the hESCs used in our studies. Recent findings suggest that a specific group of miRNAs is expressed in mESCs or hESCs.19,20,52–55 We tested whether the same group of miRNAs is enriched in the hESC lines used in this study: H1, H7 and HSF6. miRNAs expressed in these three hESC-lines were measured by qPCR with probe sets for 220 human miRNAs.56 miRNA profiling in these hESC lines revealed a set of miRNAs that was enriched in these stem cell lines (hierarchical clustering analysis Fig. S1A, a comparison between three hESC lines and human brain tissue). As expected, the ES-specific miRNAs in the miR-302 family were expressed at high levels in all three hESC lines but not in the brain sample (Fig. S1B). These analyses revealed a “Top-20” list of miRNAs expressed most abundantly in all three hESC lines (Table S3). Nine of our top 20 hESC miRNAs had been cloned previously from hESCs,20 15 had been identified by H1 hESC deep sequencing,55 and some had been shown to associate with RNA Polymerase II by CHIP-microarrays.48
We also tested which of the identified miRNAs were differentially expressed during the hESC early commitment stage by use of the four day serum forced differentiation paradigm.57 We isolated RNA from hESC lines before and after a four-day serum-forced differentiation to analyze the miRNAs in these two populations using qPCR. In particular, this short serum-induced differentiation paradigm should allow us to identify miRNAs that are altered in the earliest commitment step in hESC differentiation. To confirm that the cells have initiated differentiation, we measured changes in Nanog and Oct4 levels by qPCR analysis and showed that both stem cell markers are partially reduced in all three hESC lines (Fig. S1C). We identified 19 miRNAs that are enriched in all three hESC lines and are downregulated after four days of serum-induced differentiation (Fig. S1D). From these miRNAs, we identified a candidate set to test for their ability to rescue Dicer-knockdown cell division defects when introduced into the line as mature miRNA mimics.
miR-195 and miR-372 can partially rescue Dicer knockdown phenotypes
To analyze which mature miRNAs can rescue Dicer knockdown phenotypes, we set up a rescue assay in which we introduced mature miRNA mimic duplexes into Dicer-knockdown hESC lines by electroporation and tested the effect of the miRNAs on cell division kinetics by BrdU-incorporation. Four miRNAs from the candidate groups (Fig. S1D) were chosen for the Dicer-knockdown rescue-studies: miR-302c, miR-106a, miR-372 and miR-195, based on the fact that these miRNAs have been shown previously to control cell cycle regulators.58–60 We found that these four miRNAs were considerably downregulated in the Dicer-knockdown line (Figs. 1D and S2). Three of these miRNAs, miR-302c, -106a and -372 have a very similar seed sequence (5'-(a)aagugcu) whereas the miR-195 seed differs (5'-agcagcac). We compared the effects of mimics of these miRNAs to those of a scrambled small RNA (see Table S5). Four days after the introduction of the mature miRNA mimics, we tested the Dicer-knockdown cells for their capacity for DNA replication by use of a BrdU-assay and examined their stem cell character by analysis of Nanog, Oct4 and Sox2 mRNA levels. Some of the tested miRNAs, such as miR-302c, had no obvious rescue effect on cell cycle kinetics (Fig. 2E) or stem cell character in the Dicer-knockdown lines (Fig. S3). However, miR-195 and miR-372 partially rescued the BrdU incorporation defect (Fig. 2E). In addition, the combination of miR-195 and miR-372 had a more potent rescue capacity than either of these miRNAs alone (Fig. 2E). However, Nanog, Oct4 and Sox2 levels were similar or higher than the control levels, suggesting that stem cell characteristics were not dramatically altered in the Dicer-knockdown line by overexpression of miR-195 and miR-372 (Fig. S3A).
miR-195 is sufficient to promote hESC cell cycle
We further tested whether any of the candidate miRNAs affects the cell cycle in wild-type H1 hESCs. The same set of mature miRNAs (302c, 106a, 195 and 372) was introduced into the wild-type H1 line, BrdU-incorporation was measured and Nanog, Oct4 and Sox2 levels were analyzed by qPCR. When overexpressed in wild-type H1 hESCs, miR-372 had no effect on BrdU incorporation (Fig. S3B). However miR-195 overexpression in H1 hESCs was sufficient to increase BrdU incorporation, but had no effect on the levels of Nanog, Oct4 and Sox2 (Fig. S3C). These results are consistent with a hypothesis that miR-372 might target a cell cycle component that is already downregulated in wild type hESCs whereas miR-195 might regulate a cell cycle controller that is active in hESCs.
miR-195 regulates WEE1
To test which cell cycle stage is rescued by miR-195 in the Dicer-knockdown line, we analyzed the frequency of different cell cycle stages using the Ki67 staining in control and Dicer-knockdown cells transfected with scrambled RNA as well as in Dicer-knockdown cells transfected with mature miR-195. We found that miR-195 rescued the G2 defect in the Dicer-knockdown line (Fig. 3A). Therefore, taken together, our data on miR-195 overexpression suggest that miR-195 might directly or indirectly act on targets that normally inhibit the rate of cell cycle at the G2/M transition in hESCs. Downregulation of Cyclin/CDK complex activity either by upregulating inhibitors or downregulating ubiquitin-based degradation of cyclin are potential mechanisms controlling this process. One such potential miR-195 cell cycle target is WEE1 kinase, a negative regulator of the CycB/CDK complex in the G2/M transition that contains two predicted miR-195 binding sites in its 3'UTR (TargetScan, Fig. 3D).
