Abstract
Genotype imputation is now an essential tool in the analysis of genomewide association scans. The technique allows geneticists to accurately evaluate the evidence for association at genetic markers that are not directly genotyped. Genotype imputation increases power of genomewide association scans and is particularly useful for combining the association scan results across studies that rely on different genotyping platforms. Here, we review the history and theoretical underpinnings of the technique. To illustrate performance of the approach, we summarize results from several actual gene mapping studies. Finally, we preview the role of genotype imputation in an era when whole genome resequencing is becoming increasingly common.
Introduction
Identifying and characterizing the genetic variants that impact human traits, ranging from disease susceptibility to variability in personality measures, is one of the central objectives of human genetics. Ultimately, this aim will be achieved by examining the relationship between interesting traits and the whole genome sequences of many individuals. Although whole genome resequencing of thousands of individuals is not yet feasible, geneticists have long recognized that good progress can be made by measuring only a relatively modest number of genetic variants in each individual. This type of “incomplete” information is useful because data about any set of genetic variants in a group of individuals provides useful information about many other unobserved genetic variants in the same individuals.
The idea that data on a modest set of genetic variants measured in a number of related individuals can provide useful information about other genetic variants in those individuals forms the theoretical underpinning of genetic linkage studies and of haplotype mapping approaches in founder populations (23, 24, 50). These studies typically use <10,000 genetic markers to survey the entire human genome. These markers are used to identify stretches of chromosome inherited from a common ancestor. The shared stretches will usually span several megabases and include thousands of genetic variants. Both approaches have been incredibly successful in the identification of genes responsible for single gene Mendelian disorders (9). In contrast, both these approaches have had only limited success in the context of gene mapping studies for complex traits, although success stories do exist (40, 42, 75, 83).
More recently, technological advances have made genomewide association studies possible (39, 67, 109). Rather than genotyping <10,000 variants, these studies typically genotype 100,000 – 1,000,000 variants in each of the individuals being studied. Since >10 million common genetic variants are likely to exist (104), even these detailed studies examine only a fraction of all genetic variants. While in traditional genetic linkage and founder haplotype mapping studies, geneticists expect to identify long stretches of shared chromosome inherited from a relatively recent common ancestor, in genomewide association studies that focus on apparently unrelated individuals, geneticists expect to identify only relatively short stretches of shared chromosome. Remarkably, genotype imputation can use these short stretches of shared haplotype to estimate the effects of many variants that are not directly genotyped with great precision.
In this review, we will first attempt to provide the reader with an intuition for how genotype imputation approaches work and for their theoretical underpinnings. We will start with the relatively intuitive setting of imputing missing genotypes for a set of individuals using information on their close relatives. We will then proceed to examine how genotype imputation works when applied to more distantly related individuals. Next, we will survey results of studies that have used genotype imputation to study complex disease susceptibility. We will attempt to provide the reader with critical information to assess the merits of genotype imputation based analyses and to provide guidance to analysts attempting to implement these approaches. Finally, we will survey potential uses of imputation based analyses in the context of whole genome resequencing studies that we believe will soon become commonplace.
Genotype Imputation in Studies of Related Individuals
Family samples constitute the most intuitive setting for genotype imputation. Genotypes for a relatively modest number of genetic markers can be used to identify long stretches of haplotype shared between individuals of known relationship. These stretches of shared haplotype (or regions of “identity-by-descent”) are typically used to evaluate the evidence for linkage. Specifically, genetic linkage implies that family members who share a region of chromosome “identical-by-descent” will be more similar to each other than family members with the same degree of relatedness who do not share the region “identical-by-descent”. In the context of genotype imputation, we characterize each of these stretches in detail by genotyping additional markers in one or more individuals in the family. Genotypes for these markers can then be propagated to other family members who are only typed at a minimal set of markers.
The approach is illustrated in Figure 1. In the figure, all individuals have been genotyped for a set of genetic markers are indicated in red; a subset of individuals in the top two generations has been genotyped at additional markers indicated in black (Panel A). Genotypes for the red markers, available in all individuals, can be used to infer the segregation of haplotypes through the family (Panel B). Finally, most of the missing genotypes for individuals in the bottom generation can be inferred by comparing the haplotypes they inherited with copies of the same haplotypes that are “identical-by-descent” and present in other individuals in the family (Panel C).
Figure 1. Genotype imputation within a sample of related individuals.
Panel A illustrates the observed data which consists of genotypes at a series of genetic markers. In this case, a subset of markers have been typed in all individuals (and are marked in red), whereas the remaining markers have been typed in only a few individuals (and appear in black in individuals in the top two generations of the pedigree). Panel B illustrates the process of inferring information on “identity-by-descent” by examining markers for which genotypes are available in all individuals. Each segment of identity by descent that appears in more than one individual is assigned a unique color. For example, a segment marked in purple is shared between the first individual in the grand-parental generation at the top of the pedigree, the first individual in the parental generation, and individuals 3 and 4 in the offspring generation at the bottom of the pedigree. In Panel C, observed genotypes and identity-by-descent information have been combined to fill in a series of genotypes that were originally missing in the offspring generation.
We note that the idea that family members share long stretches of haplotype that are “identical-by-descent” underpins nearly all methods of linkage analysis. Furthermore, many early approaches for association analysis in pedigree data implicitly impute missing genotypes by considering the distribution of potential genotypes of each individual jointly with that of other individuals in the same pedigree (35, 45). The extension of this idea to the imputation of missing genotypes (as outlined above) was first described by Burdick and colleagues (12), who coined the term “in silico genotyping” to describe the idea that computational analyses could be used to replace laboratory based procedures in the determination of individual genotypes. To illustrate the potential of the approach, they re-analyzed the data of Cheung and colleagues (17). Cheung and colleagues sought to identify genetic variants associated with regulation of gene expression by examining RNA transcript levels and genotype data for individuals in the top two generations of the Centre d'Etude du Polymorphism Humain (CEPH) pedigrees (21). The CEPH pedigrees are three generation pedigrees with a structure similar to that of the cartoon pedigree in Figure 1. The top two generations of several of these pedigrees were genotyped at more than 830,000 genetic markers in the first phase of the International HapMap Project (103). Using genotypes for approximately 6,500 genetic markers genotyped by the SNP consortium in all three generations of the pedigrees (85), Burdick and colleagues proceeded to impute genotypes for most of these markers in the third generation of these pedigrees (12). They showed that this imputation based analysis was more powerful than the original analysis which examined only directly genotyped markers for each individual.
