Abstract
Brassinosteroid (BR) homeostasis and signaling are crucial for normal growth and development of plants. BR signaling through cell-surface receptor kinases and intracellular components leads to dephosphorylation and accumulation of the nuclear protein BZR1. How BR signaling regulates gene expression, however, remains unknown. Here we show that BZR1 is a transcriptional repressor that has a previously unknown DNA binding domain and binds directly to the promoters of feedback-regulated BR biosynthetic genes. Microarray analyses identified additional potential targets of BZR1 and illustrated, together with physiological studies, that BZR1 coordinates BR homeostasis and signaling by playing dual roles in regulating BR biosynthesis and downstream growth responses.
Brassinosteroids (BRs) are plant hormones that play essential roles in growth and development. Mutant plants defective in BR biosynthesis or signaling display characteristic phenotypes including dwarfism, curled leaves, male sterility, and light-grown morphology in the dark (1–4). Unlike many plant hormones that undergo long distance transport and thus have separate sites of synthesis and action, BRs appear to be synthesized and function in the same tissue or even in the same cell (5–7). With BRs acting at the site of synthesis, cells must monitor and tightly regulate their BR biosynthesis to achieve balanced cell expansion in normal plant development (7). Consequently, the expression of many BR biosynthetic genes is feedback regulated by BR signaling (8, 9). Such a mechanism of feedback regulation is therefore an integral part of BR action, and a detailed knowledge of this mechanism at the molecular level is essential for an integrated understanding of how plants use BRs to regulate growth and development.
Unlike animal steroid hormones, which regulate gene expression by binding to the nuclear receptor family of transcription factors, BRs are perceived by cell surface receptors in plants (10). Studies of BR signaling mutants and their suppressors have led to the identification of a number of BR signal transduction components. These include the cell-surface receptor kinases BRI1 and BAK1, which perceive the BR signal and initiate the signal transduction cascade (11–13); the nuclear proteins BZR1 and its homolog BZR2/BES1, which activate growth responses and are dephosphorylated and stabilized by BR signaling (14, 15); the BIN2 kinase, which negatively regulates BR responses by phosphorylating BZR1 and BZR2/BES1 and targeting them for degradation by the proteasome (15–17); and the BSU1 phosphatase, which positively regulates BR responses by dephosphorylating and stabilizing the BZR2/BES1 protein (18). These studies illustrated a signaling cascade leading from the cell surface receptors to dephosphorylation and accumulation of BZR1 and BZR2/BES1 in the nucleus. How BZR1 and BZR2/BES1 mediate BR regulation of gene expression remains a major gap in our understanding of the BR signal transduction pathway (19).
Studies of the bzr1-1D mutant suggested that BZR1 is involved in both growth promotion and feedback regulation of BR biosynthesis (14). The dominant bzr1-1D mutation increases BZR1 protein accumulation, suppresses the BR insensitive bri1 and bin2 mutants, and causes insensitivity to the BR biosynthetic inhibitor brassinazole, indicating a positive role for BZR1 in BR signaling. However, when grown in the light, bzr1-1D has reduced cell elongation, reduced levels of BR, and reduced expression of CPD, a BR biosynthetic gene feedback inhibited by BR signaling (3, 8, 14). This suggests that BZR1 can also have a negative effect on BR regulated growth, perhaps by promoting feedback inhibition of BR biosynthesis. This is in contrast to the homologous BZR2/BES1 gene, which appears to increase cell elongation under all growth conditions (15). The BR repression of CPD and several other BR biosynthetic genes was detected as early as 15 minutes after BR treatment (20), and the BR-induced BZR1 dephosphorylation and accumulation were detected within 10 min of BR treatment (17). These observations suggest that BZR1 might directly repress the BR biosynthetic genes.