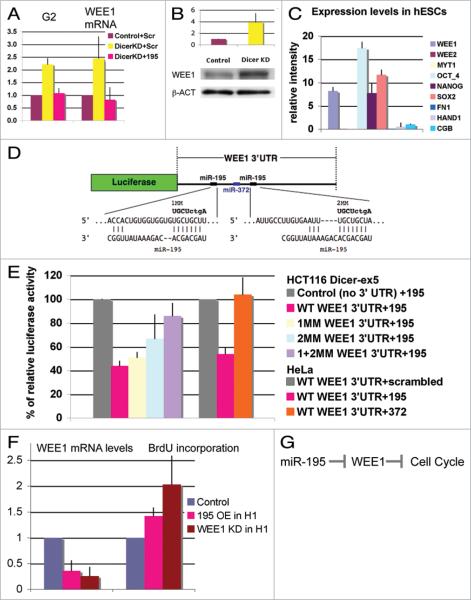
Figure 3.
miR-195 regulates normal hESC self-renewing division through the cell cycle regulator WEE1 kinase. (A) The percentage of cells in the G2 phase (Ki67 analysis) increases and WEE1 mRNA levels (qPCR analysis) are upregulated in the Dicer-knockdown line. Both of these defects can be rescued by overexpression of miR-195 in Dicer-knockdown cells by use of miR-195 duplexes. (B) Western blot analysis shows a four-fold upregulation of WEE1 protein in Dicer-knockdown H1 line #3 compared with control cell line #4. (A and B) Mean ± range was calculated from two independent experiments. (C) On the basis of microarray analysis, WEE1 is expressed at similar levels as the stem cell markers Nanog, Oct4 and Sox2, whereas WEE2 and MYT1 as well as differentiation markers are very low or undetectable. Mean ± SD was calculated from nine different hESC lines. Similar results were obtained with two different mRNA microarray platforms (Affymetrix and Agilent). (D) Diagram of the WEE1 3'UTR luciferase reporter construct. The entire WEE1 3'UTR was inserted after the luciferase open reading frame. Two miR-195 sites and one miR-372 site predicted by TargetScan are present in the WEE1 3'UTR. miR-195 and its complementary sequences in the WEE1 3'UTR as well as the mismatches (MM) made in miR-195 seed sequences are represented. (E) Cotransfection of this construct with miR-195 into HCT116 Dicer-ex5 cells results in downregulation of luciferase activity. The data are normalized to luciferase activity after cotransfection with miR-149, which has no predicted target site in WEE1 3'UTR, and, therefore, serves as a negative control. Mismatches of the miR-195 site(s) in WEE1-3'UTR release the repression of luciferase activity caused by miR-195 overexpression. The control plasmid used is SGC (SwitchGear Genomics), an empty vector with a luciferase cassette, but no inserted 3' UTR. Cotransfection of WT WEE1 3' UTR reporter construct with miR-195 in HeLa cells shows similar downregulation of luciferase activity whereas cotransfection with miR-372 has no effect. (F) The level of WEE1 mRNA is reduced upon miR-195 overexpression, in comparison with the overexpression of scrambled oligonucleotides or a miR-195 oligonucleotide bearing seed region mismatch mutations. BrdU incorporation is increased when miR-195 is overexpressed in wild-type H1 hESCs. WEE1 knockdown by siRNAs shows a similar effect, BrdU incorporation is increased 2-fold. Mean ± SD or range was calculated from three independent experiments for miR-195 OE and two independent experiments for WEE1 KD. (G) Model of miR-195 function in hESC cell cycle.
We validated microarray data suggesting that WEE1 is upregulated in Dicer- and Drosha-knockdown lines by measuring WEE1 levels using qPCR in these lines. The qPCR analysis showed that WEE1 mRNA levels were significantly upregulated in Dicer- (2.5 fold increase; Fig. 3A) and Drosha-knockdown lines (6.0-fold increase). We also observed elevated WEE1 protein levels in the Dicer-knockdown line (4-fold increase; Fig. 3B). Furthermore, the observed WEE1 upregulation in the Dicer-knockdown line could be rescued by overexpression of mature miR-195 (Fig. 3A). These data show that WEE1 levels are responsive to perturbations in the levels of mature miRNAs in hESCs and that at least one miRNA, miR-195, is capable of restoring normal growth properties to cells that divide slowly as a result of a compromised miRNA pathway.
To test whether miR-195 can regulate WEE1 directly by targeting its 3'UTR, we used a luciferase assay in a HCT116 Dicer hypomorph line.61 Importantly, we found that overexpression of miR-195 results in reduction of luciferase activity in the WEE1 3'UTR reporter construct (Fig. 3D and E). Mutations of the predicted miR-195 target sites in the WEE1 3'UTR relieve the repression of luciferase activity mediated by miR-195 (Fig. 3D and E). Although the WEE1 3'UTR also has a predicted site for miR-372, overexpression of miR-372 did not repress WEE1 3'UTR in the luciferase assay (Fig. 3D and E). Therefore these data show that miR-195 is sufficient to directly regulate the WEE1 3'UTR.
miR-195 regulates the hESC cell cycle by affecting WEE1 levels
To test whether the response of WEE1 to miR-195 is specific in hESCs with impaired miRNA processing, we analyzed WEE1 mRNA levels in wild-type hESC H1 cells after miR-195 overexpression. A significant downregulation of WEE1 levels was observed upon miR-195 overexpression (Fig. 3F) in comparison to the overexpression of scrambled oligonucleotides or a miR-195 oligonucleotide bearing seed region mismatch mutations. We also tested whether miR-195 overexpression in the wild type hESCs affected the cell cycle kinetics by BrdU incorporation analysis. Indeed, the reduction of WEE1 due to miR-195 overexpression correlated with an increase in BrdU incorporation (Fig. 3F, 195 OE in H1, BrdU).