Several formal statistical descriptions of genotype imputation procedures for association analyses in families have now been published (15, 108) and the procedures to support genotype in imputation are implemented in packages such as MERLIN (2, 3) and MENDEL (52, 53). In principle, these procedures can be implemented using the infrastructure of the Lander-Green (48) or Elston-Stewart (29) algorithms, or one of the many other pedigree analysis algorithms, including those that are based on Monte Carlo sampling (38, 96). An important observation from these more formal treatments of the problem is that even when genotypes cannot be imputed with high confidence, partial information about the identity of each of the true underlying genotypes can be productively incorporated in association analysis (15, 108). For example, when genotypes are measured directly, observed allele counts are often used in regression analyses to estimate an additive effect for each marker (1, 8, 34). These observed allele counts are discrete and indicate the number of copies of the allele of interest (0, 1 or 2) carried by each individual. When genotypes are not measured directly, these discrete counts can be replaced with an expected allele count for each marker (a real number between 0 and 2) (15).
The approach has been successfully used to study several quantitative traits in a sample of closely related individuals from four villages in Sardinia (77). Among study participants, 1,412 individuals were genotyped with the Affymetrix mapping array set (which assays ~500,000 SNPs) and a further 3,329 individuals were genotyped with Affymetrix 10K SNP mapping arrays (which assay ~10,000 SNPs) (94). The sample was then used to study the genetic architecture of a variety of quantitative traits, ranging from body mass index (94) to fetal hemoglobin levels (106) to personality traits (101). Clearly family based genotype imputation will be maximally useful in samples that include very large numbers of related individuals. In these settings, genotypes for a relatively modest number of individuals can be propagated to many other additional individuals, increasing power. Still, in our view, imputing genotypes for known relatives of the individuals included in a genomewide association scan will always increase power (15) and should be considered whenever individuals to be genotyped in a scan are selected from a larger sample of related individuals previously collected to facilitate linkage analyses or family-based association testing.
Imputation in Samples of Unrelated Individuals
Analyses of related individuals provide the intuition behind genotype imputation: whenever a particular stretch of chromosome is examined in detail in at least one individual, we learn about the genotypes of many other individuals who inherit that same stretch “identical-by-descent”. When studying samples of apparently unrelated individuals, the exact same approach can be utilized. The major difference is that, when studying apparently unrelated individuals, shared haplotype stretches will be much shorter (because common ancestors are more distant) and thus may be harder to identify with confidence. The intuition that short stretches of haplotype provide useful information about untyped genetic markers provides the justification for the potential power gains suggested for many proposed haplotype analysis strategies (22, 60, 91, 115).
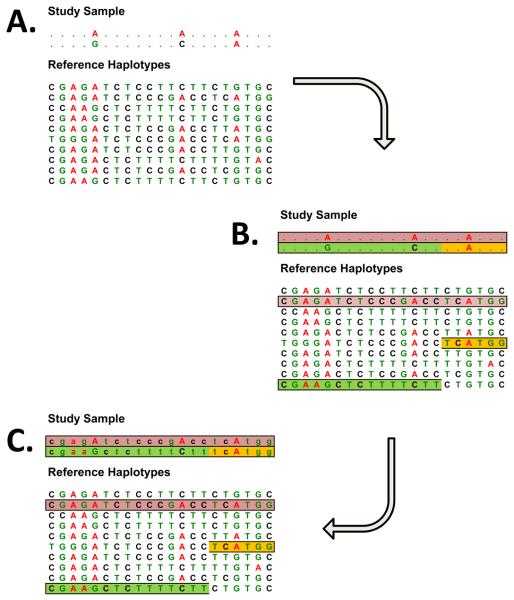
The mechanics of genotype imputation in unrelated individuals are illustrated in Figure 2. Here, study samples genotyped for a relatively large number of genetic markers (perhaps, 100,000 – 1,000,000) are compared to a reference panel of haplotypes that includes detailed information on a much larger number of markers (Panel A). To date, the HapMap Consortium database has typically served as the reference panel (104), but we expect that in the future larger sets of individuals characterized at larger numbers of markers will be available. Stretches of shared haplotype are then identified (Panel B) and missing genotypes for each study sample can be filled in by copying alleles observed in matching reference haplotypes (Panel C). In analyses of samples of European ancestry, comparisons with genotypes for the HapMap CEU panel typically yield shared haplotypes that range from about 100 – 200kb in length. Thus, in a GWAS that examines 300,000 SNP markers, these shared stretches will typically include 10 – 20 genotyped markers. When there is ambiguity about which haplotype stretch should be “copied” to fill in missing genotypes for a particular individual, imputation programs typically provide an answer that summarizes this ambiguity (for example, in 60% of reconstructions genotype A/A was observed at a specific site, whereas in the remaining 40% a different genotype A/C was observed).
Figure 2. Genotype imputation in a sample of apparently unrelated individuals.
Panel A illustrates the observed data which consists of genotypes at a modest number of genetic markers in each sample being studied and of detailed information on genotypes (or haplotypes) for a reference sample. Panel B illustrates the process of identifying regions of chromosome shared between a study sample and individuals in the reference panel. When a typical sample of European ancestry is compared to haplotypes in the HapMap reference panel, stretches of >100kb in length are typically identified. In Panel C, observed genotypes and haplotype sharing information have been combined to fill in a series of unobserved genotypes in the study sample.
In principle, any of the methods typically used to estimate missing haplotypes – whether based on a simple heuristic (18) or on a E-M algorithm (30) or on more sophisticated coalescent models (99) could be used to impute missing genotypes. In fact, most haplotyping programs will automatically “impute” missing genotypes during the haplotype estimation process. In practice, most researchers now use one of tools that have been specifically enhanced to facilitate genotype imputation based analyses. These tools typically provide convenient summaries of the uncertainty surrounding each genotype estimate or, perhaps, convenient built-in association testing. Genotype imputation tools typically fall into two categories: (i) computationally intensive tools such as IMPUTE (64), MACH (59) and fastPHASE/BIMBAM (92, 95) that take into account all observed genotypes when imputing each missing genotype and (ii) computationally more efficient tools such as PLINK (80), TUNA (71), WHAP (114) and BEAGLE (11) that typically focus on genotypes for a small number of nearby markers when imputing each missing genotype. Tools in the first category can be further sub-divided into those that compare the potential haplotypes for each individual with all other observed haplotypes (e.g. IMPUTE and MACH) and those that compare potential haplotypes for each individual to a representative set of haplotypes (e.g. fastPHASE). Typically, tools that consider all available markers and all available haplotypes can require substantially more intensive computation but do better at estimating missing genotypes, particularly for rare polymorphisms. Table 1 provides a partial list of recent genomewide association scans that used genotype imputation, together with the method(s) used for imputing missing genotypes in each scan.
