Abstract
The physiological role of human deoxycytidine kinase (dCK) is to phosphorylate deoxynucleosides required for DNA synthesis, with the exception of thymidine. Previous structural analysis of dCK implicated steric factors, specifically the thymine methyl group at the 5-position, that prevent thymidine phosphorylation by dCK. This hypothesis is supported by the observation that mutations that enlarge the active site cavity in proximity to the nucleoside 5-position endow dCK with the ability to phosphorylate thymidine. However, in conflict with this hypothesis was our discovery that the cytidine analog 5-methyl-deoxycytidine (5-Me-dC), an isostere of thymidine, can indeed be phosphorylated by wild-type (WT) dCK. To reconcile this seemingly contradicting observations, and to better understand the determinants preventing thymidine phosphorylation by WT dCK, we solved the crystal structure of dCK in complex with 5-Me-dC. The structure reveals the active site adjustments required to accommodate the methyl group at the 5-position. Combination of kinetic, mutagenesis, and structural data suggested that it is in fact residue Asp133 of dCK that is most responsible for discriminating against the thymine base. dCK variants in which Asp133 is replaced by an alanine and Arg104 by select hydrophobic residues attain significantly improved activity with 5-substituted deoxycytidine and thymidine analogs. Importantly, the ability of the designer enzymes to activate 5-substitued pyrimidines makes it possible to utilize such nucleoside analogs in suicide gene therapy or protein therapy applications that target cancer cells.
Keywords: deoxycytidine kinase, crystal structure, enzyme engineering, thymidine kinase activity
Human deoxycytidine kinase (dCK) is responsible for converting the pyrimidine deoxycytidine (dC) and the purines deoxyadenosine (dA) and deoxyguanosine (dG) into their monophosphate forms (1). In addition to its purine-pyrimidine promiscuity, dCK is endowed with the ability to phosphorylate nucleoside analogs with modifications in the base, sugar, or both moieties (2, 3). It is this characteristic that has made dCK a critical enzyme required for the pharmacological activity of numerous nucleoside analogs that are used to treat viral infections (4) and cancer (5). Yet, dCK does not phosphorylate thymidine – the role of phosphorylating this nucleoside is carried out in the cytoplasm by human thymidine kinase 1 (TK1), and in the mitochondria by human thymidine kinase 2 (TK2) (6). It is worth mentioning that, in contrast to mammals, the fruit fly contains a single nucleoside kinase that phosphorylates any type of nucleoside (7).
We are developing a targeted approach against cancer that is based on delivering a nucleoside kinase with unique activities to the cancer cells. To attain such a unique kinase we have selected to engineer human dCK (dCKEN) (8). The concept is to deliver the gene, or the gene product, of dCKEN in a specific manner to cancer cells. Subsequent administration of nucleoside analogs that are not substrates for the endogenous nucleoside kinases, but that are substrates of the delivered dCKEN, would result in the activation of the nucleoside analogs only in cells that accumulated dCKEN. This will lead to selective killing of the targeted cells.
We reasoned that dCK engineered to acquire thymidine kinase activity might provide an enzyme with a unique activity profile for combined treatment with thymidine analog prodrugs. Our goal is to design variants of dCK that can phosphorylate thymidine analogs that are not substrates of either TK1 or TK2 (9). We favored modifying dCK to become a thymidine kinase over the alternative choice of expanding the substrate repertoire of one of the two human thymidine kinases for the following reasons: in the case of the cytosolic thymidine TK1, it is known that this relatively substrate-specific kinase uses main-chain atoms to bind the nucleoside (10), which significantly complicates the enzyme engineering process. In the case of the mitochondrial TK2 (11), the essential structure information needed to guide protein engineering is not available.
As mentioned previously, the enzyme dCK currently plays a critical role in the activation of important nucleoside analogs used for the treatment of different cancer types (12) (e.g., cytarabine, gemcitabine, cladribine and fludarabine) and viral infections (13) (e.g. lamivudine and emtricitabine). If the concept is to improve the activation of established nucleoside analogs to eradicate the cancer by delivering a nucleoside-analog activating kinase to cancer cells, why not simply deliver wild-type (WT) dCK to the targeted tumor cells? The problem with such an approach is that most cells constitutively express dCK at some level, the enzyme not being cell cycle-regulated, in contrast to TK1. In fact, the clinical use of these compounds is hampered by toxic side effects on non-tumor cells (14). Therefore, delivery of additional WT dCK to tumor cells, for the purpose of promoting the phosphorylation of any of the above nucleoside analogs, would still be complicated by systemic activation of these compounds through endogenous dCK. To avoid prodrug activation in non-targeted cells, it is imperative to identify a nucleoside analog that is not phosphorylated by any human kinase, yet is a substrate of the delivered engineered enzyme.
Towards endowing dCK with thymidine kinase activity, we performed structural studies of the enzyme in complex with pyrimidine and purine substrates (3, 8, 15–19). This work suggested a mechanism of steric exclusion of the 5-methyl group of the thymine base by the active site residues Glu53 and Arg104 as the cause for the very poor thymidine kinase activity of WT dCK (20). Circumventing this steric problem by replacing Glu53 with a smaller residue was not an option since this carboxylic acid is essential for catalytic activity. Therefore, we focused on exchanging Arg104, whose positive charge has no role in catalysis, to a residue with a smaller side-chain. We simultaneously replaced Arg104 with the slightly shorter methionine, and Asp133 with an alanine, since Asp133 directly interacts with the guanidinium group of Arg104. And indeed, this double mutant (R104M-D133A) attained thymidine kinase activity (20).