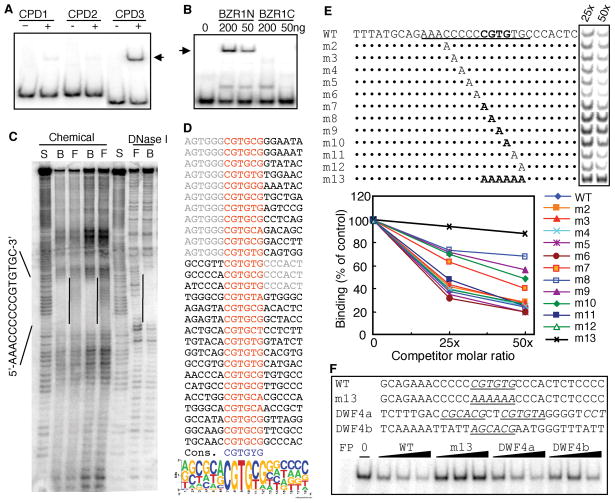
BZR1 is a member of a small gene family that shares no significant sequence similarity to any known proteins(14). To test whether BZR1 is a novel DNA-binding protein that directly binds to the CPD promoter, we performed electrophoretic mobility shift assays (EMSAs) using radio-labeled DNA fragments of the CPD promoter and bacterially expressed BZR1 proteins fused to the maltose binding protein (MBP). Figure 1A shows that the MBP-BZR1(aa 21–336) fusion protein binds to the −208 to −60 region but not to the −502 to −175 region of the CPD promoter (translational start is +1). The N-terminal domain (BZR1N, aa 21–104) but not the C-terminal domain (BZR1C, aa 90–336) of BZR1 showed DNA binding activity (Fig. 1B), indicating that the N-terminal domain of BZR1 is the DNA binding domain. The BZR1 DNA binding domain encoded by the first exon is the most conserved region of the BZR1-related proteins and is thus named the BZR domain. BZR1 and its homologs therefore represent a new kind of DNA-binding proteins that appears to be plant specific.
Fig. 1.
BZR1 is a DNA binding protein. (A) Electrophoretic mobility-shift assays (EMSAs) showing that the maltose binding protein-BZR1 (21-336 aa) fusion protein (BZR1) specifically binds to the −208 ~ −60 region (CPD3), but not the −502 ~ −339 (CPD1) or −348 ~ −175 (CPD2) regions of the CPD promoter. (B) MBP fusion proteins of the N-terminal domain (aa22–104, BZR1N) not C-terminal domain of BZR1 protein (aa 90–336, BZR1C) binds to the CPD promoter. (C) DNA footprint assays of the BZR1 binding site in the CPD promoter. Free (F) and protein-bound (B) DNAs were cleaved by either phenanthroline-copper (Chemical) in gel or DNase I in solution and separated on a sequencing gel. Lane S is the G+A chemical sequencing reaction. Footprint regions are marked by vertical lines and corresponding sequence. (D) Random binding site selection (RBSS) for optimal BZR1 binding site. Shown on the top is the alignment of all identified sequences, with the matching sequences marked in red. The non-random residues immediately flanking the matching sequences are marked in light gray and excluded from calculating the consensus sequence and logo of the alignment (bottom). (E) Oligonucleotide sequences of the CPD promoter containing wild type (WT) or mutant (m2–m13) BZR1 binding site were used as competitors at 25x or 50x fold molar ratios in EMSAs. Image and quantification of shifted bands are shown. (F) Competition of MBP-BZR1N binding to the BRRE of CPD by potential BRREs from the DWF4 (DWF4a and DWF4b) gene. Black wedges represent increasing amount of competitors (12.5x, 25x and 50x in molar excess). FP, free probe with no protein.