TABLE 1.
EXAMPLES OF GWAS THAT HAVE USED GENOTYPE IMPUTATION.
| First Author | Journal | Publication Date |
Imputation Software |
Title |
|---|---|---|---|---|
| Aulchenko (4) | Nature Genetics | 2008/12 | MACH | Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts |
| Barrett (6) | Nature Genetics | 2008/06 | MACH & IMPUTE |
Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease |
| Broadbent (10) | Hum Mol Genet | 2007/11 | MACH | Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p |
| Chambers (14) | Nature Genetics | 2008/05 | MACH | Common genetic variation near MC4R is associated with waist circumference and insulin resistance |
| Chen (16) | J Clin Invest | 2008/07 | MACH | Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels |
| Cooper (20) | Blood | 2008/06 | BIMBAM | A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose |
| Dehghan (26) | The Lancet | 2008/10 | MACH | Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study |
| Ferreira (32) | Nature Genetics | 2008/07 | PLINK & MACH | Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder |
| Hung (41) | Nature | 2008/04 | MACH | A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25 |
| Kathiresan (44) | Nature Genetics | 2008/12 | MACH | Common variants at 30 loci contribute to polygenic dyslipidemia |
| Lettre (56) | Nature Genetics | 2008/04 | MACH | Identification of ten loci associated with height highlights new biological pathways in human growth |
| Liu (61) | Hum Mol Genet | 2008/03 | IMPUTE | Genome-wide association scans identified CTNNBL1 as a novel gene for obesity |
| Loos (62) | Nature Genetics | 2008/05 | MACH & IMPUTE |
Common variants near MC4R are associated with fat mass, weight and risk of obesity |
| O'Donovan (74) | Nature Genetics | 2008/07 | IMPUTE | Identification of loci associated with schizophrenia by genome-wide association and follow-up |
| Rafiq (81) | Diabetologia | 2008/10 | MACH & IMPUTE |
Gene variants influencing measures of inflammation or predisposing to autoimmune and inflammatory diseases are not associated with the risk of type 2 diabetes |
| Raychaudhuri (82) | Nature Genetics | 2008/09 | IMPUTE | Common variants at CD40 and other loci confer risk of rheumatoid arthritis |
| Sabatti (84) | Nature Genetics | 2008/12 | WHAP | Genome-wide association analysis of metabolic traits in a birth cohort from a founder population |
| Sanders (86) | Am J Psychiatry | 2008/01 | MACH | No Significant Association of 14 Candidate Genes With Schizophrenia in a Large European Ancestry Sample: Implications for Psychiatric Genetics |
| Sandhu (87) | The Lancet | 2008/02 | IMPUTE | LDL-cholesterol concentrations: a genome-wide association study |
| Sanna (89) | Nature Genetics | 2008/01 | MACH | Common variants in the GDF5-UQCC region are associated with variation in human height |
| Scott (93) | Science | 2007/04 | MACH | A Genome-Wide Association Study of Type 2 Diabetes in Finns Detects Multiple Susceptibility Variants |
| Tenesa (100) | Nature Genetics | 2008/03 | IMPUTE | Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21 |
| Willer (111) | Nature Genetics | 2008/01 | MACH | Newly identified loci that influence lipid concentrations and risk of coronary artery disease |
| Willer (112) | Nature Genetics | 2008/12 | MACH & IMPUTE |
Six New Loci Associated with Body Mass Index Highlight a Neuronal Influence on Body Weight Regulation |
| Yuan (113) | AJHG | 2008/10 | IMPUTE | Population-Based Genome-wide Association Studies Reveal Six Loci Influencing Plasma Levels of Liver Enzymes |
| Zeggini (116) | Nature Genetics | 2008/03 | MACH & IMPUTE | Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes |
Accuracy of Genotype Imputation Based Analysis
Our first experience with genotype imputation in the context of a genetic association study occurred when fine-mapping the Complement Factor H susceptibility locus for age-related macular degeneration (58). The locus shows evidence for multiple disease associated alleles and haplotypes (58, 63). Since multi-marker association analyses are much more convenient in the absence of missing genotype data (5), we used the software PHASE (97, 98) and early version of our MACH software (59) to fill in missing genotypes in our sample. In the absence of missing data, it is much easier to compare the evidence for association at different markers and to interpret the results of conditional association analyses that sought to identify independently associated markers. To validate our imputation approach, we masked 5% of the genotypes at the locus and showed that these could be imputed correctly >99% of the time by comparing each individual with a missing genotype to other individuals who shared a common haplotype or haplotypes.
The first few applications of genotype imputation on a genomewide scale also spent considerable effort in validating the accuracy of imputed genotypes. For example, in the first published account of the performance of genotype imputation in the context of a genomewide scan, Scott et al. (93) genotyped a set of type 2 diabetes cases and controls at approximately 300,000 SNPs. They then imputed genotypes at an additional >2 million SNPs to facilitate comparisons with the results of two other genomewide association scans for type 2 diabetes that relied on a different genotyping platforms (90, 117). To evaluate the accuracy of imputed genotypes, they contrasted imputed genotypes generated “in silico” with experimental genotypes generated in the lab for >500 SNPs, including 16 SNPs with imputation based p-values of <10−5 (see online supplementary material in ref. 93). Their results showed excellent concordance between genotype calls, estimated allele frequencies and test statistics for both types of data with an overall allelic discrepancy rate of <1.50% between genotyped and imputed SNPs.
Similar comparisons with newer genotyping platforms, which can provide better coverage of the genome because they include larger numbers of tag SNPs, show that imputed genotypes can achieve even greater accuracy. For example, in the GAIN psoriasis study (69) imputed and experimentally derived genotypes were compared at >660,000 SNPs in 90 individuals with an overall allelic discrepancy rate of <0.90% and an r2 correlation between observed and imputed allele counts that averaged 0.93. The r2 correlation coefficient is a particular useful summary of the impact of genotype imputation on power: in the context of the GAIN psoriasis study we expect that, on average, imputing genotypes for one of the 660,000 evaluated markers in 1,000 individuals would provide a similar amount of information as could be obtained by genotyping the same marker in 930 individuals (69).