To test whether the downregulation of WEE1 is causal for the BrdU defect observed after miR-195 overexpression in wild type H1 hESCs, we specifically downregulated WEE1 by RNAi and analyzed potential cell cycle defects by BrdU incorporation. We show that BrdU incorporation is elevated two-fold in the WEE1-knockdown H1 hESCs (Fig. 3F). Similar results have been observed previously in primary fibroblasts with WEE1 silenced by RNAi.62 These data suggest that WEE1 levels are critical for normal cell cycle progression in hESCs. WEE1 is one of the three negative kinases that regulate G2 CycB/CDK complex activity in mammalian cells. The other negative CycB/CDK kinases are WEE2 and MYT1. It has been shown that MYT1 acts mainly in developing and adult cells whereas WEE1 acts mainly at a very early stage of development in Drosophila and mouse.63–65 Accordingly, MYT1 as well as WEE2 levels are extremely low in hESCs, whereas WEE1 is expressed to levels similar to those of the key stem cell markers (Fig. 3C). These data suggest that the only functional negative kinase for CycB/CDK in hESCs is WEE1. This supports the previously observed function of WEE1 in preimplantation stages of development.63 In summary, these data are consistent with the model that miR-195 represses WEE1 and therefore affects hESC cell cycle (Fig. 3G).
miR-372 affects p21 levels
As discussed above, miR-372 can partially rescue Dicer-knockdown phenotypes but is not sufficient to affect wild-type hESC cell cycle kinetics. These data could be explained by very high levels of miR-372, very low levels of the critical target in hESCs or a combination of both. We searched for a tumor suppressor that contains potential miR-372 binding sites and is highly repressed in the hESCs. Importantly, previous studies have already identified one such tumor suppressor, p21. p21 is expressed at a low level in hESCs.67 Furthermore, Luciferase assays have revealed that the 3'UTR of p21 can be regulated by miR-106, miR-17 and miR-372.58 Accordingly, as shown in Figure 4A, overexpression of miR-372 reduced p21 levels in HeLa cells. Furthermore, p21 protein was upregulated four-fold in Dicer-knockdown hESC line and this upregulation could be reduced by overexpression of miR-372 (Fig. 4A). Thus p21 shows the characteristics of a cell cycle regulator controlled by miR-372; it is highly downregulated in wild type hESCs; it is upregulated in the Dicer-knockdown hESC line H1; and it can be downregulated by miR-372 in HeLa cells and in Dicer-knockdown hESCs (Fig. 4A).
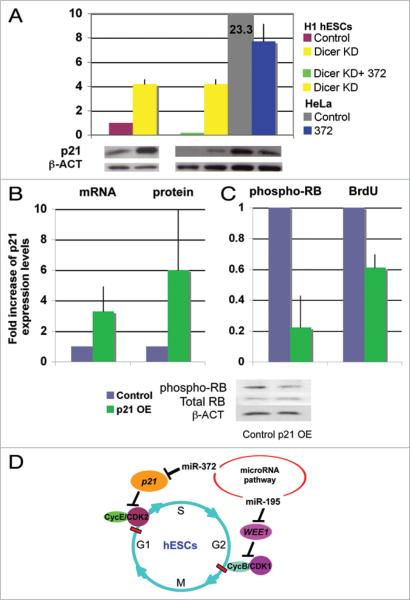
Figure 4.
miR-372 regulates the cell cycle regulator p21 to control hESC division. (A) Western blot analysis shows a four-fold upregulation of p21 protein in Dicer-knockdown H1 line #3 compared to control cell line #4. HeLa cells show 23-fold higher expression of p21 than the control hESCs. Overexpression of miR-372 results in reduction of p21 protein levels in HeLa cells (US2-372 OE transient transfection) and in Dicer-knockdown hESCs (Lenti-372 OE stable cell line). Mean ± range is shown from two independent experiments after quantification and normalization to β-ACTIN expression level. Overexpression of p21 in wild-type H1 hESCs (B) reduced phospho-RB levels and resulted in reduced BrdU incorporation (C). Mean ± range is shown from two independent experiments. (D) Model for miRNA function in hESC cell cycle regulation. The data suggest that miRNAs are important for proper cell cycle control of both G1 and G2 checkpoints in hESCs. miR-195 is required for proper hESC division. miR-195 inhibits the CycB/CDK1 inhibitor, WEE1, and, therefore, indirectly controls the G2/M transition. miR-372 suppresses the CDK inhibitor p21 and, therefore, indirectly controls the G1/S transition.
The low levels of p21 in hESCs are consistent with the hypothesis that hESCs have limited DNA damage-induced G1/S checkpoint control.68 To test whether a forced overexpression of p21 can affect the hESC cell cycle, we overexpressed p21 from a retroviral vector69 and analyzed effects on phosphorylated Rb levels and BrdU incorporation. Interestingly, six-fold p21 overexpression correlated with reduction of phospho-Rb levels to 23% and BrdU incorporation to 60% of normal (Fig. 4B and C). These data suggest that a moderate (six-fold) increase in p21 levels in hESCs can partially affect the cell cycle kinetics. Therefore, it is possible that p21 levels are low in hESCs partially due to the repressive effect of miR-372 and that in the Dicer-knockdown line, reduction of miR-372 might result in an increase in p21 that can affect the cell cycle.
Discussion
We show that knockdown (KD) of Dicer or Drosha dramatically attenuates cell division in hESCs. By use of qPCR analysis we identified a group of miRNAs that are enriched in hESCs compared with differentiated cells and tested whether any of these miRNAs could restore normal cell division in cells with a compromised miRNA pathway. We discovered that two miRNAs, miR-195 and miR-372, could partially reverse the cell division delay in the Dicer-knockdown hESC lines. Furthermore, we show that miR-372 regulates the G1/S checkpoint CDK inhibitor p21, whereas miR-195 regulates the G2/M checkpoint inhibitory kinase WEE1. In wild-type hESCs, miR-195, but not miR-372, overexpression increased the rate of stem cell division. These effects of miR-195 on cell cycle kinetics correlate with changes in levels of WEE1. Furthermore, we show that miR-195 directly regulates the WEE1 3'UTR, and that WEE1 knockdown in the hESC line H1 increases BrdU incorporation. These observations suggest that WEE1 plays a regulatory role in this cell type. In hESCs, p21 levels are normally low and phospho-RB levels are high. However, forced overexpression of p21 can reduce hESC phospho-RB levels and BrdU incorporation. These data suggest that miR-195 and miR-372 regulate hESC division by eliminating/regulating the gap-phase checkpoints (Fig. 4D).