However, if steric reasons were the sole factor behind the lack of thymidine kinase activity of dCK, then the isostere of thymidine, the dC-analog 5-methyl-deoxycytidine (5-Me-dC), should also be excluded by dCK. Here we report the unexpected observation that 5-Me-dC yet is a substrate of WT dCK. To understand how WT dCK accommodates 5-Me-dC we solved the crystal structure of the enzyme in complex with 5-Me-dC at the nucleoside acceptor site and ADP at the phosphoryl donor site. The structure exposes the precise active site adjustments needed to accept this 5-modified deoxycytidine analog by WT dCK. Importantly, it also prompted us to modify our previous explanation for the exclusion of thymidine by dCK to include both steric and electrostatic factors. From electrostatic considerations, the presence of an aspartic residue at position 133 acts to favor pyrimidines with a cytosine base and exclude those with a thymine/uracil base. Size remains a major factor in discriminating against 5-substituted pyrimidines. Increasing the cavity size in proximity of the base 5-position by varying the residue at position 104 – concomitant with the D133A mutation - resulted in dCK variants with increased activity towards 5-substituted dC and thymidine analogs.
Materials and Methods
Materials
General laboratory reagents were purchased from Fisher (Pittsburgh, PA) and Sigma & Aldrich (St. Louis, MO). Enzymes were purchased from New England BioLabs (Ipswich, MA) unless indicated otherwise. DNA samples were purified using the EndoFree Plasmid Maxi Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocols. Primers were purchased from Sigma. L-dU, L-dT, and the 5-substitued dC analogs were purchased from ChemGenes Corporation, Wilmington, MA. All other nucleosides and nucleotides were obtained from Sigma.
Site-directed mutagenesis
Point mutations were generated using the QuikChange Site Directed mutagenesis kit (Stratagene, La Jolla, CA). Gene sequences were confirmed by DNA sequencing.
Protein expression and purification
C41 (DE3) Escherichia coli was transformed with the wildtype (WT) dCK or the dCK R104-D133 double mutants in the pET14b vector, grown in 2YT media, and induced with 0.1 mM IPTG over 4 h at 37 °C. Cells were harvested and the pellet lysed by sonication. Lysates were cleared by centrifugation at 30,000 × g for 1 hour at 4 °C and subjected to purification with HisTrap™ HP (GE Healthcare) following the supplier’s protocol. After elution with 250 mM imidazole, the protein was further purified by gel filtration using an S-200 column. Protein fractions were pooled, concentrated, aliquoted, frozen in liquid nitrogen, and stored until use at −80 °C.
Human TK2 was cloned into a pET14b vector by PCR using a commercial brain cDNA library. For improved expression, we truncated the seven N-terminal residues, substituted cysteine 222 with a glycine, and expressed TK2 as a fusion protein with SUMO. This version of TK2 (pET14b-SUMO-TK2[37N; C222G]) was purified using a HisTrap™ HP column, with the eluted protein subject to SUMO-tag cleavage by SUMO protease. TK2 was further purified by gel filtration, concentrated to 10 mg/ml, and kept frozen until use.
Kinetic assay
The activities of WT dCK and mutants, and of TK2 were determined using an NADH-dependent enzyme-coupled assay and a Cary UV spectrophotometer (8, 21). All measurements were performed in triplicate at 37 °C in a buffer containing 100 mM Tris, pH 7.5, 100 mM KCl, 10 mM MgCl2 and 1 mM ATP. For data in which kobs is given, a single nucleoside concentration of 200 μM was used. The enzyme concentration was 0.33 μM. For data in which Km and kcat values are given, the nucleoside concentration was varied with all other components being fixed. Data were fit to the Michaelis-Menten equation using SigmaPlot.
Crystallization, X-ray data collection and refinement
Crystals of WT dCK complexed with 5-Me-dC+ADP were grown at room temperature using the vapor diffusion method from hanging drops (1 μl of protein at 8 mg/ml, 5 mM 5-Me-dC and 5 mM ADP mixed with 1 μl of reservoir buffer) using a reservoir solution that contained 0.9–1.5M trisodium citrate di-hydrate and 100 mM Tris, pH 7.5. Crystals were cryoprotected with mineral oil, and diffraction data were collected from a single frozen crystal using a RAXIS-IV++ detector mount on a Rigaku RH-200 rotating anode x-ray generator. Data were processed with XDS (22). The structure was solved by molecular replacement using the program MOLREP (23) and the dCK structure 1P5Z as search model. Refinement was carried out with REFMAC (24). Data collection and refinement statistics are presented in Table 2. Arginine 128 is the sole residue in the disallowed region of the Ramachandran Plot, a feature constant in all human dCK structures.
Table 2.