Power of Genotype Imputation Based Analyses
One obvious use of genotype imputation based analysis is to accelerate fine-mapping studies. Once an association signal has been identified and confirmed, genotype imputation can be used to evaluate the evidence for association at each of several nearby SNPs and help focus the search for potential causal variants. An example of the approach occurs in the fine-mapping study of Orho-Melander et al. (76). In order to fine-map an association signal linking SNPs in the glucokinase regulatory protein (GCKR) gene and triglyceride levels in blood, Orho-Melander examined evidence for association with genotyped and imputed SNPs in the region and showed that an imputed common missense variant in the GCKR gene was more strongly associated with triglyceride levels than any other nearby SNP, a result that was subsequently confirmed by direct genotyping (76).
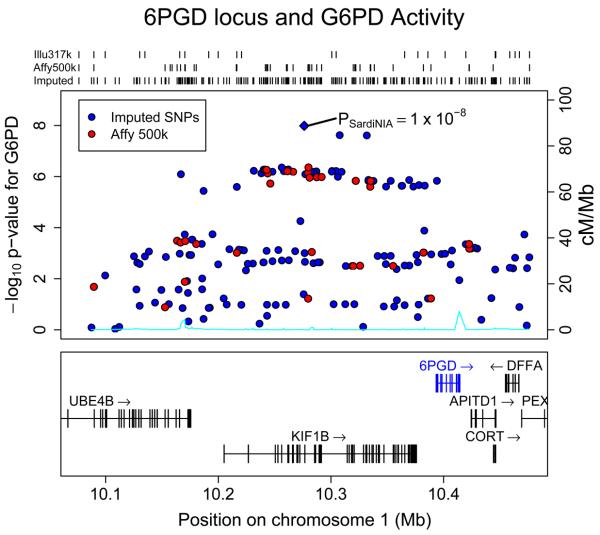
Although we agree that examining evidence for association at imputed markers can be extremely useful in the context of fine-mapping association signals, it is important to note that genotype imputation is also expected to increase the power of genomewide association studies. For example, Willer et al. (111) and Kathiresan et al. (43) showed that rs6511720, a common variant in the low density lipoprotein receptor gene (LDLR), was strongly associated with blood low density lipoprotein (LDL) cholesterol levels (Figure 3). The association signal was missed in an initial analysis that considered only genotyped SNPs because rs6511720 is not included in the Affymetrix arrays used to scan the genome in the majority of their samples and is only poorly tagged by individual SNPs on the chip (the best single marker tag is rs12052058 with pairwise r2 of only 0.21). Another example we have encountered concerns the genomewide association analysis of G6PD activity levels in a sample of Sardinian individuals (77, 94). There, analysis of directly genotyped SNPs revealed two sets of SNPs strongly associated (p < 5×10−8) with G6PD activity levels, one near the G6PD gene locus on chromosome X and another near the HBB locus on chromosome 11. Genotype imputation revealed a strong additional signal (also with p < 5×10−8) upstream of the 6PGD locus on chromosome 1 (Manuela Uda, Serena Sanna, David Schlessinger, personal communication; Figure 4). The three signals (near G6PD, HBB and 6PGD) all fit with our understanding of the biological basis of measurements of G6PD activity: the role of variants near G6PD in the regulation of G6PD activity in Sardinia and elsewhere is well established (25), variants in the HBB locus can influence the lifespan and rate of turnover of red blood cells and it is well established that G6PD activity is higher in younger cells (70) and, finally, it is well known than 6PGD activity levels impact commonly used assays for G6PD activity (13, 31).
Figure 3. Association of genetic variants near LDLR with LDL-cholesterol levels.
The figure illustrates evidence for association between genetic variants near LDLR and LDL-cholesterol levels using data from the SardiNIA (94) and Diabetes Genetics Initiative (DGI, 90) studies reported in Willer et al (111). Evidence for association at each SNP, measured as −log10 P-value, is represented along the y-axis. The placement of each SNP along the X axis corresponds to assigned chromosomal location in the current genome build. Results for directly genotype SNPs are colored in red, imputed SNPs are colored in blue. Note that rs6511720, the SNP showing strongest association in the region, is not well tagged by any of the variants on the Affymetrix genotyping arrays use in the SardiNIA and DGI studies. Evidence for association at the SNP increases to p < 10−25 after follow-up in >10,000 individuals where the SNP was genotyped directly (111).
Figure 4. Association of genetic variants near 6PGD with measurements of G6PD activity.
The figure illustrates evidence for association between genetic variants near 6PGD and measurements of G6PD activity using data from the SardiNIA study (94). Evidence for association at each SNP, measured as −log10 P-value, is represented along the y-axis. The placement of each SNP along the X axis corresponds to assigned chromosomal location in the current genome build. Results for directly genotype SNPs are colored in red, imputed SNPs are colored in blue. Note that although there is evidence for association in the region prior to imputation, the signal increases substantially, to reach genomewide significance, after imputation. The connection between 6PGD activity and measurements of G6PD activity is long established (13).
Overall, the LDLR and 6PGD loci, together with many other anecdotal examples, suggest that genotype imputation can improve the power of genomewide association analyses. Nevertheless, accurately estimating the impact of genotype imputation on the power of a genomewide association studies is more challenging. We have tried to accurately quantify this potential power gain in two ways: first, by generating and analyzing simulated datasets; and second, by analyzing datasets that combine genomewide genotype data and large scale surveys of gene expression. The second approach is especially attractive because true positive associations between genetic variants and transcript levels are easy to identify (they often map to the locus encoding the transcript). Both approaches suggest that genotype imputation can increase the power of gene-mapping studies, particularly when the associated variants have frequencies <10-20%. When we imputed genotypes and then reanalyzed the gene expression data of Dixon et al. (28) we mapped, on average, 10% more genomewide association peaks to the locus surrounding each transcript than before imputation (Liang , Cookson and Abecasis, unpublished data).