Reduction of miRNA levels in hESCs results in attenuated cell division
We show that although silencing of Dicer and Drosha results in normal stem cell morphology, the cells are defective in stem cell division (Fig. 2A). The cells also appear to have elevated levels of the stem cell regulators Nanog, Oct4 and Sox2, suggesting that they have maintained a stem-cell like nature.
miRNAs targeting cell cycle checkpoint regulators modulate hESC cell division
Two miRNAs can partially rescue the Dicer-knockdown cell division defects in hESCs, miR-372 and miR-195. miR-372 and other miRNAs in the miR-106b family have been shown recently to regulate p21/CDKN1A.70 Furthermore, we show that miR-372 is sufficient to downregulate p21 in HeLa cells. Because p21 levels are upregulated in the Dicer knockdown line, and overexpression of miR-372 can rescue this effect, we propose that miR-372 regulates p21/CDKN1A in hESCs and thereby contributes to the reduction of p21 to a level at which it cannot act as an effective G1 checkpoint component to regulate the activity of the CycE/CDK complex (Fig. 4D). Consistent with our hypothesis is our inability to observe any obvious cell cycle defect when miR-372 was overexpressed in the wild type hESCs H1-line. We hypothesize that the normal level of miR-372 is sufficient to repress p21 levels in hESCs.9 However, the 10-fold reduction of miR-372 observed in the Dicer knockdown line (Fig. 1D) translates to a 4.5-fold increase in p21 levels (Fig. 4A) causing a cell cycle defect that responds to re-introduction of mature miR-372. Interestingly, p21/CDKN1A has been shown previously to be regulated by miRNAs in two other stem cell types, Drosophila germ line stem cells and mouse embryonic stem cells.23,28,32 In mouse, members of the miR-290 family regulate p21. Here, we show that miR-372 can regulate p21 in hESCs. This shared role for these miRNAs across species supports the hypothesis that the members of miR-290 and miR-372 are functional orthologs.
A significant effect on cell division was observed with alterations of miR-195 levels. Transfection of mature miR-195 duplexes partially rescued cell division defects in hESCs line with silenced Dicer. Additionally, overexpression of miR-195 in the wild type hESC H1-line significantly accelerated BrdU incorporation. We show further that the levels of a miR-195 target, WEE1, a negative regulator of G2/M phase, inversely correlate with miR-195 levels in hESCs. These data suggest that miR-195 and its target WEE1 are critical components in the regulation of hESC division. However, further studies are required to address whether miR-195 also targets hESC cell cycle regulators other than WEE1. One such candidate is CHK1, a kinase that has been shown to activate both the G1/S and G2/M checkpoints in response to blocks in DNA replication and DNA damage.71 Similarly other cell cycle targets for miR-372 may play a role in hESC division control.81 One such candidate is Rbl2, whose expression is increased two-fold in Dicer-knockdown hESC lines (Fig. S5).
The results presented here are intriguing in light of previous reports that hESCs lack a DNA damage checkpoint and that members of the miR-16 family act as negative regulators of the G1/S transition. We show that in hESCs, increased levels of p21 correlate with at least partial cell cycle defects arguing that G1/S transition in hESCs can be controlled by forced expression of p21. In the normal situation, p21 levels are very low and irradiation-induced G1 checkpoint activation is not observed (Fig. S4).72 We propose that the low, repressed p21 levels in hESCs might, at least partially, be maintained by miRNAs. Furthermore, we show that a member of miR-16 family, miR-195 acts mainly on G2/M, not G1/S transition in hESCs. miR-195 downregulates WEE1, a tumor suppressor that functions in the G2/M transition. Because miR-195 is sufficient to regulate G1/S cell cycle controllers in other cell types,59 it is possible that hESCs primarily make use of regulatory mechanisms in the S and G2 phases, rendering them more sensitive to the alterations in G2/M checkpoint proteins. These data are in agreement with the hypothesis that both G1-and G2-phase checkpoints in hESCs are at least partly regulated by miRNAs (Fig. 4D).
Other defects in Dicer KD hESCs
We have shown that Dicer-and Drosha-knockdown results in the formation of stem cells with high levels of stem cell regulators. As shown before,31,45 high levels of stem cell regulators correlates with delayed differentiation (Fig. S5). Furthermore, the delayed expression of differentiation markers (Fig. S5B) could be due to the delay in downregulation of Oct4 levels we observe in the Dicer- and Drosha-knockdown lines (Fig. S5A). Previous studies in mESCs have shown that delay in Oct4 downregulation correlates with defective DNA methylation resulting from upregulation of a repressor, the miR-290 family target Rbl2.31,73,74 Similarly, we have observed upregulation of Rbl2 in hESCs with reduced miR-372 levels in microarray data and qPCR analysis (Fig. S6). Further, human Rbl2 is a predicted target for miR-372 and we show that miR-372 upregulation correlates with Rbl2 downregulation in HeLa cells (Fig. S6). These findings suggest that the proposed human ortholog for the mouse miR-290 family, miR-372, might regulate human Rbl2. Further research is required to determine whether Dicer-knockdown hESCs have defects in global DNA-methylation.
Summary
This study defines the control of self-renewing division as a process that is sensitive to miRNA levels in hESCs. In addition, by identifying two miRNAs and their targets in hESC division, this work paves the way to future control of hESC activity through manipulation of the critical miRNAs for stem cell division.