Using chemical and DNase I footprint assays, the BZR1 binding site was further localized in the −90 to −75 region of the CPD promoter containing a AAACCCCCCGTGTGC sequence (Fig. 1C). To further define the optimal DNA sequence that BZR1 binds to, a random binding site selection (RBSS) experiment was performed (21). The BZR1-binding DNA was selected from a pool of DNA fragments containing 16-base random sequences in the middle using the MBP-BZR1N protein immobilized on amylose agarose beads. As shown in Fig. 1E, all 28 sequences identified contain the CGTG sequence. Alignment of these sequences identified CGTG(T/C)G as the optimal binding site for BZR1 (Fig. 1D), which in fact is present in the footprinted region of the CPD promoter (Fig. 1C). Mutating individual residues in the CGTG sequence dramatically reduced competition for BZR1 binding (Fig. 1E, m7, 8, 9, 10), indicating that BZR1 binding requires a CGTG sequence. Mutation of the CGTGTG element in the CPD promoter to AAAAAA completely abolished BZR1 binding (Fig. 1E, m13). These results demonstrate that BZR1 is a novel DNA-binding protein that binds to the CGTG(T/C)G sequence. In support of this, we found BZR1-binding sequences in the promoters of all four feedback-regulated BR biosynthetic genes (CPD, DWF4, ROT3, BR6OX) (20), indicating a general role of BZR1 in feedback regulation. Fig. 1F shows that the promoter of DWF4 contains two BZR1-binding sequences and either of them can bind to BZR1.
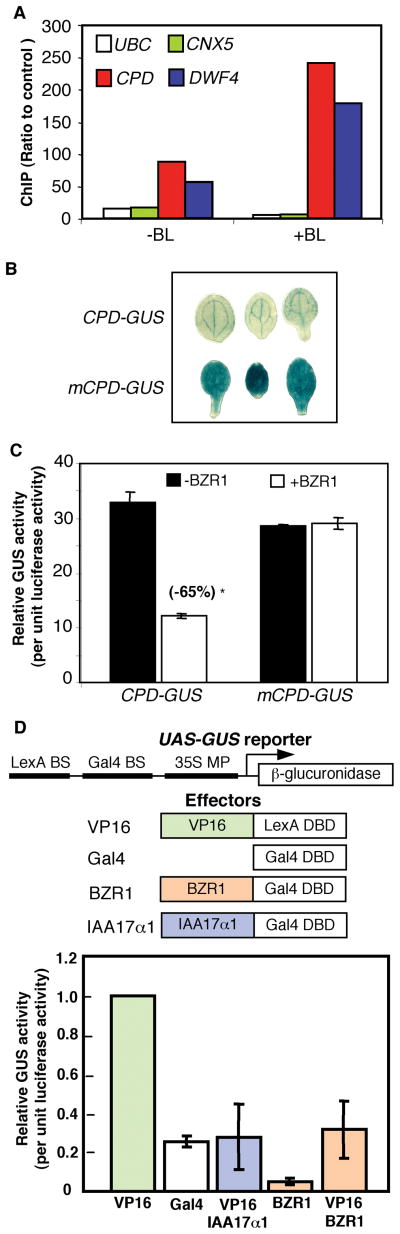
To determine whether BZR1 binds to DNA in vivo, chromatin immunoprecipitation (ChIP) experiments were performed. The BZR1-CFP fusion protein expressed in transgenic plants using the BZR1 native promoter (14) was immunoprecipitated using an anti-GFP antibody. Genomic DNA fragments that co-immunoprecipitate with BZR1-CFP were analyzed by real-time quantitative PCR. As shown in Fig 2A, the amount of the CPD and DWF4 promoter DNAs immunoprecipitated from the BZR1-CFP transgenic plants was over twenty seven fold higher than that precipitated from the non-transgenic control plants and about five fold higher than the two control genes that are not responsive to BR and contain no BRRE (Molybdopterin synthase sulphurylase CNX5, At5g55130; and ubiquitin-conjugating enzyme, UBC, At5g56150). BR treatment significantly increased the co-immunoprecipitation of the CPD and DWF4 DNA but not of the control genes (Fig. 2A). These results provide direct evidence that BZR1 binds to the promoters of CPD and DWF4 genes in vivo and that BR signaling increases BZR1 binding.