Meta-Analysis of Genomewide Association Scans
Perhaps the most dramatic illustration of the utility of genotype imputation has been the ability of researchers to conduct meta-analysis of genomewide association scans even in samples that were originally genotyped using several different platforms. Genotype imputation was first used to combine genomewide association scans for blood lipid levels (43, 111) and height (89) and soon thereafter to combine data across genomewide scans for type 2 diabetes (116), body-mass index (62) and Crohn's disease (6). The success of these meta-analysis can be quite dramatic: in the case of blood lipid levels (43, 111) a meta-analysis of three studies with relatively modest findings (each identifying one to three strongly associated loci), resulted in a total of 19 strongly associated loci including 7 loci not previously implicated in regulating cholesterol and lipoprotein levels in humans. Because it greatly simplifies issues related to examining data collected on multiple different platforms, genotype imputation also makes it simple for researchers to compare results of genomewide association studies that target related traits. In this way, it has been possible to contrast results from genetic studies of blood lipid levels (111) to those of previous studies of coronary artery disease (105), to compare results of studies of blood glucose levels in non-diabetic individuals (79) to those of previous case-control studies of type 2 diabetes (116), and to compare results of studies of height (89) to those of previous studies of osteoarthritis (68). We expect that these sorts of contrasts between the results of genomewide studies for different traits will become ever more commonplace and that they will ultimately provide useful insights about the genetic basis of many complex human traits.
Imputation Based Analysis in Non-European Samples
While most genomewide association studies completed to date have focused on populations of European ancestry (see Table 1 for examples), we expect that genomewide association scans will be conducted in much more diverse groups of samples. The success of genotype imputation depends critically on the choice of reference population from which densely characterized haplotypes are drawn. For studies of European ancestry samples, it is now clear that the HapMap CEU samples (102-104) usually constitute an appropriate reference panel. Similarly, we expect the HapMap CHB+JPT (102-104) samples will constitute a good reference for imputing genotypes in samples of East Asian ancestry and that the HapMap YRI (102-104) samples will constitute a good reference for imputing genotypes in populations of West African ancestry.
Studies of populations that are genetically more distinct from those examined by the HapMap consortium will require more careful consideration in the design of strategies for genotype imputation. For example, we expect that when imputing missing genotypes in Middle Eastern samples, Native American samples or even samples from the Indian sub-continent, it will be advantageous to use a reference panel that includes all HapMap haplotypes, rather than just the CEU, just the YRI or just the CHB+JPT haplotypes. Fortunately, whenever the choice of reference panel is unclear it is possible to mask a subset of the available genotype data, run genotype imputation using each of the different reference panels being considered, and finally contrast imputed and masked genotypes to identify the strategy that provides the most accurate genotypes. Table 2 summarizes the results of a recent analysis (59) that sought to identify the most appropriate reference panel for a series of samples in the Human Genome Diversity Panel (19).
Table 2.
Recommended Choices of HapMap Reference Panel Haplotypes for Imputing Genotypes in Human Genome Diversity Panel Different Samples …
| These Reference Panel Haplotypes… | … Are Best for Imputing Genotypes in these Human Genome Diversity Panel Samples: | |
|---|---|---|
| CEU | Europe: | Orcadian, Basque, French, Italian, Sardinian |
| Middle East: | Druze | |
| CHB+JPT | East Asia: | Han, Han-Nchina, Dai, Lahu, Miao, Oroqen, She, Tujia, Tu, Xibo, Yi, Mongola*, Naxi,Japanese |
| YRI | Africa: | Bantu, Yoruba, San, Mandenka, MbutiPygmy, BiakaPygmy |
| Combined (CEU, CHB, JPT, YRI) | Europe: | Adygei, Russian, Tuscan |
| Middle East: | Mozabite, Bedouin, Palestinian | |
| Asian: | Balochi, Brahui, Makrani, Sindhi, Pathan, Burusho, Hazara, Uygur, Kalash | |
| East Asia: | Daur, Hezhen, Mongola*, Cambodian, Yakut | |
| Oceania: | Melanesian, Papuan | |
| Americas: | Colombian, Karitiana, Surui, Maya, Pima | |
Tie. The Human Genome Diversity Panel Mongola samples are equally well imputed using either the combined HapMap samples (CEU, CHB, JPT and YRI) or just the CHB+JPT samples as a reference. This analysis summarized in this table is adapted from (59). The analysis used estimated haplotypes from (104) and genotype data from (19).
An alternative to using the HapMap samples as a reference is to genotype a subset of study samples for additional markers of interest and then use these as templates for genotype imputation in the remaining samples. This approach was used by Chambers et al (14) to combine data across three different platforms in a recent study of the genetics of obesity focused on individuals of South Asian ancestry. Compared to approaches that use the HapMap as a reference, this strategy can greatly reduce imputation error (14).
Practical Considerations
In this review, we have tried to provide readers with an intuition about why genotyping imputation methods work, describe their history in the context of genomewide association studies, and to summarize some examples of current uses of genotype imputation. For readers that are encouraged to attempt genotype imputation in their own samples, we would like to spend a few paragraphs summarizing important practical issues to consider when carrying out genotype imputation based analyses. In particular, we will focus on issues we have encountered when developing, implementing and supporting our Markov Chain Haplotyping (MACH) software package for haplotype estimation and genotype imputation. As with other analyses of genetic association data, we recommend that a standard set of quality filters should be used to exclude markers with poor quality genotypes. These quality filters typically flag markers that have low call rates, significant evidence for deviations from Hardy-Weinberg equilibrium, a large rate of discrepancies between duplicate genotypes, or evidence for non-Mendelian inheritance (67).
When using an external reference panel as a template for imputation, the most important challenge for successfully imputing genotypes in genome scan samples is ensuring that alleles are labeled consistently (that is, on the same strand) in the reference sample being used and in the samples where missing genotypes will be imputed. MACH checks that allele frequencies are similar in the reference panel and in the samples being imputed, but it cannot catch all errors. In practice, we have found it extremely useful to genotype a small number of HapMap samples as part of each genomewide scan – this helps evaluate genotyping error rates but also ensures that consistency of allele labels can be easily checked.