Summary of data collection and refinement statistic for the WT dCK 5-Me-dC+ADP complex
| Data Collection statistics | |
| X-ray source | Rotating anode |
| Wavelength (Å) | 1.54 (Cu anode) |
| Temperature (K) | 100 |
| Resolution Range (Å) | 1.96–30.0 |
| Reflections | |
| Observed | 210295 |
| Unique | 39888 |
| Completeness (%)a | 99.0 (95.5) |
| Rsym(%) | 4.6 (35.7) |
| I/σ(I) | 23.0 (4.1) |
| Space group | C2221 |
| Unit cell (Å) | |
| a | 52.74 |
| b | 132.95 |
| c | 156.89 |
| Molecules per a.u. | 2 |
| Refinement statistics | |
| Rcryst (%) | 19.34 |
| Rfree (%) | 23.90 |
| Number of atoms | |
| Protein | 3862 |
| Nucleoside | 34 |
| ADP | 54 |
| Water | 257 |
| R.m.s. deviation | |
| Bond length (Å) | 0.013 |
| Bond angles (°) | 1.501 |
| Average B-factors (Å2) | |
| Overall Chain A | 29.38 |
| Main chain A | 28.72 |
| Side chain A | 30.01 |
| Overall Chain B | 26.32 |
| Main chain B | 25.66 |
| Side chain B | 26.97 |
| ADP | 20.90 |
| Nucleoside | 20.90 |
| Waters | 34.22 |
| Ramachandran plot (residues in): | |
| most favored region (%) | 92.3 |
| additionally allowed regions (%) | 6.6 |
| generously allowed regions (%) | 0.7 |
| disallowed regions (%) | 0.4 |
values for the highest resolution shell is in parenthesis
Results and Discussion
Endowing dCK with thymidine kinase activity
Previously we reported that the dCK double mutant R104M-D133A acquired thymidine kinase activity (20), with both the physiological D-form of dT, and with its enantiomeric L-form – see Table 1, section A. This finding is congruent with our earlier observations on the structural features that allow dCK to phosphorylate both the D- and L-form of nucleosides (3, 15). We next examined which other dCK modifications at position 104 would be consistent with thymidine kinase activity. Based upon size and environment considerations, we tested select large hydrophobic and non-charged residues (Table 1). The best double mutant for this purpose, R104L-D133A acquired an observed rate (kobs) of ~2.9 sec−1 with D-dT as substrate, a 350-fold increase over the thymidine kinase rate of WT dCK (kobs is the rate observed at 200 μM nucleoside, 1 mM ATP, 37 °C). This thymidine kinase rate is 4-fold higher than that measured with TK2 (Table 1, A), and comparable to the rate with TK1 (10).
Table 1.
Observed rate (kobs) of WT dCK, R104X-D133A double mutants, and human TK2 (sec−1)
| Nucleoside | WT dCK | R104M-D133A | R104L-D133A | R104I-D133A | R104Q-D133A | TK2 | |
|---|---|---|---|---|---|---|---|
| A | D-dC | 0.040a±0.001b | 1.84±0.04c | 0.21±0.09c | 0.06±0.01c | 0.08±0.01c | 0.44±0.02 |
| D-dT | 0.008±0.001c | 1.15±0.01c | 2.86±0.09c | 0.40±0.01c | 0.10±0.06c | 0.70±0.04 | |
| L-dT | 0.017±0.002c | 1.95±0.09c | 1.07±0.09c | 0.19±0.02c | 0.15±0.02c | 1.50±0.02 | |
| D-dU | 0.021±0.002 | 0.370±0.005 | 1.84±0.08 | 0.55±0.07 | 0.100±0.005 | 0.57±0.01 | |
| L-dU | 0.027±0.001 | 0.59±0.02 | 2.38±0.13 | 0.51±0.06 | 0.140±0.001 | 0.90±0.02 | |
| B | 5-Me-dC | 0.080±0.004 | 0.280±0.013 | 0.180±0.014 | 0.027±0.001 | 0.110±0.022 | 0.260±0.014 |
| 5-Bromo-dC | 0.050±0.001 | 0.250±0.001 | 0.076±0.017 | 0.012±0.005 | 0.060±0.004 | 0.250±0.010 | |
| 5-Iodo-dC | 0.031±0.007 | 0.160±0.013 | 0.086±0.002 | 0.023±0.001 | 0.070±0.003 | 0.145±0.209 | |
| 5-Propynyl-dC | 0.014±0.002 | 0.380±0.001 | 0.115±0.006 | 0.023±0.002 | 0.027±0.002 | 0.093±0.009 | |
| C | BVdU | 0.010±0.001 | 0.57±0.06 | 1.16±0.08 | 0.190±0.040 | 0.100±0.020 | 0.180±0.013 |
kobs are for a nucleoside concentration of 200 μM, plus 1 mM ATP
Standard deviation
Data from Hazra et al, Biochemistry, 2009.