Fig. 2.
BZR1 mediates BR-repression of gene expression in vivo. (A) Chromatin immunoprecipitation (ChIP) of the CPD, DWF4, UBC and CNX5 promoters from BZR1-CFP transgenic plants using anti-GFP antibodies. Results were presented as the ratio of the amount of DNA immunoprecipitated from BZR1-CFP samples treated with BL (+BL) or mock solution (−BL) to that from non-transgenic control plants, determined by quantitative real-time PCR. The average and standard error from six PCR analyses of two biological repeats are shown. (B) Expression of the CPD-GUS and mCPD-GUS reporter genes in bzr1-1D plants. Cotyledons of 10-day-old seedlings from 3 independent T1 transgenic lines were histochemically stained for GUS enzyme activity. (C) Expression of CPD-GUS or mCPD-GUS reporter gene in Arabidopsis protoplasts transformed without (−) or together with (+) the BZR1 overexpression construct (35S-BZR1), relative to the 35S-luciferase internal control. (D) Transient assays of transcriptional activity of BZR1. BY-2 protoplasts were transformed with the reporter (UAS-GUS) and effecter constructs (upper panel), and the reporter gene expression is shown in the bottom panel. UAS-GUS, reporter construct containing Gal4 and LexA binding sites and a 35S minimal promoter upstream of the coding sequence of GUS; VP16, VP16 fused to the LexA DNA binding domain (DBD); Gal4, Gal4 DBD; IAA17α1, the transcription repression domain of IAA17 fused to the Gal4 DBD; BZR1, full length BZR1 fused to Gal4 DBD. 35S-luciferase was co-transformed as internal control, and the GUS reporter gene expression was normalized to the luciferase activity and presented as values relative to the VP16 control.
To determine whether BZR1 binding is required for its regulation of CPD expression, the mutation that abolishes BZR1 binding (m13, Fig. 1E) was introduced into a CPD promoter-β-glucuronidase (CPD-GUS) reporter gene construct (mCPD-GUS) and its effect on gene expression was examined in transgenic Arabidopsis plants and in transiently transformed protoplasts. As shown in Fig. 2B, Arabidopsis transformed with mCPD-GUS expressed significantly higher GUS activity than those transformed with the wild type CPD-GUS reporter gene, indicating that the BZR1 binding site is required for repression of CPD expression in vivo. In protoplast transient assays, overexpression of BZR1 significantly reduced CPD-GUS expression, confirming that BZR1 negatively regulates CPD expression. By contrast, overexpression of BZR1 did not have a significant effect on the expression of the mCPD-GUS reporter gene (Fig. 2C). These results indicate that BZR1 binding is essential for the feedback inhibition of CPD gene expression. The BZR1 binding sequence (CGTG(T/C)G) is thus named BR response element (BRRE).
To determine whether BZR1 is sufficient to repress transcription, we performed transcriptional reporter gene assays using a promoter-reporter gene that contains binding sequences for the LexA and Gal4 DNA binding proteins (Fig. 2D). This reporter gene is expressed at high levels when co-transformed into protoplasts with the LexA-VP16 fusion construct, which contains the coding sequences for the LexA DNA binding domain fused in frame with the coding sequence of the VP16 transcriptional activation domain (22). As reported previously (22), the Gal4 fusion with the transcriptional repressor domain of IAA17 (IAA17α1) reduced the activation of the reporter gene by LexA-VP16 (Fig. 2D). Similarly, co-transformation of the reporter gene with a construct for overexpression of the BZR1-Gal4 fusion protein (35S-BZR1-Gal4) significantly reduced the reporter gene expression both in the absence and presence of the LexA-VP16 construct (Fig. 2D), indicating that BZR1 is a potent transcriptional repressor. Together the above results demonstrate that BZR1 is a transcriptional repressor that, upon activation by BR signaling, binds to the BRRE of the BR biosynthetic genes and confers feedback inhibition of BR biosynthesis.