Once this first hurdle has been surpassed, the next step is to impute missing genotypes for each sample. As noted in Table 2 and in the previous discussion, a key step is to select an appropriate set of reference haplotypes. Different choices of reference panel can be assessed by masking a subset of the available genotypes and checking whether these can be recovered accurately. After a reference panel has been selected and imputation is complete a key issue is deciding which markers to take forward for analysis. Typically, not all markers can be well imputed and several different measures have been proposed to help identify well imputed markers. The simplest of these measures focus on the average probability that an imputed genotype call is correct – in this context, one might look for markers where genotypes are imputed with >90% certainty or so. We don't recommend these types of measures because they are not very meaningful when comparing markers with different allele frequencies (for example, if a marker has an allele frequency of <5%, it should be possible to achieve 90% accuracy by simply assigning the most common genotype to every individual). Instead, we typically recommend measures that try to capture the correlation between imputed genotype calls and the true underlying genotypes – typically expressed as an r2 coefficient. Most often these measures are calculated by comparing the variance in a set of imputed allele counts to theoretical expectations based on Hardy-Weinberg equilibrium (because imputed allele counts for poorly imputed markers show less variability than expected based on allele frequency).
The final step in the analysis of imputed data is to analyze the resulting imputed “genotypes”. MACH and other genotype imputation programs summarize imputation results in a variety of forms. Most often, imputed genotypes are not discrete but, instead, probabilistic. For example, a particular individual might have a 90% probability of carrying genotype A/A and a 10% probability of carrying genotype A/C at a specific marker – corresponding to 1.9 expected copies of allele A. We do not recommend transforming these “probabilistic” genotype calls into discrete genotypes as that can result in a substantial loss of information – especially so for less common alleles. Most often, imputed allele counts for each allele (e.g. 1.9 expected copies of allele A) can conveniently be tested for association with quantitative or discrete traits using an appropriate regression model. Of course, as in other genetic association analyses, adequate adjustment for potential population stratification is essential (27, 36, 78). If ancestry informative principal components are estimated from genetic data (78), we recommend that these should be estimated before imputation.
If results from multiple studies are to be combined, we recommend that each study should be analyzed individually and that results should then be meta-analyzed across studies using standard approaches – for examples, see (89, 111, 112, 116). We never recommend pooling data across studies, especially when these have been genotyped using different platforms.
Challenges for the Future
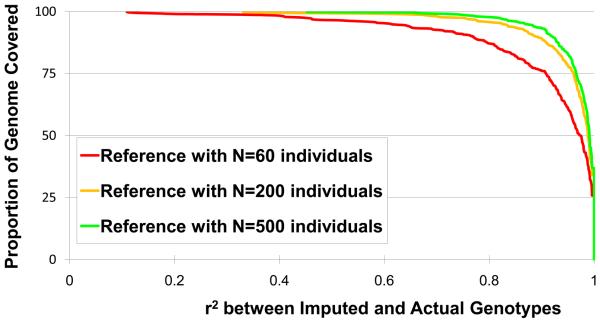
The technologies used in human genetic studies are rapidly improving. We expect several enhancements to genetic imputation technologies. First, we expect that as better characterized reference panels are developed, it will become possible to use genotype imputation methods to study not only single nucleotide polymorphisms but also other types of genetic variants, such as copy number variants (33, 66) or classical HLA types (55). Second, we expect that improved algorithms for genotype imputation will continue to be developed, motivated by the desire of geneticists to tackle ever more complex problems. Similar pressures previously motivated constant development of methods for pedigree analysis, both for large pedigrees (29, 51, 54, 73) and for smaller ones (2, 37, 46-48, 65). Still, the most useful advance that we expect, in the context of genotype imputation based analyses, is the development of larger reference panels. As illustrated in Figure 5, the accuracy of genotype imputation based analyses should increase substantially as the size of reference panels increases. This increase in accuracy occurs because haplotype stretches shared between study samples and samples in the reference panel increase in length and are easier to identify unambiguously with a larger reference panel.
Figure 5. Genome coverage as a function of reference panel size.
The accuracy of imputation increases with the number of individuals in the reference panel. To generate the figure, we analyzed genotyped data from the FUSION study (93). For any given r2 threshold, the results illustrate the proportion of markers whose genotypes can be imputed with equal or greater accuracy. The results illustrate how the proportion of markers whose genotypes are recovered accurately (with high r2 between imputed and actual genotypes) increases with larger reference panels.
Imputation and Genomewide Resequencing Data
So far, we have focused our discussion on the analysis of genotype data. However, it is also clear that genome sequencing technologies are improving extremely rapidly. While the first two human whole genome assemblies took years to complete (49, 107), several additional genomes have been assembled just in the past 18 months (7, 57, 110). Many of the advances in whole genome sequencing have been the result of the deployment of massive throughput sequencing technologies. These technologies differ from standard Sanger based sequencing (88) in many ways. For example, the data produced by these new technologies typically has somewhat higher error rates (on the order of 1% per base). Since these technologies produce very large amounts of data, one typically accommodates these error rates by re-sequencing every base of interest many times to achieve a high-quality consensus.
We expect that the continued deployment of these technologies will change how genotype imputation is used in many different ways. An example of these changes is given by the 1,000 Genome Project (see www.1000genomes.org). The 1,000 Genomes Project aims to deliver whole genome sequences for >1,000 individuals from several different populations in next 12-18 months. To deliver these sequences in a cost effective manner, the 1,000 Genomes Project is using a strategy that combines massively parallel shotgun sequencing technology with the same statistical machinery used to drive genotype imputation based analyses. Specifically, the project is collecting a relatively modest amount of shotgun sequence data for each of the individuals being sequenced: each of the target bases will be re-sequenced only 2-4x on average (statistical fluctuations around this average mean that many bases will not be covered even once), rather than the 10-20x used in previous applications of these technologies to whole genome resequencing. To accurately call polymorphisms in each genome, the Project will then use imputation based techniques to combine information across individuals who share a particular haplotype stretch. Using simulations, we have predicted that when 400 diploid individuals are sequenced at only 2x depth (1x per haploid genome) and the data is analyzed using approaches that combine data across individuals sharing similar haplotype stretches, polymorphic sites with a frequency of >2% can be genotyped with >99.5% accuracy (Li and Abecasis; unpublished data). Note that the same 2x average depth would not be useful for genotype calling when examining a single individual – since, by chance, ~50% of alleles would not be sampled. For another example of how genotype imputation can be combined with sequence data, see (72).
