Abstract
This report describes Vibrio Seventh Pandemic Island II (VSP-II) and three novel variants revealed by comparative genomics of 23Vibrio cholerae strains and their presence among a large and diverse collection of V. cholerae isolates. Three VSP-II variants were previously reported and our results demonstrate the presence of three novel VSP-II in clinical and environmental V. cholerae marked by major deletions and genetic rearrangements. A new VSP-II cluster was found in the seven pandemic V. cholerae O1 El Tor strain CIRS101, which is dominant (95%) among recent (2004-2007) seven pandemic V. cholerae O1 El Tor isolates from two endemic sites, but was not found in older strains from the same region. Two other variants were found in V. cholerae TMA21 and RC385, two environmental strains from coastal Brazil and the Chesapeake Bay, respectively, the latter being prevalent among environmental V. cholerae non-O1/non-O139 and V. mimicus. Results of this study indicate that the VSP-II island has undergone significant rearrangement through a complex evolutionary pathway in V. cholerae. Interestingly, one of the new VSP-II revealed the presence of “old” and “new” V. cholerae O1 El Tor pandemic clones circulating in some of the areas where cholera is endemic.
Introduction
Vibrio cholerae, an autochthonous aquatic bacterium, is the causative agent of cholera, a severe, watery, life-threatening diarrheal disease. Cholera bacteria are serogrouped based on the variable somatic O antigen, with more than 200 serogroups identified (Chatterjee et al., 2003). Although strains of most serogroups of V. cholerae are capable of causing a mild gastroenteritis or sporadic local outbreaks of cholera, only toxigenic strains of V. cholerae O1 and O139 have been linked to epidemics and pandemics. Genes encoding for cholera toxin, ctxAB, and other pathogenic factors have been shown to reside in various mobile genetic elements.
The epidemic potential of cholera has been realized throughout human history with seven recorded pandemics and the disease is persistent in many developing countries. Isolates from the sixth pandemic are almost exclusively Classical biotype. However, the seventh, current pandemic has been dominated by V. cholerae O1 El Tor (Kaper et al., 1995). Isolates of all previous pandemics originated in the Indian subcontinent, whereas those associated with the seventh pandemic have their origin in the Indonesian island of Sulawesi, with subsequent isolation from Asia, Africa and Latin America. In 1992, a new serogroup, V. cholerae O139, was identified as the cause of cholera outbreaks in India and Bangladesh (Ramamurthy et al., 1993).
Two gene clusters associated with seventh pandemic strains were identified by comparative genomics using microarray analysis and named Vibrio Seventh Pandemic (VSP) I and II [4]. These clusters were absent in Classical and pre-pandemic V. cholerae El Tor strains and showed an unusual G+C content (40%), compared with the entire V. cholerae genome (47%) (Dziejman et al., 2002). VSP-II was originally identified as a 7.5-kb island, spanning genes VC0490 to VC0497 in V. cholerae O1 El Tor N16961 (Dziejman et al., 2002), and, subsequently, found to include a larger 26.9-kb region, spanning from VC0490 to VC0516 (O’Shea et al., 2004). Its site of integration is a tRNA-methionine locus, VC0516.1. As described in V. cholerae O1 El Tor N16961, VSP-II encodes type IV pilin, two methyl-accepting chemotaxis proteins, an AraC-like transcriptional regulator, a DNA repair protein and a P4-like integrase (VC0516) at the 3′ end of the island. Murphy and Boyd (Murphy and Boyd 2008) found that VSP-II excises from the chromosome, forming an extra-chromosomal circular intermediate through a site-specific recombination mediated by the integrase encoded in the island.
To date, two variants of VSP-II have been described in the literature, one in a V. cholerae non-O1 strain from Bangladesh and one in a V. cholerae O1 El Tor strain isolated in Peru during 1991-2003; moreover the cluster was detected in several V. cholerae non-O1 non-O139 strains (Dziejman et al., 2002; Dziejman et al., 2005; Nusrin et al., 2009).
In this study, comparative genomic analysis was employed to determine the presence and the genetic composition of VSP-II islands among 23 strains of V. cholerae. In our analysis, we re-annotated the VSP-II present in V. cholerae O1 El Tor N16961 and analyzed the VSP-II previously described in V. cholerae O37 MZO-3 (Dziejman et al., 2005). Further, three new variants with significant genetic polymorphisms were discovered and their distribution among a large V. cholerae collection was assessed. From this study it is concluded that VSP-II is not as conserved as has been reported and can be considered a molecular tag in epidemic V. cholerae.