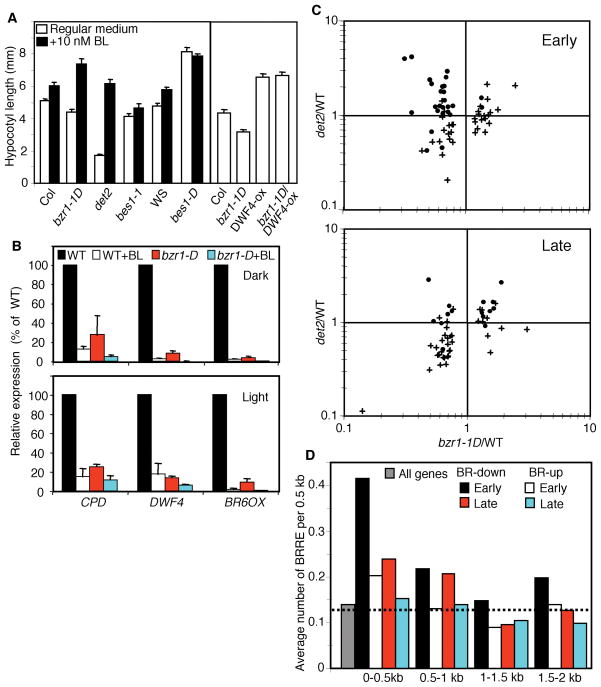
Such a direct role of BZR1 in feedback inhibition of BR biosynthetic genes can potentially explain the dwarf phenotype of light-grown bzr1-1D mutant plants. While bzr1-1D mutation increases cell elongation in the dark or in the absence of upstream BR signaling, bzr1-1D mutant plants grown in the light show a weak dwarf phenotype with reduced hypocotyl and petiole lengths and dark green curled leaves (14). These light grown bzr1-1D plants resemble BR deficient and insensitive mutants. Such opposite growth phenotypes of bzr1-1D in the dark and light suggest that BZR1 is regulated by light. Light either switches BZR1 from a positive regulator to a negative regulator of BR signaling or alters the contributions of BZR1 to feedback inhibition of BR biosynthesis and to growth responses (14). To determine whether the reduced BR level is responsible for the dwarf phenotypes of the light-grown bzr1-1D, we tested the effects of both exogenous BR application and increasing endogenous BR synthesis on the dwarf phenotype of bzr1-1D. As shown in figure 3A, the hypocotyls of the bzr1-1D mutant seedlings are shorter than wild type on regular medium but longer than wild type on medium containing 10 nM brassinolide (BL, the most active brassinosteroid), indicating that the bzr1-1D mutant is hypersensitive to BR. By contrast, the bes1-D mutant seedlings have long hypocotyls that are not affected by BL (Fig. 3A). Consistent with the gain-of-function phenotypes of bes1-D, the T-DNA knockout bes1-1 mutant showed slightly reduced hypocotyl length and weaker response to BR treatment. The short hypocotyl phenotype of bzr1-1D was also suppressed by a transgenic line that overexpresses the BR biosynthetic gene DWF4 (DWF4-ox), because the bzr1-1D/DWF4-ox double homozygous plants have similar hypocotyl lengths as the DWF4-ox plants (Fig. 3A). These results indicate that the dwarf phenotype of light-grown bzr1-1D is due to BR deficiency caused by increased feedback inhibition of BR biosynthesis and that bzr1-1D has increased BR sensitivity.
Fig. 3.