Experimental Procedures
Cell culture
The hESC lines H1, H7 and HSF6 were cultured as previously described.57 Briefly, the cells were cultured on a feeder layer. Prior to the experiments the cultures were transferred to Matrigel (BD) growth conditions without feeders. When grown on feeders, hESCs were cultured in Dulbecco's modified Eagle's medium (DMEM)/Ham's F-12 medium containing GlutaMax supplemented with 20% serum replacer (SR), 1 mM sodium pyruvate, 0.1 mM non-essential amino acids, 50 U/ml penicillin, 50 ug/ml streptomycin, 0.1 mM β-mercapto-ethanol (Sigma-Aldrich, St. Louis, http://www.sigmaaldrich.com), and 2 ng/ml basic fibroblast growth factor (Peprotech, Rocky Hill, NJ, http://www. peprotech.com) (hESC medium). Feeders used were primary mouse embryonic fibroblasts (MEFs) from E12.5 CD-1 mice. Briefly, heads and red tissue (heart and liver) were discarded, and the rest of the fetus was disrupted by passage through a 25-gauge needle and plated as passage 0. Passage 4 to 6 MEFs were γ-irradiated with 3,000 rads and plated at 104 cells per cm2. If the hESCs were cultured off of feeders, Matrigel was used as the substrate matrix and the cells were cultured in MEF conditioned medium (CM). To make conditioned medium, MEF cells were irradiated as for feeders and seeded at 55,000 cells/cm2 in MEF medium (DMEM with GlutaMax + 10% FBS + penicillin and streptomycin). After 24 hours, the medium was exchanged with hESC medium (0.5 ml/cm2) and conditioned medium collected after 3 days. MEF cells were fed with hESC medium every 3 days and used for 7 days for CM collection. To passage, 1.2 units/ml dispase in PBS supplemented with 10% fetal bovine serum is used. All the hESC lines used in this study were cultured in colonies and can not grow from single cells. All reagents are from Invitrogen (Carlsbad, CA, http://www.invitrogen.com) unless otherwise specified.
Cell growth was monitored by counting the mean cell number multiplied by the mean colony area. Each trial was plated in triplicates. Cells were allowed to grow for 72 hours and counted under microscope for total colony number (n) on the plate and measured the diameter of the first 10 colonies across the mid line of each plate to calculate a mean diameter (D). The cell colony/size index was calculated by the mean colony number (n) multiplied by the mean colony area: n × πD2/4.
Differentiation of hESC
For serum forced, un-directed differentiation of hESCs, cultures were removed from the plate using dispase and plated on either a Matrigel or a 0.1% gelatin (Sigma) coated plate in differentiation media (80% DMEM with Glutamax, 20% FBS (ES qualified, Invitrogen), penicillin and streptomycin) for 4 days.
Selection and validation of lentiviral shRNA constructs
We prescreened Dicer and Drosha lentiviral vectors purchased from Open Biosystems or designed and constructed at Rosetta Inpharmatics/Merck by transiently transfecting midi-prepped plasmid DNA into HeLa cells. We extracted cellular RNA 24 hours after transfection and assayed the target gene knock-down by qPCR. We then used the best performing vectors for each gene to prepare virus for use in hESCs (virus were packaged in 293T cells). The best KD constructs were selected from multiple constructs with different promoters (U6, H1 or CMV). We used at least two constructs per gene and in two different hESC l lines H1 and H7.
Lentivirus infection to hESCs
The validated lentivirus-constructs were used to infect hESCs. Drosha knockdown constructs #1 and #2 were in pLKO.1 vector and the Dicer-knockdown constructs #3 was in pLKO.1 vector and #5 in pCMV-GIN vector using the empty vectors as controls (#4 pLKO.1 and #7 pCMVGIN respectively), shRNA sequences are shown in Figure 1A and B. H1 or H7 cells were grown on 35 mm plate with 2 ml medium (105 cells), the fluid were changed with 0.5 ml medium +0.5 ml virus and virus was incubated with the cells overnight. To get a functional Dicer or Drosha knockdown, atypically high MOI (>500) is required. Similar MOIs were used for the control and knockdown viruses. The cells were passaged the next day. The following day, infected cells were selected (1 μg/ml Puromycin or 0.5 μg/ml Blasticidin) until discrete clusters of cells appeared. The cells were re-exposed to antibiotics if there were questions about the stability of expression. The Dicer or Drosha knockdown lines were stored frozen and only thawed and grown for 5–10 passages for experiments since longer continuous culture caused modifications.
For Lenti-372 OE Dicer-knockdown hESC line, Pri-miR-372 sequence was amplified from HeLa genomic DNA using forward primer (5'-TCA ACC TGC GGA GAA GAT AC-3') and reverse primer (5'-CGC CCT CTG AAC CTT CTC TT-3') and then cloned between Age I and EcoR I sites of pLKO.1 TRC vector (Open Biosystems) under human U6 promoter. 293FT cells were plated one day before transfection and pLKO.1-pri-miR-372 was co-transfected with packaging vectors (psPAX2 and pMD2.G) using Lipofectamine 2000 (Invitrogen). Medium was changed 24 hours later and the lentiviruses were harvested 48 hours after transfection. For virus transduction, Dicer-knockdown H1 hESCs were incubated with diluted lentivirus in the presence of hexadimethrine bromide (Polybrene) for 24 hours.
For US2-372 OE construct, the same sequence of primiR-372 was used and cloned into EcoR1/Xba1 sites in the pUS2 plasmid.82
Transient transfection
One million hESC cells were transfected with short dsRNA duplexes (1 μM) designed and synthesized miRNA sequences of interest for overexpression using Amaxa Nucleofector system (mouse ES solution and program A23 were used). Cells were passaged 48 hours after transfection: for BrdU incorporation assays, cells were passed on matrigel coated 96-well plate; for immunostaining, cells were seeded on feeder cells; and for qPCR, cells were grown on 10 cm Matrigel plates. All the cells were harvested and assayed 4 days after transfection.