The ability to combine relatively modest amounts of sequence data across many individuals to generate high-quality sequence data for all may become one of the most common uses of imputation technologies in the next several years. For a given sequencing effort, genotype imputation based analyses may allow an increase in the number of individuals to be sequenced by 5 to 10-fold with minimal loss of accuracy in individual genotypes. This sort of increase in sample size is critical when attempting gene-mapping for complex diseases. Of course, even before massively parallel sequencing technologies are deployed more widely, one immediate change will occur with the completion of the 1,000 Genome Project (see www.1000genomes.org). Specifically, we expect these data will include accurate genotype information on >10 million common variants and quickly replace the HapMap Consortium genotypes as the reference panel of choice for imputation studies. Two immediate consequences will be that imputation based analyses will be able to examine even more genetic markers and that each of these markers will, on average, be imputed much more accurately.
Conclusions
Just in the past two years, genotype imputation based analyses have become a key tool for the analysis of human genetic data. They have been used to aid fine-mapping studies, to increase the power of genome wide association studies, to extract maximum value from existing family samples, and to facilitate meta-analysis of genomewide association data. In the next few years, we expect these imputation based analysis will become a key tool in the analysis of massively parallel shotgun sequence data, enabling geneticists to rapidly deploy these technologies to analyze large samples and dissect the genetic basis of complex disease.
Acknowledgements
We thank S. Kathiresan, K. Mohlke, D. Schlessinger and M. Uda for the example relating common variants near LDLR and LDL-cholesterol levels. We thank D. Schlessinger and M. Uda for the example relating variants near 6PGD to G6PD activity levels. Finally, we thank M. Boehnke, K. Mohlke and FUSION colleagues for the data used to generate Figure 5. Yun Li was supported in part by a Rackham Merit fellowship. Cristen Willer was supported in part by an American Diabetes Association Fellowship. Gonçalo Abecasis is a Pew Scholar for the Biomedical Sciences. This research was supported in part by research grants HG-2651, HL-84729 and MH-84698.
REFERENCES
- 1.Abecasis GR, Cardon LR, Cookson WOC. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–92. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 3.Abecasis GR, Wigginton JE. Handling Marker-Marker Linkage Disequilibrium: Pedigree Analysis with Clustered Markers. American Journal of Human Genetics. 2005;77:754–67. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aulchenko YS, Ripatti S, Lindqvist I, Boomsma D, Heid IM, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2008 doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–91. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 6.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50(Pt 2):181–94. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 9.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(Suppl):228–37. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 10.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–14. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 11.Browning SR. Multilocus association mapping using variable-length Markov chains. Am J Hum Genet. 2006;78:903–13. doi: 10.1086/503876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nat Genet. 2006;38:1002–4. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catalano EW, Johnson GF, Solomon HM. Measurement of erythrocyte glucose-6-phosphate dehydrogenase activity with a centrifugal analyzer. Clin Chem. 1975;21:134–8. [PubMed] [Google Scholar]
- 14.Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–8. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 15.Chen WM, Abecasis GR. Family Based Association Tests for Genome Wide Association Scans. American Journal of Human Genetics. 2007;81:913–26. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen WM, Erdos MR, Jackson AU, Saxena R, Sanna S, et al. Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest. 2008;118:2620–8. doi: 10.1172/JCI34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–9. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark AG. Inference of haplotypes from PCR-amplified samples of diploid populations. Molecular Biology and Evolution. 1990;7:111–22. doi: 10.1093/oxfordjournals.molbev.a040591. [DOI] [PubMed] [Google Scholar]
- 19.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 20.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–7. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–7. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 22.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 23.de la Chapelle A. Disease gene mapping in isolated human populations: the example of Finland. J Med Genet. 1993;30:857–65. doi: 10.1136/jmg.30.10.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Chapelle A, Wright FA. Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc Natl Acad Sci U S A. 1998;95:12416–23. doi: 10.1073/pnas.95.21.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Vita G, Alcalay M, Sampietro M, Cappelini MD, Fiorelli G, Toniolo D. Two point mutations are responsible for G6PD polymorphism in Sardinia. Am J Hum Genet. 1989;44:233–40. [PMC free article] [PubMed] [Google Scholar]
- 26.Dehghan A, Kottgen A, Yang Q, Hwang SJ, Kao WL, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–61. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 28.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, et al. A genome-wide association study of global gene expression. Nature Genetics. 2007;39:1202–07. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 29.Elston RC, Stewart J. A general model for the genetic analysis of pedigree data. Hum Hered. 1971;21:523–42. doi: 10.1159/000152448. [DOI] [PubMed] [Google Scholar]
- 30.Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–7. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 31.Fan YH, Lazenbery L, Foster E, Duelm F, Grant E., Jr Improved quantitative method for G6PD deficiency detection. J Clin Lab Anal. 2007;21:107–13. doi: 10.1002/jcla.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008 doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Franke L, de Kovel CG, Aulchenko YS, Trynka G, Zhernakova A, et al. Detection, imputation, and association analysis of small deletions and null alleles on oligonucleotide arrays. Am J Hum Genet. 2008;82:1316–33. doi: 10.1016/j.ajhg.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fulker DW, Cherny SS, Sham PC, Hewitt JK. Combined linkage and association analysis for quantitative traits. Am J Hum Genet. 1999;64:259–67. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.George VT, Elston RC. Testing of association between polymorphic markers and quantitative traits in pedigrees. Genetic Epidemiology. 1987;4:193–201. doi: 10.1002/gepi.1370040304. [DOI] [PubMed] [Google Scholar]
- 36.Guan W, Liang L, Boehnke M, Abecasis GR. Genotype-Based Matching to Correct for Population Stratification in Large-Scale Case-Control Genetic Association Studies. Genetic Epidemiology. 2009 doi: 10.1002/gepi.20403. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–3. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 38.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–60. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirschhorn JN, Daly MJ. Genome-Wide Association Studies for Common Diseases and Complex Traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 40.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 41.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 42.Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–97. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kathiresan S, Willer CJ, Peloso G, Demissie S, Musunuru K, et al. Common DNA Sequence Variants at Thirty Genetic Loci Contribute to Polygenic Dyslipidemia. Nature Genetics. 2009 doi: 10.1038/ng.291. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keavney B, McKenzie CA, Connell JM, Julier C, Ratcliffe PJ, et al. Measured haplotype analysis of the angiotensin-I converting enzyme gene. Hum Mol Genet. 1998;7:1745–51. doi: 10.1093/hmg/7.11.1745. [DOI] [PubMed] [Google Scholar]