BZR1 has dual roles in regulating BR biosynthesis and growth responses. (A) Hypocotyl lengths of seedlings grown in continuous light for 7 days on medium containing 0 or 10 nM BL. Col, wild type control for bzr1-1D, det2, and bes1-1 mutants. WS, wild type control for the bes1-D mutant. DWF4-ox, a transgenic line that overexpresses DWF4. (B) Real-time PCR analysis of expression of BR biosynthetic genes in the bzr1-1D mutant and wild type (WT) seedlings grown in the dark (upper) or light (lower panel), and after 3 hr BL treatment (+BL). Average and standard deviation of 3 experiments are shown. (C) Microarray analysis of BR regulated genes in det2, bzr1-1D, and wild type (WT) Arabidopsis. Y axis: ratio of expression between det2 and wild type; X axis: ratio of expression between bzr1-1D and wild type. Upper panel: early-BR-responsive genes; Lower panel: late-BR-responsive genes. Dots: BR-induced genes; crosses, BR-repressed genes. (D) Average number of BRREs in the promoter regions of genes that are up- (BR-up) or down- (BR-down) and early- or late-regulated by BR treatment. Dashed line, calculated probability of occurrence of the BRRE sequence in any 500 bp Arabidopsis intergenic sequence. Grey bar, average number of BRREs in the 500 bp upstream regions of all genes in the Arabidopsis genome.
The BR hypersensitivity of bzr1-1D is consistent with its partial suppression of bri1 in the light (14) and supports a direct positive effect of bzr1-1D on downstream growth responses in the light. To determine whether BZR1 also mediates feedback regulation in the dark, we performed quantitative expression analysis of the BR biosynthetic genes CPD, DWF4, and BR6OX in both light and dark conditions using real-time PCR. The results indicate that the expression of these genes in bzr1-1D was reduced to similar extent in the dark as in the light (Fig. 3B). Thus, BZR1 appears to mediate both growth promotion and feedback regulation of BR biosynthesis in both light and dark conditions. The opposite hypocotyl phenotypes of bzr1-1D in the dark and light suggest that the direct growth promoting effect of bzr1-1D is sufficient to overcome the negative effect of BR deficiency in the dark but not in the light. The dwarf phenotype of light-grown bzr1-1D thus suggests that a BZR1-independent BR pathway contributes to growth promotion in the light and is inactivated due to BR deficiency in bzr1-1D.
To identify additional BZR1 regulated genes and to understand the BR regulated transcriptional pathways, the effects of bzr1-1D and det2 mutations on the expression of BR regulated genes were studied using the Arabidopsis full-genome oligo microarray (Affymetrix). Based on previous studies and public microarray data (20, 23, 24), 324 BR-early responsive genes (> 2 fold change within 3 hr of BR treatment, full-genome array) and 235 late-BR-responsive genes (>2 fold change at 12 or 24 hr but not 3 hr of BR treatment, 8300-gene array) (23) were selected for our analysis. These genes were further classified into BR-induced and BR-repressed genes, and those genes affected by bzr1-1D (differ from wild type by more than 20%, P<0.2) were analyzed in detail (Fig 3C). Unlike det2, which consistently affects most genes in the opposite way to BR treatment, bzr1-1D appears to have different effects on different sets of BR regulated genes (Table 1 and Fig. 3C). First, expression of nearly all the bzr1-1D affected early-BR- repressed genes was reduced in bzr1-1D, which is consistent with BZR1 directly regulating BR-early-repressed genes. Second, more of the late-BR-induced genes were down-regulated than up-regulated (72% vs 28%) in bzr1-1D, which could explain the dwarf phenotype of light-grown bzr1-1D. For the early-BR-induced and late-BR-repressed genes, the number of genes activated is similar to that reduced in bzr1-1D. In agreement with physiological studies (Fig. 3A), these results support that bzr1-1D has positive effects on BZR1-downstream components and negative effects on BZR1-independent BR pathways by reducing BR levels. Genes affected similarly by bzr1-1D and det2 but oppositely by BR treatment are likely regulated by the BZR1-independent pathways that are negatively affected by BR deficiency in bzr1-1D. Conversely, genes affected similarly by bzr1-1D and BR treatment are likely downstream targets of BZR1; of those, the early-BR-repressed genes are potentially direct targets of BZR1, and the late-BR responsive genes are most likely indirectly regulated by BZR1. The increased expression of 17 early-BR-induced genes in bzr1-1D suggests that BZR1 might also function as a transcriptional activator; however, genes that respond within 3 hr of BR treatment could also be indirectly regulated by BZR1.