Materials and Methods
Strains and media. Twenty-three V. cholerae strains were included in a comparative genomics analysis were screened for VSP-II, along with 188 well characterized laboratory collection strains and 190 V. cholerae isolates from two cholera-endemic regions of Bangladesh. All strains were grown in Luria-Bertani medium (Difco/BD, Sparks, MD) and stored at −80°C in LB broth amended with 25% glycerol. Comparative genomics. Genome comparisons of the 23 sequenced genomes was done as described by Chun et al. (2009). New VSP-II variants were discovered and annotated by RAST and their genetic organization analyzed and compared using MUMmer (Delcher et al., 1999) and Artemis Comparative Tool (ACT) (Carver et al., 2005). Individual gene polymorphisms were analyzed by ClustalX alignments and homology was attributed after BLASTN search in the non-redundant database (Larkin et al., 2007). Primer design and PCR conditions. Conserved and group-specific regions of VSP-II were identified by examining aligned and unaligned sequences, using ClustalX software (Larkin et al., 2007). PCR primers for group-specific targets were designed using FastPCR Molecular Biology Software (Kalendar et al., 2009). PCR primers are listed in Table 3 and PCR was done using those primers to screen 398 isolates of V. cholerae for the five VSP-II variants.
Table 3.
Primer pairs used in this study
| VSP-II variant amplicon length (bp) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Primer | Sequence | N16961 | MZO3 | TMA21 | CIRS101 | RC385 | No island | Reference |
| pVSP2-IFcacctgtcatgttatgaggtgca | 361 | 678 | - | - | - | This study | ||
| pVSP2-IRaacaggtctcttatcggctttgc | This study | |||||||
| pVSP2-IIFgcacaacttgtaagatagccttgc | 570 | 570 | 570 | 570 | - | This study | ||
| pVSP2-IIRacgcaagacaaaactacagcttgc | This study | |||||||
| pVSP2-IIIFccagcaaacggtcattcgct | 451 | 451 | 451 | - | 451 | This study | ||
| pVSP2-IIIRtggttggaaggtgggttgtgt | This study | |||||||
| p489Fagatcaactacgatcaagcc | - | - | - | - | - | 3532 | O’Shea et al., 2004 | |
| p517Rgcagtcacagcttaaac | O’Shea et al., 2004 | |||||||
Results
Genomic analysis
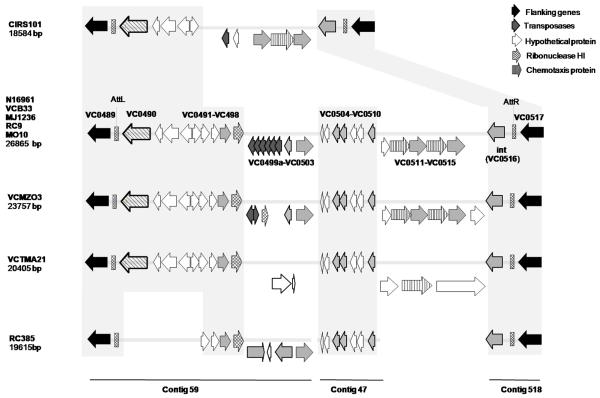
From RAST annotation, the 26.9 Kb VSP-II found in the V. cholerae N16961 encompasses 30 ORFs, compared with 24 ORFs previously annotated (O’Shea et al., 2004). Specifically, six putative transposases were newly annotated by RAST (Fig. 1).
Fig. 1. Genetic organization of the five variants of VSP-II in V. cholerae.
The direction of transcription of the ORFs is indicated by direction of the arrows. The numbers refer to the genetic organization of genes along the genome of V. cholerae N16961 (O’Shea et al., 2004). Genes are pattern-coded, according to function. Homologous regions are indicated by grey shadow. For V. cholerae RC385, the contigs number where the VSP-II island sequence resides are indicated.