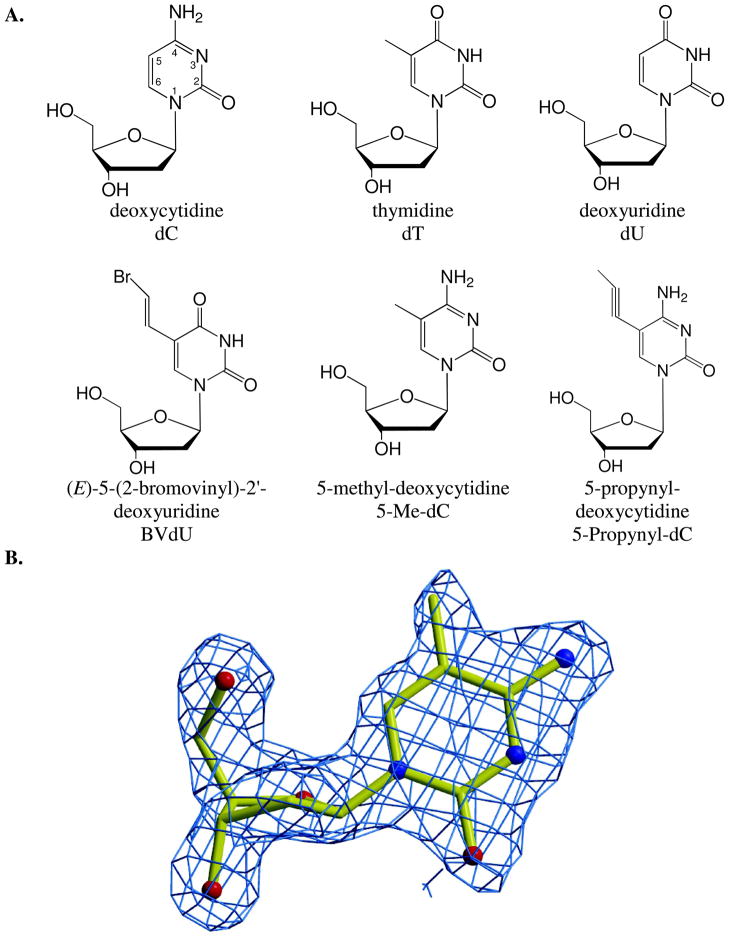
If steric reasons were the sole factors behind the exclusion of thymidine by WT dCK, it would be predicted that 5-methyl-deoxycytidine (5-Me-dC) would also not be a substrate of dCK (5-Me-dC is isosteric with dT). Conversely, deoxyuridine (dU) would be predicted to be a good substrate since it lacks the 5-methyl group present on thymidine (dU is isosteric with dC). Both of those predictions were incorrect, as shown by the kinetic data with WT dCK (Table 1). In fact, under the conditions tested, 5-Me-dC was phosphorylated by WT dCK at a kobs 2-fold faster, and dU was phosphorylated at a kobs 2-fold slower, than dC. These results revealed that steric factors alone do not account for the very low thymidine phosphorylation rate of WT dCK. The chemical structure of these and other nucleosides discussed in this report are presented in Figure 1.
Figure 1.
A. Chemical structure of the nucleoside analogs examined. In case of dT and dU, both the physiological D-form and non-physiological L-form were tested. B. Electron density map (blue) for 5-Me-dC, shown as ball-and-stick representation.
Structure of WT dCK in complex with 5-Me-dC and ADP
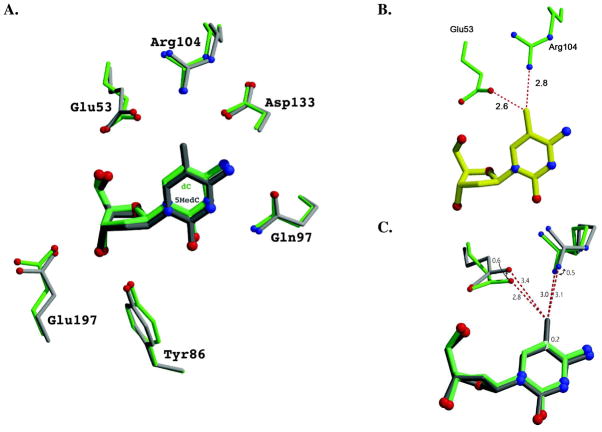
Why, in contrast to thymidine, is 5-Me-dC a substrate of WT dCK? To address this question, we solved the crystal structure of WT dCK in complex with 5-Me-dC at the nucleoside binding site and ADP at the nucleotide binding site. Data collection and refinement statistics for this complex are provided in Table 2. The overall structure of WT dCK in complex with 5-Me-dC+ADP is almost identical to that observed with other pyrimidine nucleosides (rmsd to the D-dC+ADP complex [PDB ID 1P5Z] is 0.56 Å over 226 atoms). The electron density at the nucleoside binding site clearly shows the presence of the methyl group at the 5 position of 5-Me-dC (Figure 1B). 5-Me-dC is bound in the anti conformation, thereby conserving the orientation seen with dC (8). Overlay of the 5-Me-dC+ADP and dC+ADP structures reveals that, in order to accommodate the 5-methyl group, there is slight adjustment in the position of the nucleoside in combination with repositioning of side chains of Arg104 and of Glu53 (Figure 2A). For comparison, the position of 5-Me-dC modeled as if it were to bind at the exact position of dC is shown in Figure 2B. To minimize steric hindrance with Glu53 and Arg104, the base of 5-Me-dC shifts ~0.2 Å relative to the position observed with D-dC (Figure 2C). In a counter movement, the side chains of Glu53 and Arg104 adjust their position. The combination of these slight—0.2 to 0.6 Å— changes in positions suffice to make room for the methyl group at the 5-position. We realize that the conformational change of the nucleoside is close to the coordinate error at our resolution. However, we note that since the change in nucleoside position encompasses the whole molecule and not a single atom, this slight shift seems to be real. The changes in conformation of the enzyme side-chains in response to 5-Me-dC binding are >0.5 Å, so those changes are well resolved in our structures.