Table 1.
Summary of microarray analysis of the bzr1-1D mutant. Numbers of genes regulated by BR treatment and altered in the bzr1-1D mutant.
| BR-response | BR-regulate genes | Altered in bzr1-1D | Reduced in bzr1-1D | Increased in bzr1-1D |
|---|---|---|---|---|
| Early down | 101 | 26 (25.7%) | 24 (92.3%) | 2 (7.7%) |
| Early up | 223 | 33 (14.8%) | 16 (48.5%) | 17 (51.5%) |
| Late down | 63 | 17 (26.7%) | 8 (47.1%) | 9 (52.9%) |
| Late up | 172 | 36 (20.9%) | 26 (72.2%) | 10 (27.8%) |
The presence of the BRRE in the promoters of various BR regulated genes was analyzed. Consistent with BZR1 being a BR-activated transcriptional repressor, the BRRE is significantly over-represented in the proximal promoter regions of the BR-early-repressed genes (Fig. 3D), compared to all genes of the Arabidopsis genome or to the BR-induced genes. Most of the BR-repressed genes containing BZR1 binding sites are also repressed in the bzr1-1D mutant, further supporting that they are likely direct target genes of BZR1 (Supplementary Table S1). These genes include the BR biosynthetic genes CPD, DWF4, ROT3, and BR6OX (20).
Although bzr1-1D mutation has positive effects on many BR induced genes (Fig. 3b), the fact that the BZR1 binding site sequence is not significantly enriched in BR-induced genes suggests that BZR1 does not activate BR induced genes directly. It is possible that BZR1 activates gene expression by interacting with other DNA binding proteins that bind directly to BR-induced promoters. It is more likely that BZR1 activates gene expression indirectly by repressing the expression of other transcriptional repressors. Previous microarray studies have shown that most BR induced genes respond to BR treatment very slowly compared to BR-repressed or auxin-induced genes (20, 23), suggesting that BR induces these genes indirectly through transcriptional relay. This observation is consistent with auxin and BR regulating gene expression by promoting (25) and preventing the degradation of transcriptional repressors, respectively. Potential transcriptional repressors that are repressed by BZR1 include two Myb-related DNA binding proteins (At4g01680, and At1g71030) and the BZR1 homolog BZR3 (At4g36780) (Supplementary Table S1). These genes contain the BRRE in their promoters and are repressed by BR and bzr1-1D. They are therefore potential direct targets of BZR1. Functional studies of these BZR1-regulated transcription factors will further illustrate the circuit for BR regulation of gene expression.
This study illustrates the molecular mechanisms by which BR signaling regulates gene expression and defines BZR1 as a central regulator for coordinating BR signaling, BR biosynthesis, and growth responses. We demonstrate that BR-regulated gene expression is mediated directly by the novel DNA-binding protein BZR1 and likely its homologs, which are stabilized by upstream BR signaling. Together with previous studies, our results support a model for the function of BZR1 in the BR signal transduction pathway as diagrammed in supplementary figure S1. When BR level is low, BZR1 is phosphorylated by BIN2 and then degraded by the proteasome (17), and the lack of BZR1 would in turn allow expression of BR biosynthetic genes and increase of the BR level. When BR level is high, BR activation of BRI1 and BAK1 receptor kinases leads to dephosphorylation of BZR1 by either inhibiting BIN2 kinase or activating BSU1 phosphatase or both (12, 18, 19). Dephosphorylation of BZR1 prevents its degradation by the proteasome and increases its accumulation in the nucleus (17). BZR1 then binds to the promoters and inhibits the expression of BR biosynthetic genes to reduce the BR level. BZR1 also promotes growth responses by regulating downstream BR responsive genes. Such dual roles of BZR1 in regulating BR biosynthesis and growth responses ensure the optimal levels of BR action for plant development, making BZR1 a central regulator of the BR pathway. The BZR1 feedback loop is thus the key mechanism for maintaining the equilibrium of BR action. How plant cells reset this equilibrium in response to developmental and environmental signals is one of the most interesting questions to be answered in future studies.