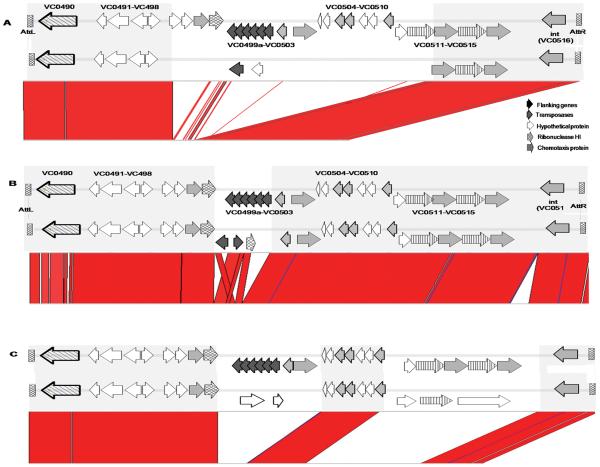
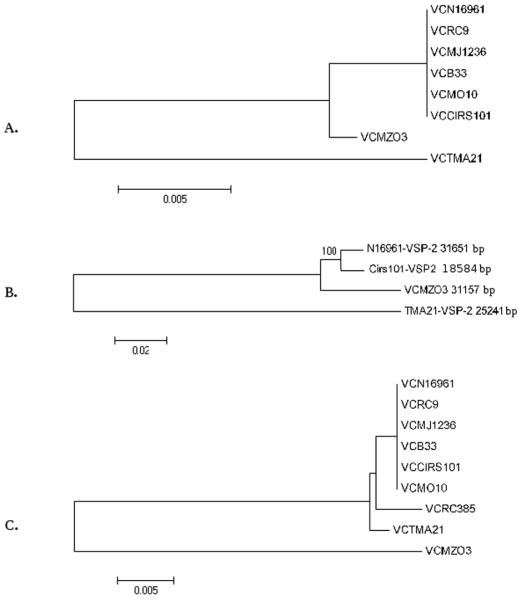
Results of comparative genomics, using 23 complete and draft genomes of V. cholerae and the V. cholerae O1 El Tor N16961 VSP-II sequence as reference, revealed the presence of a VSP-II island with 99% nucleotide sequence similarity in four of the V. cholerae 7th pandemic strains: V. cholerae O1 El Tor B33; V. cholerae O1 El Tor MJ-1236; V. cholerae O139 MO10; and V. cholerae O1 El Tor RC9 (Fig.1). Results of a phylogenetic analysis of the 23 V. cholerae studied showed that these five strains formed a monophyletic clade, termed the seventh phylopandemic clade (Chun et al., 2009). Interestingly, a sixth strain included in this clade, V. cholerae O1 El Tor CIRS101 (Nair et al., 2006), isolated in 2002 in Bangladesh, carries yet another variant of VSP-II (Fig. 2). The VSP-II cluster found in V. cholerae CIRS101 is 18.5Kb long and 99% similar over the 13Kb homologous region (Figs. 1 and 2) to the V. cholerae N16961 VSP-II, with a 14.4Kb deletion at nt 118 of VC0495, spanning ORFs VC0495 to VC0512 (Fig. 2). Inserted downstream of VC0494 in VSP-II of V. cholerae CIRS101 is a 1260 nt transposase (Fig.2). The 3′ region of the V. cholerae CIRS101 VSP-II island is identical to the prototypical seven pandemic VSP-II (Fig. 2).
Fig. 2. Comparative analysis of prototypical VSP-II and other VSP-II.
Alignment generated by Artemis Comparative Tool (ACT) of (A) prototypical VSP-II and V. cholerae CIRS101 VSP-II; (B) prototypical VSP-II and V. cholerae MZO-3 VSP-II; and (C) of prototypical VSP-II and V. cholerae TMA21 VSP-II.
VSP-II genes were present in V. cholerae strains other than the seven pandemic. As previously reported, V. cholerae MZO-3 O37 has a 26.5Kb VSP-II inserted at the same locus as in V. cholerae N16961 (Figs.1 and 2) (Dziejman et al., 2005). Our analysis and annotation showed this island contained 28 ORFs (Fig.2) and regions VC0490-VC0497 and VC0502-VC0516 of the island were 98% similar to the VSP-II in V. cholerae N16961 (Table 1, Fig.2). However, along the island are found two major regions of sequence discontinuity and/or rearrangement (Fig.1): two transposases are inserted within the VC0498 gene and a putative transposase is located between the VC0515 gene and the integrase at the 3′ end of the island (Fig.2), which has 99% similarity with a putative transposase in V. cholerae Vibrio Pathogenicity Island I (VPI-I) (Fig.2) (Karaolis et al., 2006). Despite significant sequence similarity, from a phylogenetic point of view, the VSP-II variant found in V. cholerae O37 MZO-3 appears to have diverged with respect to the VSP-II evolutionary path (Fig.3). All three phylogenetic trees generated using the entire island, three conserved concatenated genes and two flanking genes of the island concluded that V. cholerae MZO-3 VSP-II lies outside the VSP-II of the seventh pandemic clade (Fig.3).