Figure 2.
A slight shift in position of the nucleoside, plus adjustment of the side chains of Glu53 and Arg104, allow WT dCK to accommodate the additional methyl group present in 5-Me-dC versus dC. (A) Superposition of the WT dCK D-dC+ADP complex (green; PDB ID 1P5Z) on the WT dCK 5-Me-dC+ADP complex (gray). 5-Me-dC is positioned slightly further from Arg104. (B) A theoretical model of 5-Me-dC (yellow) were it to bind at the same position as D-dC. Such a binding mode would produce unfavorable close contacts with Glu53 (2.6 Å) and Arg104 (2.8 Å). (C) The adjustments made by the active site residues due to the methyl group of 5-Me-dC include a shift of the side chain of Glu53 by 0.6 Å, Arg104 by 0.5 Å, and the nucleoside itself by 0.2 Å. The shift of Glu53 increases the distance to the methyl group from otherwise 2.8 Å to 3.4 Å.
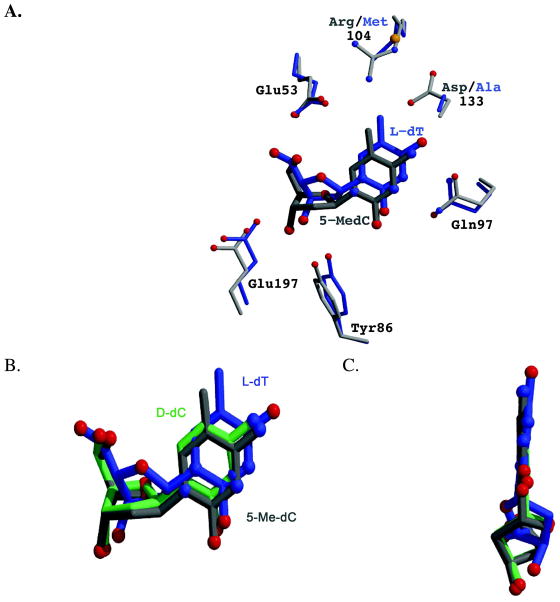
In a previous study of the binding of L-dT to the R104M-D133A dCK double mutant (20), we observed that the nucleoside binds more deeply in the active site relative to the positioning of dC bound to WT dCK (Figure 3). The deeper positioning of L-dT is made possible by the space generated due to the double mutations. The need to limit steric repulsion between the 5-methyl group of dT and the side chain of Glu53 provides the driving force for this deeper positioning. However, in the case of 5-Me-dC complexed with the WT enzyme, a similar shift by the nucleoside is not possible. In this case, we observe that 5-Me-dC binds in a similar position as dC, and it is predominantly the enzyme that compensates for the presence of the 5-methyl group by repositioning the side chains of Glu53 and Arg104 (Figure 2). An overlay of 5-Me-dC, dC, and L-dT shows the relative positioning of the three nucleosides (Figure 3, panels B & C).
Figure 3.
Comparison of the binding of the nucleosides L-dT (blue), 5-Me-dC (gray) and D-dC (green) to dCK. (A) The L-dT complex was solved with the R104M/D133A variant of dCK. The substitution of Arg104 by a methionine and of Asp133 by alanine increases the active site cavity. Exploiting the increased cavity, L-dT binds deeper into the active site. This repositioning of the nucleoside increases the distance between the 5-methyl group of L-dT and the side-chain of Glu53, thereby eliminating an unfavorable interaction. An analogous movement deeper into the active site is not possible for 5-Me-dC in the context of WT dCK. (B) Overlay of the nucleosides as observed in the structure reported here of WT dCK with 5-Me-dC+ADP (gray), WT dCK with D-dC+ADP (green; PDB ID 1P5Z), and R104M/D133A dCK with L-dT+ADP (blue; PDB ID 3HP1). L-dT binds deeper in the active site due to the larger cavity generated by the R104M/D133A mutations. 5-Me-dC binds slightly lower in this perspective in order to minimize the negative interaction between the 5-methyl group and the side-chains of Glu53 and Arg104. (C) View parallel to the face of the base. The base in L-dT is tilted relative to the base as seen in D-dC and 5-Me-dC. Such base tilting is observed whenever dCK binds nucleosides of the L-chirality.
This analysis of the structure of WT dCK in complex with 5-Me-dC+ADP revealed that some positional flexibility of the Glu53 and Arg104 side chains is tolerated. This was not obvious previously, since Glu53 requires precise positioning for dCK activity. Glu53 functions to catalyze phosphoryl transfer by promoting the deprotonation of the nucleoside 5′-OH group (mutation of Glu53 to either an alanine or glutamine abolishes activity, data not shown). This deprotonation increases the nucleophilicity of the nucleoside, thereby promoting its reaction with the nucleotide γ-phosphate. To retain activity, any repositioning of Glu53, due to the presence of a substituent at the 5-position of a cytosine base, must maintain its ability to fulfill its catalytic role. The enzyme being able to tolerate a methyl group, we asked what other 5-position substituents would be tolerated by dCK. Halogens such as bromine or iodine at this position were still compatible with dCK activity (Table 1, section B). Surprisingly, even the larger propynyl group (—C≡C-CH3) is accepted, albeit with a penalty in rate, compared to compounds with a smaller substituent. These results reveal the unexpected and surprising adaptability of WT dCK to dC analogs with substituents at the 5-position.