Short RNA oligos were ordered from IDT with the sequences of human miRNA-106a, -195, -302c, -372 and their antisense strands. The miRNA and their antisense oligos were annealed (100°C for 5 min, cool down to room temperature in a water bath) and the resulting short dsRNA duplexes were used for Amaxa nucleofection assays. We also used annealed oligo duplexes ordered from Sigma for miR-195 and 106a and saw similar results with their counterparts from IDT as shown in Figure 2E. One million hESCs were used for each Amaxa nucleofection reaction (single cuvette). Briefly, cells were dispersed and washed twice with PBS. The cells were resuspended into 100 μl of the mouse ES solution (Amaxa) with 1 μM short dsRNA duplexes and electroporated (Nucleofector I device, Amaxa, program A23). After electroporation, the reaction was transferred into a 1.5 ml eppendorf tube with 1 ml of pre-warmed ES cell media and incubated at 37°C for 15 min and thereafter plated on Matrigel. Forty-eight hours or 4 days after the Amaxa electroporation, the cells were washed twice with PBS, harvested and subjected to miRNA isolation (mirVana kit, Ambion) or BrdU analysis.
For p21 overexpression or WEE1 knockdown, H1 cells were plated on 6-well Matrigel plates 2 days before transfection and grown in conditioned media (CM) to attain around 50% confluency. For transfection, Gene Juice Transfection Reagent (Novagen) was used according to manufacturer's protocol. Briefly, 3 ul of Gene Juice was added to 100 ul of serum free media along with 2 ug of vector, for the p21 overexpression experiment, control pLNSX75 or the p21.pLNSX overexpression construct69 plus 1 ug of GFP (US2.gfp) per well; for WEE1 knockdown, a stem-loop structure oligonucleotide containing a WEE1-targeting sequence (5'-CCG GCT AGA AAG AGT GCA GAA CAA TCT CGA GAT TGT TCT GCA CTC TTT CTA GTT TTT-3') was designed and cloned under the control of the human U6 promoter between Sal I and Xba I sites in the pLUG plasmid and a scrambled shRNA sequence was used as control.76 The resulting transfection mixture was added on top of the cells with 1 ml of fresh CM. After 24 hrs, the transfection media was replaced with fresh CM. For US2-372 OE transient transfection, HeLa cells in 35 mm plates were transfected with pUS2-372 OE plasmid (4 μg) using Lipofectamine 2000 (Qiagen). Cells were harvested and assayed 48 hours after transfection.
Immunostaining
hESC cells were seeded in cell chamber slides with feeder cells and grown 48 hours before fixing with 4% PFA at room temperature for 15 min. Then fixed cells were washed three times with PBT (1× PBS, 0.2% Triton) room temperature for 10 min and then left in blocking buffer PBTB (1× PBS, 0.2% Triton, 5% Goat Serum, 0.2% BSA) for 1 hour. The cells were then incubated with primary antibodies at 4°C overnight and washed 3 times with PBT. Secondary antibodies were incubated with the cells for 1 hour at room temperature and and the slides washed three times with PBT before mounting with Prolong Gold antifade reagent (Invitrogen). The antibodies used were: anti-Ki67 mouse monoclonal antibody (Vector laboratories) at 1:100, anti-phospho-Histone H3 (Ser10), rabbit polyclonal IgG (Upstate cell signaling solutions, catalog #06-570) at 1:300, and anti-Oct4 goat antibody at 1 μg/ml (R&D systems, cat# AF1759). Goat anti-rabbit (1:6,500), rabbit anti-goat (1:5,000), and goat anti-mouse (1:5,000) IgG (H + L)-HRP conjugated secondary antibodies from Bio-Rad.
Western blot
Dicer-knockdown and control hESC cells were grown on Matrigel in 10 cm dishes and harvested after two washes with PBS 3 days after passage. In general, 20 μg of protein was resolved by 4–20% SDS-PAGE (Bio-Rad), transferred onto a nitrocellulose membrane, and probed overnight with antibodies against OCT4 (anti-goat, R&D systems), p21 (anti-rabbit), WEE1 (anti-rabbit), phospho-RB (anti-rabbit) and total RB (anti-mouse), all used at a 1:1,000 dilution. All antibodies were purchased from Cell Signaling unless otherwise specified. The blot was washed and incubated with a horseradish peroxidaseconjugated secondary antibody (Pharmacia Biotech) for 1 hour. Immunoreactive proteins were detected by ECL (Pharmacia Biotech). The membranes were stripped for 30 min in stripping buffer (Pierce) and re-probed with anti-β-actin antibody (anti-rabbit, Sigma Aldrich) as described above. For quantification, Adobe Photoshop software was used.
BrdU incorporation assay
hESC colonies were cultured for two days in a 96-well plate that was coated with matrigel the day before use. The BrdU ELISA assays were done using a BrdU labeling and detection kit III (Roche) according to the manufacturer's protocol. Briefly, cells were labeled with 10 nM BrdU for 4 hours, washed twice with PBS containing 10% serum and fixed with 70% Ethanol in 0.5 M HCl at −20°C for 30 min. Cells were washed three times with PBS containing 10% serum and treated with 100 ul of nuclease working solution at 37°C for 30 min. Cells were washed three times with PBS containing 10% serum followed by incubation with 100 ul of antibody conjugate (anti-BrdU-POD, Fab fragments) working solution at 37°C for 30 min. After removal of the antibody and three washes, 100 ul of peroxidase substrate with substrate enhancer was added. After incubation at room temperature until positive samples showed a green color, the extinction of the samples was measured in a microplate reader at 405 nm with a reference wavelength at 492 nm. p-values were calculated using a one-tailed Student's t test.
mRNA and miRNA qPCR analysis
miR-10a, -195, -218, -372, -106a and -302c Taqman assays (Applied Biosystems) were used for miRNA qPCRs. In brief, total RNAs were extracted with mirVana miRNA isolation kit (Ambion) or trizol (invitrogen) and DNase-treated with DNA-free kit (Ambion). Following the reverse transcription reaction using Omniscript RT kit (Qiagen), Dicer and Drosha, as well as Gapdh or β-actin (loading control) was tested by qPCR with SyberGreen master mix (Applied Biosystems) on the ABI 7300 Real-time PCR system. The reactions were incubated in a 96 well plate at 95°C for 10 min, followed by 40 cycles of 94°C for 15 s and 54°C for 30 s and 72°C for 60 s. All reactions were run in triplicates. Human TaqMan miRNA assays were used for miRNA qPCR according to the manufacturer's protocol using RNU66 snoRNA as a loading control. p-values were calculated using a one-tailed Student's t test.