Table 1.
Nucleotide-nucleotide comparison between the seventh pandemic VSP-II island and other variants described in this study, including flanking genes. Shaded regions indicate absent ORFs. 7P: Prototypical seventh pandemic VSP-II
| N16961 | 7P | CIRS101 | TMA21 | MZO3 | RC385 | ||||
|---|---|---|---|---|---|---|---|---|---|
| locus | start | End | size | gene | % similarity | ||||
| VC0489 | 522394 | 520634 | 1761,− | Putative hemolysin | 100 | 100 | 99 | 82 | 99 |
| VC0490 | 525117 | 523156 | 1962,− | Plasmid-related protein | 100 | 100 | 99 | 100 | n/a |
| VC0491 | 525654 | 525118 | 537,− | hypothetical protein | 100 | 100 | 100 | 100 | n/a |
| VC0492 | 526789 | 525623 | 1167,− | hypothetical protein | 100 | 100 | 99 | 100 | n/a |
| VC0493 | 527920 | 527045 | 876,− | hypothetical protein | 100 | 100 | 99 | 99 | n/a |
| VC0494 | 528305 | 528949 | 645,+ | hypothetical protein | 100 | 100 | 95 | 99 | 55 |
| VC0495 | 529011 | 529685 | 675,+ | hypothetical protein | 100 | 55 | 92 | 92 | 92 |
| VC0496 | 529739 | 530338 | 600,+ | hypothetical protein | 100 | n/a | 98 | 99 | 98 |
| VC0497 | 530402 | 530602 | 201,+ | Transcriptional regulator | 100 | n/a | 98 | 99 | 99 |
| VC0498 | 530684 | 531124 | 441,+ | Ribonuclease HI, Vibrioparalog | 100 | n/a | 97 | 76 | 97 |
| VC0499a | 531411 | 531205 | 207,− | transposaseOrfAB, subunit B | 100 | 78 | n/a | 78 | n/a |
| VC0499b | 532035 | 531436 | 600,− | transposaseOrfAB, subunit B | 100 | 52 | n/a | 80 | n/a |
| VC0500a | 532217 | 532071 | 147,− | transposaseOrfAB, subunit a | 100 | 79 | n/a | 91 | n/a |
| VC0500b | 532396 | 532244 | 153,− | Transposase | 100 | n/a | n/a | 85 | n/a |
| VC0501a | 533387 | 532899 | 489,− | Transposase | 100 | 63 | 99 | 99 | 56 |
| VC0501b | 533809 | 533360 | 450,− | Transposase | 100 | 72 | 98 | 99 | n/a |
| VC0502 | 534722 | 534198 | 525,− | type IV pilin, putative | 100 | n/a | n/a | 99 | n/a |
| VC0503 | 535418 | 536698 | 1281,+ | Cell wall endopeptidase, | 100 | n/a | n/a | 99 | n/a |
| VC0504 | 537103 | 536876 | 228,− | hypothetical protein | 100 | n/a | 89 | 100 | 89 |
| VC0505 | 537519 | 537151 | 369,− | hypothetical protein | 100 | n/a | 92 | 100 | 94 |
| VC0506 | 538284 | 537550 | 735,− | Transcriptional factor MdcH | 100 | 46 | 95 | 99 | 96 |
| VC0507 | 538599 | 538423 | 177,− | hypothetical protein | 100 | n/a | 92 | 100 | 96 |
| VC0508 | 539046 | 538603 | 444,− | hypothetical protein | 100 | 64 | 94 | 99 | 94 |
| VC0509 | 539540 | 539097 | 444,− | hypothetical protein | 100 | n/a | 92 | 96 | 90 |
| VC0510 | 540004 | 539531 | 474,− | DNA repair protein RadC | 100 | 59 | 92 | 93 | 93 |
| VC0511 | 540216 | 540335 | 120,+ | hypothetical protein | 100 | n/a | n/a | 100 | n/a |
| VC0512 | 541319 | 542908 | 1590,+ | Methyl-accepting chemotaxis protein | 100 | 52 | n/a | 100 | n/a |
| VC0513 | 544362 | 545177 | 816,+ | AraC-domain-containing protein | 100 | 100 | n/a | 99 | n/a |
| VC0514 | 545174 | 547054 | 1881,+ | Methyl-accepting chemotaxis protein | 100 | 100 | n/a | 100 | n/a |
| VC0515 | 547158 | 548390 | 1233,+ | EAL domain protein | 100 | 100 | n/a | 99 | n/a |
| VC0516 | 550021 | 548780 | 1242,− | Phage integrase | 100 | 100 | 89 | 98 | 92 |
| VC0517 | 552284 | 550407 | 1878,− | RNA polymerase sigma factor RpoD | 100 | 100 | 99 | 99 | 99 |
Fig. 3. Phylogenetic analysis of VSP-II variants.