We also determined the observed rates of 5-Me-dC and that of its isostere thymidine with the dCK double mutants that we generated. The highest kobs were seen with the mutants that had either a methionine or a leucine at position 104. As expected, the increased space due to these smaller residues (relative to arginine in WT dCK) in proximity to the 5-position resulted in an increased phosphorylation rate with the double mutants in comparison to that attained by WT dCK. For 5-Me-dC, the enzyme variant with the highest observed phosphorylation rate had the methionine at position 104 (Table 1, B). In contrast, for the isosteric D-dT, the highest kobs was seen for the enzyme that had a leucine at position 104. Interestingly, while in both cases the phosphorylation rates of the D-dT and 5-Me-dC were much improved by the mutant enzymes compared to the rates with WT dCK, the observed rate of 5-Me-dC was 10-fold lower than that with D-dT. As D-dT and 5-Me-dC are isosteres, this suggests that while the size of the cavity plays a role in determining the rate of phosphorylation, other factors are also important.
The kinetic data in Table 1 was obtained at a fixed nucleoside concentration of 200 μM. Since phosphorylation efficiency is a function of both rate and Km values, we also performed a full kinetic analysis for many of the nucleosides discussed here with WT and mutant dCK – see Supplementary Table S1. When comparing the catalytic rate (kcat) and Km values for the isosteric nucleosides D-dT and 5-Me-dC, measured with the methionine-containing double mutant, we observe a lower kcat with 5-Me-dC (0.36 versus 1.74 sec−1) that is compensated by a much lower Km value (4 versus 144 μM). Thus, in terms of catalytic efficiency (i.e., kcat/Km), 5-Me-dC is ~7-fold better than D-dT. We note that a common feature observed with dCK is the correlation between a low Km value and a low phosphorylation rate (8).
Rationalizing the lack of thymidine kinase activity by WT dCK
The observation that even bulky substituents at the cytosine base 5-position are accepted as substrates by WT dCK begs the questions: What impedes dT from being phosphorylated, and why is dU, with only a hydrogen at the 5-position, such a poor substrate? Re-examining all available kinetic and structural data suggested that the answer lies in the nature of the residue at position 133. In WT dCK, this residue is an aspartic acid. When dC binds at the active site, its 4-amino group forms an attractive interaction with the side chain of Asp133 (Figure 2A). The same would apply for 5-Me-dC. However, in the case of dT or dU, which have a carbonyl group at position 4, a repulsive interaction occurs, which discriminates against their productive binding at the active site of WT dCK. In the case of the double dCK mutants that are described here, Asp133 is replaced by an alanine. This eliminates the repulsive interaction, allowing efficient phosphorylation of both dT and dU (Table 1, section A).
The conclusion of these studies is that the main factor that prevents WT dCK from being a thymidine kinase is the negative interaction between Asp133 and dT’s carbonyl group at the 4-position. In the absence of this conflict, as the structure of WT dCK in complex with 5-Me-dC demonstrates, the methyl group at the 5-position could be accommodated by repositioning of the Glu53 and Arg104 side chains. Does this suggest that the single mutation of D133A would suffice to convert human dCK to a thymidine kinase? Previously, using a thymidine kinase-deficient E. coli strain, it was observed that dCK harboring the single D133A mutation could not complement for the deficiency (25). This suggests that the D133A variant is either devoid of thymidine kinase activity, is unstable, or both. We favor the latter explanation, since, in the context of the single mutation D133A, the arginine at position 104 is left without its charge-stabilizing partner. Thus, to add thymidine kinase activity to dCK, both Asp133 and Arg104 need to be changed.
Our structural analysis of WT dCK in complex with 5-Me-dC explains how a dC analog with a 5-subsituent can be accommodated as a substrate. Not surprisingly, as the substituent size is increased, the observed rate decreases. For example, with the small methyl group at the 5-position (i.e. 5-Me-dC), the kobs is 0.08 sec−1, whereas with the larger 5-propynyl group at this position the rate is reduced to 0.014 sec−1 (Table 1, B). In contrast, with the R104M-D133A variant of dCK, where the cavity near the 5-substituent is enlarged, bulkier substituents can be accepted without penalty (Table 1, B). Thus, in the case of this dCK variant, 5-propynyl-dC has a rate of 0.38 sec−1 (a 27-fold increase over WT dCK), whereas 5-Me-dC has a rate of 0.28 sec−1 (only a 3.5-fold increase over WT dCK).
Identifying thymidine analogs that are phosphorylated efficiently by the engineered dCK variants
The potential translational aspect of this work is to employ the engineered dCK variants to activate 5-modified pyrimidine analogs, in the context of suicide gene therapy or protein therapy strategies. The desired properties of such analogs are that they are not, or only poor substrates for human nucleoside kinases, yet efficiently phosphorylated by dCKEN. In comparison to WT dCK, the R104M-D133A dCK variant exhibits a modest 2-fold increased catalytic efficiency with AraC, but a ~2-fold decrease with gemcitabine (Table S1). Since these dC-analogs compete with dC, the ideal mutant dCK enzyme would have negligible dC efficiency. This is not the case for the R104M-D133A dCK variant. In fact, the efficiency for dC increased by ~25-fold in the mutant (Table S2). As a result, the ratio between the efficiency for the physiological substrate dC versus that for the dC analogs has actually increased for the mutant enzyme. This suggests that the R104M-D133A dCK variant is not promising for the enhancement of dC-analog activation. Of note, the increased Km value for the substrates dA and dG increased considerably for the mutant dCK, thus significantly reducing competition of purine analogs with these natural dCK substrates.