Dual luciferase assay
The 3'UTR from human WEE1 was cloned into a vector containing the Luciferase ORF (pSGG_3UTR, SwitchGear Genomics). Seed regions were mutated to remove complementarity to miR-195 by using the Quickchange II XL Mutagenesis Kit (Stratagene). Specifically, two miR-195-complementary sites, TGCTG CTA, were changed to TGCTctgA. HCT116 Dicer-ex5 cells61 or HeLa cells were co-transfected with reporter construct and RNA duplexes for miR-149, 195 and 372 using Lipofectamine 2000 (Invitrogen). Cells were lysed 24 hrs after transfection, and ratios between firefly luciferase and Renilla luciferase activity were measured with a dual luciferase assay (Promega). The data for each construct are normalized to luciferase activity after cotransfection with miR-149.
mRNA microarray for nine hESC lines
The hESC lines H1 (WA01), H7 (WA07), H9 (WA09), H13 (WA13), H14 (WA14), HSF-6, BG01, BG02 and BG03 were used for mRNA profiling experiments. Cells were grown for three passages in feeder-free conditions prior to RNA extraction. Total RNA for mRNA microarray analysis was isolated using RNeasy kit (Qiagen) as per manufacturer's protocol. Samples were amplified and labeled using a custom automated version of the RT/IVT protocol and reagents provided by Affymetrix. Hybridization, labeling and scanning were completed following the manufacturer's recommendations (Affymetrix). Sample amplification, labeling and microarray processing were performed by the Rosetta Inpharmatics Gene Expression Laboratory in Seattle, WA. Rosetta Error Modeling was used to analyze the data.
Supplementary Material
Acknowledgements
We thank members of the Ruohola-Baker lab for helpful discussions, Dr. Merav Bar for reading the paper, Jennifer Hesson for assistance at the UW hESC core and Andy Shih and Shomir Chaudhuri for preparing MEF cells and MEF-conditional media. We thank the Rosetta Gene Expression Laboratory for processing microarray experiments. We thank Drs. Prabha Sampath and Charles Murry for providing H7 hESCs samples for microarray analysis. This work was supported by the Tietze Award and grants from NIH and MOD for H.R.-B. and NIH/NIGMS F32 postdoc fellowship for J.Q.
Footnotes
Note Supplementary materials can be found at: www.landesbioscience.com/supplement/QiCC8-22-Sup.pdf
References
- 1.Cheng T. Cell cycle inhibitors in normal and tumor stem cells. Oncogene. 2004;23:7256–66. doi: 10.1038/sj.onc.1207945. [DOI] [PubMed] [Google Scholar]
- 2.Bucher N, Britten CD. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br J Cancer. 2008;98:523–8. doi: 10.1038/sj.bjc.6604208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fluckiger AC, Marcy G, Marchand M, Negre D, Cosset FL, Mitalipov S, et al. Cell cycle features of primate embryonic stem cells. Stem Cells. 2006;24:547–56. doi: 10.1634/stemcells.2005-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shcherbata HR, Hatfield S, Ward EJ, Reynolds S, Fischer KA, Ruohola-Baker H. The MicroRNA pathway plays a regulatory role in stem cell division. Cell Cycle. 2006;5:172–5. doi: 10.4161/cc.5.2.2343. [DOI] [PubMed] [Google Scholar]
- 5.Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–93. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 6.Hong Y, Stambrook PJ. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc Natl Acad Sci U S A. 2004;101:14443–8. doi: 10.1073/pnas.0401346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aladjem MI, Spike BT, Rodewald LW, Hope TJ, Klemm M, Jaenisch R, et al. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8:145–55. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 8.Prost S, Bellamy CO, Clarke AR, Wyllie AH, Harrison DJ. p53-independent DNA repair and cell cycle arrest in embryonic stem cells. FEBS Lett. 1998;425:499–504. doi: 10.1016/s0014-5793(98)00296-8. [DOI] [PubMed] [Google Scholar]
- 9.Zeng X, Rao MS. Human embryonic stem cells: long term stability, absence of senescence and a potential cell source for neural replacement. Neuroscience. 2007;145:1348–58. doi: 10.1016/j.neuroscience.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 10.Andang M, Hjerling-Leffler J, Moliner A, Lundgren TK, Castelo-Branco G, Nanou E, et al. Histone H2AX-dependent GABA(A) receptor regulation of stem cell proliferation. Nature. 2008;451:460–4. doi: 10.1038/nature06488. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Neganova I, Przyborski S, Yang C, Cooke M, Atkinson SP, et al. A role for NANOG in G1 to S transition in human embryonic stem cells through direct binding of CDK6 and CDC25A. J Cell Biol. 2009;184:67–82. doi: 10.1083/jcb.200801009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–52. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 13.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 14.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 15.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–25. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatfield S, Ruohola-Baker H. microRNA and stem cell function. Cell Tissue Res. 2008;331:57–66. doi: 10.1007/s00441-007-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stadler BaR-BH. Small RNAs: Keeping Stem Cells in Line. Cell. 2008;132:563–6. doi: 10.1016/j.cell.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barroso-delJesus A, Romero-Lopez C, Lucena-Aguilar G, Melen GJ, Sanchez L, Ligero G, et al. Embryonic stem cell-specific miR302-367 cluster: human gene structure and functional characterization of its core promoter. Mol Cell Biol. 2008;28:6609–19. doi: 10.1128/MCB.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–8. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 20.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–98. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:18097–102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–8. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 24.Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr Biol. 2007;17:539–44. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 25.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–6. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 26.Shcherbata HR, Ward EJ, Fischer KA, Yu JY, Reynolds SH, Chen CH, et al. Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell Stem Cell. 2007;1:698–709. doi: 10.1016/j.stem.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Duan R, Chen D, Wang J, Chen D, Jin P. Fragile X mental retardation protein modulates the fate of germline stem cells in Drosophila. Hum Mol Genet. 2007;16:1814–20. doi: 10.1093/hmg/ddm129. [DOI] [PubMed] [Google Scholar]