(A) Neighbor-Joining tree based on three VSP-II concatenated genes: plasmid related protein, hypotetical protein, and phage integrase; (B) Neighbor-Joining tree based on the whole VSP-II and (C) Neighbor-Joining tree based on concatenated VSP-II flanking genes: putative hemolysine and Rpo sigma factor.
A VSP-II variant was identified in V. cholerae non-O1/non-O139 TMA21, isolated from a sewage sample collected in Brazil in 1982 (Table 1, Fig.1). The cluster found in this strain is 20.4 Kb long, integrated at the same locus and shares 99% sequence similarity over homologous regions with the prototypical 7th pandemic VSP-II island (Fig.2). As in the case of the V. cholerae MZO-3 variant, significant genetic rearrangement was detected in the region downstream of VC0498 where ORFs VC0499a-VC0500b and VC0502-VC0503 are deleted. In contrast, at this locus, we annotated two ORFs encoding hypothetical proteins not found in the prototypical 7th pandemic island. These ORFs have 92% and 85% nucleotide sequence similarity with two hypothetical proteins in Vibrio vulnificus YJ016, VV0516-VV0517, in the same arrangement (dbj|BA000037.2|). As reported by O’Shea and colleagues, the 5′ region of the prototypical V. cholerae VSP-II shows homology to the 5′ end of the 43.4 Kb V. vulnificus island-I (VVI-I), but ORFs VC0499-VC0503 of VSP-II are absent in VVI-I (O’Shea et al., 2004). Therefore, in this region, V. cholerae TMA21 VSP-II appears to have an organization identical to VVI-I, i.e., ORFs VC0499-VC0503 are substituted by two hypothetical proteins (Fig. 5). Another major genetic rearrangement in V. cholerae TMA21 VSP-II occurs downstream of ORF VC0511, which is a deletion encompassing ORFs VC0512 to VC0516 substituted with three ORFs encoding two hypothetical proteins and a nucleotidyltransferase (Table 1, Fig.2). Interestingly, the same deletion was observed in the VSP-II variant found in V. cholerae O1 El Tor strains from Peru (Nusrin et al., 2009). Two of the ORFs present in V. cholerae TMA21 VSP-II have 69% sequence similarity with two ORFs encoding hypothetical proteins in Nitrosomonaseuropaea ATCC 19718 (emb|AL954747.1|), arranged in the same order. The third ORF did not share significant similarity with any sequence in GenBank.
A fourth variant of the VSP-II island was found in the genome of V. cholerae RC385 O135, an isolate from Chesapeake Bay, MD, USA (Fig.1). Because of low sequence coverage resulting in a large number of contigs in this draft genome, we were only able to reconstruct the 5′ region of the island. VSP-II sequences were present in three contigs: ctg 59; ctg 47; and ctg 518. The 5′ region of the island resides on contig 59 and, according to the sequence in this contig, the island is inserted in the same location as all other VSP-II islands described in this study. The rest of the contig comprises 19,615 bp (Fig.1). There is a significant deletion in this region, conserved in the prototypical 7th pandemic VSP-II, V. cholerae MZO3 and TMA21 variants. ORFs VC0490 to VC0494 are absent in VSP-II of V. cholerae RC385 (Fig.1). Furthermore, three new ORFs are inserted after the VC0498 gene, indicating that this locus represents a hot spot for recombinational events within the island. Genes VC0504 to VC0510 and the integrase are conserved, as had been found in the other VSP-II variants (Fig.1).
PCR screening
To assess the distribution of the VSP-II variants identified by comparative genomics, a well-characterized collection of 188 clinical and environmental isolates of V. cholerae representing different serogroups and biotypes and featuring diverse virulence pattern and 190 recent isolates from two cholera endemic sites in Bangladesh, were screened by PCR.