In contrast, BVdU and the L-nucleoside analogs L-dT and L-dU appear to be prospective compounds for this approach. Foremost, WT dCK has very low efficiency with these compounds, whereas the mutant has good catalytic efficiency (Table 1, D). Additionally, the advantage of L-nucleosides is that they are not phosphorylated by human TK1 since this enzyme is enantioselective for the D-form (26). On the other hand, both compounds have a considerable rate with human TK2 (Table 1, A). In fact, the L-enantiomers of dT and dU are phosphorylated at a ~2-fold higher kobs than their D-counterpart. Yet L-dT (Telbivudine) is already used clinically to treat hepatitis B infection (27). The low reported toxicity of L-dT (28) suggests that this TK2-mediated phosphorylation does ultimately not provide sufficient activated L-dT to kill mammalian cells.
An additional candidate thymidine analog is (E)-5-(2-bromovinyl)-2′-deoxyuridine (BVdU, Brivudine). Due to the steric and charge reasons discussed above, it is not surprising that BVdU is a very poor substrate for WT dCK (Table 1, section C). However, since the dCK double mutants lack the repulsion to a thymine base caused by Asp133 and have increased space near the 5-position, we predicted that BVdU can be phosphorylated by these variants. This prediction was confirmed, with kobs for BVdU phosphorylation being 0.57 and 1.16 sec−1 for the R104M-D133A and R104L-D133A variants, respectively (Table 1, C). The latter rate is >6-fold higher than that measured with TK2 (Table 1, C).
Conclusion
Structural analysis of human dCK in complex with various pyrimidine derivatives reveals the reasons that prevent the enzyme from phosphorylating thymidine— a combination of charge repulsion and steric constraints. The fact that 5-Me-dC is substrate of WT dCK suggests that the main reason preventing thymidine, a 5-Me-dC isostere, from being a substrate is the repulsive interaction between the side chain of Asp133 and the carbonyl group at the 4-position of the thymine base. The size of the cavity near the 5-position, a function of the nature of the residue at position 104, also influences the phosphorylation rate. However, the relationship between cavity size and catalytic rate is not a simple one: For example, for 5-Me-dC, the optimal rate is achieved with a methionine at position 104, but for D-dT the leucine substitution is preferred. This implies that the nature of the residue at this position affects not only the size of the cavity near the nucleoside 5-position, but also some rate-limiting conformational change of the enzyme, and that this conformational change is nucleoside-dependent. By pairing a 5-subsitutued pyrimidine analog with the appropriate dCK double mutant, it may now be possible to ascertain the pharmacological effect of these compounds on the proliferation of human cells.
Supplementary Material
Acknowledgments
Funding: This work was supported by a National Institutes of Health grant (S.H., S.O., and A.L.) and the Max-Planck-Society (M.K.).
Abbreviations
- dCK
deoxycytidine kinase
- dCKEN
engineered version of dCK
- 5-Me-dC
5-methyl-deoxycytidine
- WT
wild-type
Footnotes
ACCESSION NUMBERS: Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3KFX.
Supporting Information Available: Kinetic data (kcat and Km values) for several of the discussed nucleosides measured with WT dCK and the R104M-D133A double mutant. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Eriksson S, Munch-Petersen B, Johansson K, Eklund H. Structure and function of cellular deoxyribonucleoside kinases. Cell Mol Life Sci. 2002;59:1327–1346. doi: 10.1007/s00018-002-8511-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arner ES, Eriksson S. Mammalian deoxyribonucleoside kinases. Pharmacol Ther. 1995;67:155–186. doi: 10.1016/0163-7258(95)00015-9. [DOI] [PubMed] [Google Scholar]
- 3.Sabini E, Hazra S, Konrad M, Burley SK, Lavie A. Structural basis for activation of the therapeutic L-nucleoside analogs 3TC and troxacitabine by human deoxycytidine kinase. Nucleic Acids Res. 2007;35:186–192. doi: 10.1093/nar/gkl1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Clercq E. Antivirals and antiviral strategies. Nat Rev Microbiol. 2004;2:704–720. doi: 10.1038/nrmicro975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonate PL, Arthaud L, Cantrell WR, Jr, Stephenson K, Secrist JA, 3rd, Weitman S. Discovery and development of clofarabine: a nucleoside analogue for treating cancer. Nat Rev Drug Discov. 2006;5:855–863. doi: 10.1038/nrd2055. [DOI] [PubMed] [Google Scholar]
- 6.Arner ES, Spasokoukotskaja T, Eriksson S. Selective assays for thymidine kinase 1 and 2 and deoxycytidine kinase and their activities in extracts from human cells and tissues. Biochem Biophys Res Commun. 1992;188:712–718. doi: 10.1016/0006-291x(92)91114-6. [DOI] [PubMed] [Google Scholar]