- 46.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: A unified multipoint approach. American Journal of Human Genetics. 1996;58:1347–63. [PMC free article] [PubMed] [Google Scholar]
- 47.Kruglyak L, Lander ES. Faster multipoint linkage analysis using Fourier transforms. J Comput Biol. 1998;5:1–7. doi: 10.1089/cmb.1998.5.1. [DOI] [PubMed] [Google Scholar]
- 48.Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci U S A. 1987;84:2363–7. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 50.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–48. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 51.Lange K, Boehnke M. Extensions to pedigree analysis. V. Optimal calculation of Mendelian likelihoods. Human Heredity. 1983;33:291–301. doi: 10.1159/000153393. [DOI] [PubMed] [Google Scholar]
- 52.Lange K, Sinsheimer JS, Sobel E. Association testing with Mendel. Genet Epidemiol. 2005;29:36–50. doi: 10.1002/gepi.20073. [DOI] [PubMed] [Google Scholar]
- 53.Lange K, Weeks D, Boehnke M. Programs for Pedigree Analysis: MENDEL, FISHER, and dGENE. Genet Epidemiol. 1988;5:471–2. doi: 10.1002/gepi.1370050611. [DOI] [PubMed] [Google Scholar]
- 54.Lathrop GM, Lalouel JM, Julier C, Ott J. Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet. 1985;37:482–98. [PMC free article] [PubMed] [Google Scholar]
- 55.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–91. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–54. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y, Ding J, Abecasis GR. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. American Journal of Human Genetics. 2006;79:S2290. [Google Scholar]
- 60.Lin S, Chakravarti A, Cutler DJ. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat Genet. 2004;36:1181–8. doi: 10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- 61.Liu YJ, Liu XG, Wang L, Dina C, Yan H, et al. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum Mol Genet. 2008;17:1803–13. doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maller J, George S, Purcell S, Fagerness J, Altshuler D, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–9. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 64.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 65.Markianos K, Daly MJ, Kruglyak L. Efficient multipoint linkage analysis through reduction of inheritance space. Am J Hum Genet. 2001;68:963–77. doi: 10.1086/319507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCarroll SA. Extending genome-wide association studies to copy-number variation. Hum Mol Genet. 2008;17:R135–42. doi: 10.1093/hmg/ddn282. [DOI] [PubMed] [Google Scholar]
- 67.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 68.Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529–33. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 69.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, et al. Genomewide Scan Reveals Association of Psoriasis with IL-23 and NF-kB Pathways. Nature Genetics. 2009 doi: 10.1038/ng.311. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nathan DG, Stossel TB, Gunn RB, Zarkowsky HS, Laforet MT. Influence of hemoglobin precipitation on erythrocyte metabolism in alpha and beta thalassemia. J Clin Invest. 1969;48:33–41. doi: 10.1172/JCI105972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nicolae DL. Testing untyped alleles (TUNA)-applications to genome-wide association studies. Genet Epidemiol. 2006;30:718–27. doi: 10.1002/gepi.20182. [DOI] [PubMed] [Google Scholar]
- 72.Nyholt DR, Yu CE, Visscher PM. On Jim Watson's APOE status: genetic information is hard to hide. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O'Connell JR, Weeks DE. The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet. 1995;11:402–8. doi: 10.1038/ng1295-402. [DOI] [PubMed] [Google Scholar]
- 74.O'Donovan MC, Craddock N, Norton N, Williams H, Peirce T, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008 doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 75.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 76.Orho-Melander M, Melander O, Guiducci C, Perez-Martinez P, Corella D, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57:3112–21. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pilia G, Chen WM, Scuteri A, Orru M, Albai G, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:e132. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 79.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, et al. Variants in the melatonin receptor 1B gene (MTNR1B) influence fasting glucose levels and risk of type 2 diabetes. Nature Genetics. 2009 doi: 10.1038/ng.290. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al. PLINK: a toolset for whole genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rafiq S, Melzer D, Weedon MN, Lango H, Saxena R, et al. Gene variants influencing measures of inflammation or predisposing to autoimmune and inflammatory diseases are not associated with the risk of type 2 diabetes. Diabetologia. 2008;51:2205–13. doi: 10.1007/s00125-008-1160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–23. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rivera A, Fisher SA, Fritsche LG, Keilhauer CN, Lichtner P, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–36. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 84.Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2008 doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–33. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 86.Sanders AR, Duan J, Levinson DF, Shi J, He D, et al. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008;165:497–506. doi: 10.1176/appi.ajp.2007.07101573. [DOI] [PubMed] [Google Scholar]
- 87.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–91. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanna S, Jackson AU, Nagaraja R, Willer CJ, Chen WM, et al. Common variants in the GDF5 region are associated with variation in human height. Nature Genetics. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 91.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–44. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–5. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scuteri A, Sanna S, Chen W-M, Uda M, Albai G, et al. Genome Wide Association Scan shows Genetic Variants in the FTO gene are Associated with Obesity Related Traits. PLoS Genetics. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007 doi: 10.1371/journal.pgen.0030114. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–37. [PMC free article] [PubMed] [Google Scholar]
- 97.Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–93. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- 98.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–62. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Terracciano A, Sanna S, Uda M, Deiana B, Usala G, et al. Genome-wide association scan for five major dimensions of personality. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 103.The International HapMap Consortium The International HapMap Project. Nature. 2005;437:1299–320. [Google Scholar]
- 104.The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A. 2008;105:1620–5. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 108.Visscher PM, Duffy DL. The value of relatives with phenotypes but missing genotypes in association studies for quantitative traits. Genet Epidemiol. 2006;30:30–6. doi: 10.1002/gepi.20124. [DOI] [PubMed] [Google Scholar]
- 109.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–18. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 110.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–6. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 111.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, et al. Genome-Wide Association Scans Identify Novel Loci That Influence Lipid Levels and Risk of Coronary Artery Disease. Nature Genetics. 2008;40:161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Willer CJ, Speliotes EK, Loos RJF, Li S, Lindgren CM, et al. Six New Loci Associated with Body Mass Index Highlight a Neuronal Influence on Body Weight Regulation. Nature Genetics. 2009 doi: 10.1038/ng.287. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yuan X, Waterworth D, Perry JR, Lim N, Song K, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520–8. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zaitlen N, Kang HM, Eskin E, Halperin E. Leveraging the HapMap correlation structure in association studies. Am J Hum Genet. 2007;80:683–91. doi: 10.1086/513109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- 116.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–41. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]