- 28.Yu JY, Reynolds SH, Hatfield SD, Shcherbata HR, Fischer KA, Ward EJ, et al. Dicer-1-dependent Dacapo suppression acts downstream of Insulin receptor in regulating cell division of Drosophila germline stem cells. Development. 2009;136:1497–507. doi: 10.1242/dev.025999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102:12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–5. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–83. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Assou S, Le Carrour T, Tondeur S, Strom S, Gabelle A, Marty S, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–73. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–46. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 36.Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 37.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 38.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 39.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–4. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 41.Matin MM, Walsh JR, Gokhale PJ, Draper JS, Bahrami AR, Morton I, et al. Specific knockdown of Oct4 and beta2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells. 2004;22:659–68. doi: 10.1634/stemcells.22-5-659. [DOI] [PubMed] [Google Scholar]
- 42.Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 2005;23:299–305. doi: 10.1634/stemcells.2004-0252. [DOI] [PubMed] [Google Scholar]
- 43.Fong H, Hohenstein KA, Donovan PJ. Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells. 2008;26:1931–8. doi: 10.1634/stemcells.2007-1002. [DOI] [PubMed] [Google Scholar]
- 44.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–75. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 45.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–58. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 46.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 47.Darr H, Mayshar Y, Benvenisty N. Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development. 2006;133:1193–201. doi: 10.1242/dev.02286. [DOI] [PubMed] [Google Scholar]
- 48.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–29. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 50.Smith A. The battlefield of pluripotency. Cell. 2005;123:757–60. doi: 10.1016/j.cell.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 51.Solovei I, Schermelleh L, Albiez H, Cremer T. Detection of the cell cycle stages in situ in growing cell populations. In: Celis J, editor. Celis J Cell Biology Handbook: A Laboratory Manual; San Diego. Academic Press, NYP; 2005. [Google Scholar]
- 52.Tzur G, Levy A, Meiri E, Barad O, Spector Y, Bentwich Z, et al. MicroRNA expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS ONE. 2008;3:3726. doi: 10.1371/journal.pone.0003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lakshmipathy U, Love B, Goff LA, Jornsten R, Graichen R, Hart RP, et al. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007;16:1003–16. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laurent LC, Chen J, Ulitsky I, Mueller FJ, Lu C, Shamir R, et al. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–16. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 55.Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. Rna. 2005;11:1737–44. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ware CB, Nelson AM, Blau CA. A comparison of NIH-approved human ESC lines. Stem Cells. 2006;24:2677–84. doi: 10.1634/stemcells.2005-0452. [DOI] [PubMed] [Google Scholar]
- 58.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–74. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–38. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA. let-7 Overexpression leads to an increased fraction of cells in G2/M, direct downregulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J Biol Chem. 2009;284:6605–9. doi: 10.1074/jbc.C900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tominaga Y, Li C, Wang RH, Deng CX. Murine Wee1 plays a critical role in cell cycle regulation and preimplantation stages of embryonic development. Int J Biol Sci. 2006;2:161–70. doi: 10.7150/ijbs.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price DM, Jin Z, Rabinovitch S, Campbell SD. Ectopic expression of the Drosophila Cdk1 inhibitory kinases, Wee1 and Myt1, interferes with the second mitotic wave and disrupts pattern formation during eye development. Genetics. 2002;161:721–31. doi: 10.1093/genetics/161.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin Z, Homola E, Tiong S, Campbell SD. Drosophila myt1 is the major cdk1 inhibitory kinase for wing imaginal disc development. Genetics. 2008;180:2123–33. doi: 10.1534/genetics.108.093195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–21. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maimets T, Neganova I, Armstrong L, Lako M. Activation of p53 by nutlin leads to rapid differentiation of human embryonic stem cells. Oncogene. 2008;27:5277–87. doi: 10.1038/onc.2008.166. [DOI] [PubMed] [Google Scholar]
- 68.Filion TMQM, Ghule PN, Mandeville M, van Wijnen AJ, Stein JL, Lian JB, et al. Survival responses of human embryonic stem cells to DNA damage. J Cell Physiol. 2009;220:586–92. doi: 10.1002/jcp.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheaff RJ, Singer JD, Swanger J, Smitherman M, Roberts JM, Clurman BE. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell. 2000;5:403–10. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 70.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–74. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 2000;14:1439–47. [PMC free article] [PubMed] [Google Scholar]
- 72.Filion TM, Qiao M, Ghule PN, Mandeville M, van Wijnen AJ, Stein JL, et al. Survival responses of human embryonic stem cells to DNA damage. J Cell Physiol. 2009 doi: 10.1002/jcp.21735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–67. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 74.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–79. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller AD, Rosman GJ. Improved retroviral vectors for gene transfer and expression. Biotechniques. 1989;7:980–2. [PMC free article] [PubMed] [Google Scholar]
- 76.Wang L, Schulz TC, Sherrer ES, Dauphin DS, Shin S, Nelson AM, et al. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood. 2007;110:4111–9. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tyson JJ, Novak B. Temporal organization of the cell cycle. Curr Biol. 2008;18:R759–68. doi: 10.1016/j.cub.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Blelloch R. Cell cycle regulation by microRNAs in embryonic stem cells. Cancer Res. 2009;69:4093–6. doi: 10.1158/0008-5472.CAN-09-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nimmo RA, Slack FJ. An elegant miRror. microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–18. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–81. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 81.Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, et al. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acid Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.