Supplementary Material
Acknowledgments
We thank Dr. M. Szekeres for providing the CDP-GUS constructs; Dr. T. J. Guilfoyle for the UAS-GUS, VP16-LexA and IAA17α1-Gal4 constructs; V. Walbot for the 35S-Luciferase vector; W. Frommer for the BY2 cells, B-H. Hou and T. Hamman for assistance with microarray analysis; Y. Yang and N. Marinova for technical assistance; Y. Lou for assistance with promoter cis-element analysis; W. Briggs, D. Bergmann, D. Ehrhardt, Z. He, A. Grossman, and C. Somerville for helpful comments on the manuscript. This work was supported by the grants from the NIH (R01 GM66258-01), DOE (DE-FG02-04ER15525), and the Carnegie Institution of Washington to Z-Y. Wang, and training grant from NIH to J.M. Gendron (5T32GM007276).
References and notes
- 1.Clouse SD, Langford M, McMorris TC. Plant Physiol. 1996;111:671. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altmann T. Trends Genet. 1998;14:490. doi: 10.1016/s0168-9525(98)01598-4. [DOI] [PubMed] [Google Scholar]
- 3.Szekeres M, et al. Cell. 1996;85:171. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Nam KH, Vafeados D, Chory J. Plant Physiol. 2001;127:14. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop GJ, Harrison K, Jones JD. Plant Cell. 1996;8:959. doi: 10.1105/tpc.8.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimada Y, et al. Plant Physiol. 2003;131:287. doi: 10.1104/pp.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Symons GM, Reid JB. Plant Physiol. 2004;135:2196. doi: 10.1104/pp.104.043034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathur J, et al. Plant J. 1998;14:593. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- 9.Bancos S, et al. Plant Physiol. 2002;130:504. doi: 10.1104/pp.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thummel CS, Chory J. Genes Dev. 2002;16:3113. doi: 10.1101/gad.1042102. [DOI] [PubMed] [Google Scholar]
- 11.Li J, et al. Cell. 2002;110:213. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 12.Li J. Curr Opin Plant Biol. 2003;6:494. doi: 10.1016/s1369-5266(03)00088-8. [DOI] [PubMed] [Google Scholar]
- 13.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. Nature. 2001;410:380. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 14.Wang ZY, et al. Dev Cell. 2002;2:505. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 15.Yin Y, et al. Cell. 2002;109:181. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Nam KH. Science. 2002;295:1299. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 17.He JX, Gendron JM, Yang Y, Li J, Wang ZY. Proc Natl Acad Sci U S A. 2002;99:10185. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mora-Garcia S, et al. Genes Dev. 2004;18:448. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ZY, He JX. Trends Plant Sci. 2004;9:91. doi: 10.1016/j.tplants.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Plant Physiol. 2002;130:1319. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackwell TK, Weintraub H. Science. 1990;250:1104. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 22.Tiwari SB, Hagen G, Guilfoyle TJ. Plant Cell. 2004;16:533. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goda H, et al. Plant Physiol. 2004;134:1555. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goda H, Yoshida S, Shimada Y Arabidopsis Information Resource (TAIR) http://arabidopsis.org/servlets/TairObject?type=hyb_descr_collection&id=1007966053.
- 25.Dharmasiri N, Estelle M. Trends Plant Sci. 2004;9:302. doi: 10.1016/j.tplants.2004.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.