- 7.Munch-Petersen B, Piskur J, Sondergaard L. Four deoxynucleoside kinase activities from Drosophila melanogaster are contained within a single monomeric enzyme, a new multifunctional deoxynucleoside kinase. J Biol Chem. 1998;273:3926–3931. doi: 10.1074/jbc.273.7.3926. [DOI] [PubMed] [Google Scholar]
- 8.Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat Struct Biol. 2003;10:513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 9.Eriksson S, Kierdaszuk B, Munch-Petersen B, Oberg B, Johansson NG. Comparison of the substrate specificities of human thymidine kinase 1 and 2 and deoxycytidine kinase toward antiviral and cytostatic nucleoside analogs. Biochem Biophys Res Commun. 1991;176:586–592. doi: 10.1016/s0006-291x(05)80224-4. [DOI] [PubMed] [Google Scholar]
- 10.Segura-Pena D, Lutz S, Monnerjahn C, Konrad M, Lavie A. Binding of ATP to TK1-like Enzymes Is Associated with a Conformational Change in the Quaternary Structure. J Mol Biol. 2007;369:129–141. doi: 10.1016/j.jmb.2007.02.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Munch-Petersen B, Herrstrom Sjoberg A, Hellman U, Bergman T, Jornvall H, Eriksson S. Human thymidine kinase 2: molecular cloning and characterisation of the enzyme activity with antiviral and cytostatic nucleoside substrates. FEBS Lett. 1999;443:170–174. doi: 10.1016/s0014-5793(98)01711-6. [DOI] [PubMed] [Google Scholar]
- 12.Ruiz van Haperen VW, Peters GJ. New targets for pyrimidine antimetabolites for the treatment of solid tumours. 2: Deoxycytidine kinase. Pharm World Sci. 1994;16:104–112. doi: 10.1007/BF01880661. [DOI] [PubMed] [Google Scholar]
- 13.Spadari S, Maga G, Verri A, Focher F. Molecular basis for the antiviral and anticancer activities of unnatural L-beta-nucleosides. Expert Opin Investig Drugs. 1998;7:1285–1300. doi: 10.1517/13543784.7.8.1285. [DOI] [PubMed] [Google Scholar]
- 14.Lewis W, Dalakas MC. Mitochondrial toxicity of antiviral drugs. Nature Medicine. 1995;1:417–422. doi: 10.1038/nm0595-417. [DOI] [PubMed] [Google Scholar]
- 15.Sabini E, Hazra S, Konrad M, Lavie A. Nonenantioselectivity property of human deoxycytidine kinase explained by structures of the enzyme in complex with L- and D-nucleosides. J Med Chem. 2007;50:3004–3014. doi: 10.1021/jm0700215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabini E, Hazra S, Konrad M, Lavie A. Elucidation of different binding modes of purine nucleosides to human deoxycytidine kinase. J Med Chem. 2008;51:4219–4225. doi: 10.1021/jm800134t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabini E, Hazra S, Ort S, Konrad M, Lavie A. Structural basis for substrate promiscuity of dCK. J Mol Biol. 2008;378:607–621. doi: 10.1016/j.jmb.2008.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godsey MH, Ort S, Sabini E, Konrad M, Lavie A. Structural basis for the preference of UTP over ATP in human deoxycytidine kinase: illuminating the role of main-chain reorganization. Biochemistry. 2006;45:452–461. doi: 10.1021/bi0518646. [DOI] [PubMed] [Google Scholar]
- 19.McSorley T, Ort S, Hazra S, Lavie A, Konrad M. Mimicking phosphorylation of Ser-74 on human deoxycytidine kinase selectively increases catalytic activity for dC and dC analogues. FEBS Lett. 2008;582:720–724. doi: 10.1016/j.febslet.2008.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazra S, Sabini E, Ort S, Konrad M, Lavie A. Extending thymidine kinase activity to the catalytic repertoire of human deoxycytidine kinase. Biochemistry. 2009;48:1256–1263. doi: 10.1021/bi802062w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agarwal KC, Miech RP, Parks RE., Jr Guanylate kinases from human erythrocytes, hog brain, and rat liver. Methods Enzymol. 1978;51:483–490. doi: 10.1016/s0076-6879(78)51066-5. [DOI] [PubMed] [Google Scholar]
- 22.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;24:795–800. [Google Scholar]
- 23.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Iyidogan P, Lutz S. Systematic exploration of active site mutations on human deoxycytidine kinase substrate specificity. Biochemistry. 2008;47:4711–4720. doi: 10.1021/bi800157e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Chattopadhyaya J, Eriksson S. The enantioselectivity of the cellular deoxynucleoside kinases. Nucleosides Nucleotides. 1999;18:807–810. doi: 10.1080/15257779908041567. [DOI] [PubMed] [Google Scholar]
- 27.Han SH. Telbivudine: a new nucleoside analogue for the treatment of chronic hepatitis B. Expert Opin Investig Drugs. 2005;14:511–519. doi: 10.1517/13543784.14.4.511. [DOI] [PubMed] [Google Scholar]
- 28.Bridges EG, Selden JR, Luo S. Nonclinical safety profile of telbivudine, a novel potent antiviral agent for treatment of hepatitis B. Antimicrob Agents Chemother. 2008;52:2521–2528. doi: 10.1128/AAC.00029-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.