Three primers pairs were designed and incorporated into a multiplex PCR to distinguish the five VSP-II variants. Amplification patterns associated with specific VSP-II variants are shown in Table 3. Furthermore, the insertion site of the island was confirmed by amplification of a primer pair designed using flanking genes (Table 3). Positive amplification with the primer pair was considered evidence of an intact insertion site or absence of the island.
As expected, all the V. cholerae O1 Classical and El Tor pre-seven pandemic isolates from the laboratory collection did not contain the VSP-II island (Table 2). Twenty-nine of 31 seven pandemic V. cholerae O1 El Tor strains (93.5%) harbored the prototypical VSP-II island. In addition to V. cholerae CIRS101, only one other strain, a clinical isolate from Bangladesh, yielded an amplification pattern corresponding to the V. cholerae CIRS101 VSP-II variant, (Table 2); both harbored the typical 7th pandemic VSP-I (Grim et al., 2010). In contrast, 91% of V. cholerae O1 of environmental origin did not contain VSP-II and only two strains showed the V. cholerae RC385 VSP-II island amplification pattern: one isolated from a sewage sample collected in Brazil in 1978 and a second strain from Mexico. All were negative for VSP-I (Grim et al., 2010).
Table 2.
Distribution of VSP-II variants in two V. cholerae culture collections. Results are from PCR screening
| Source of isolate | VSP-II variants | ||||||
|---|---|---|---|---|---|---|---|
| N16961 | CIRS101 | MZO3 | TMA21 | RC385 | Negative | Total | |
| Lab culture collection | 188 | ||||||
|
| |||||||
| V. choleraeO1 classical | 0 | 0 | 0 | 0 | 0 | 8 | 8 |
| V. cholerae O1 El Tor seventh pandemic | 29 | 2 | 0 | 0 | 0 | 0 | 31 |
|
V. cholerae O1 El Tor pre-seventh pandemic |
0 | 0 | 0 | 0 | 0 | 3 | 3 |
| V. cholerae O1, environmental | 0 | 0 | 0 | 0 | 2 | 21 | 23 |
| V. cholerae O139 | 16 | 0 | 0 | 0 | 1 | 0 | 17 |
| V. cholerae non-O1/non-O139 | 0 | 0 | 1 | 1 | 8 | 81 | 91 |
| V. mimicus | 0 | 0 | 0 | 1 | 4 | 11 | 15 |
|
| |||||||
| Bangladesh isolates | 190 | ||||||
|
| |||||||
| V. cholerae O1 clinical | 1 | 96 | 0 | 0 | 1 | 3 | 101 |
| V. cholerae O1 environmental | 0 | 16 | 0 | 0 | 0 | 1 | 17 |
| V. cholerae O139 clincal | 10 | 0 | 0 | 0 | 0 | 0 | 10 |
| V. cholerae O139 environmental | 15 | 0 | 0 | 0 | 0 | 0 | 15 |
|
V. cholerae non-O1/non-O139 environmental |
0 | 0 | 0 | 2 | 12 | 33 | 47 |
The V. cholerae O139 strains in our collection all had typical 7th pandemic VSP-II except for one strain carrying the RC385 variant (Table 2), an environmental isolate, and the only V. cholerae O139 not carrying the VSP-I island (Grim et al., 2010). Furthermore, 89% of the V. cholerae non-O1/non-O139 were negative for VSP-II. Since this collection contained strains used for comparative genomics, V. cholerae RC385, TMA21, and MZO-3 gave expected amplification patterns. In addition, seven carried the V. cholerae RC385 VSP-II (Table 2). Among these, two were isolated from Chesapeake Bay, MD, USA, same location as V. cholerae RC385, and one also carried a new variant of VSP-I (Grim et al., 2010). Of the remainder, one was isolated from a sewage sample collected in Brazil, one was from Czechoslovakia, two were from Japan, and one was from Bangladesh. It should be noted that four of 15 V. mimicus strains also were positive for the V. cholerae RC385 VSP-II variant.
Interesting results emerged from screening the collection of V. cholerae isolates from two cholera endemic sites in Bangladesh, collected from 2004 to 2007. Among the clinical V. cholerae O1 El Tor, a total of 96 carried the V. cholerae CIRS101 VSP-II variant and only one harbored the typical 7th pandemic VSP-II (Table 2). Moreover three isolates did not contain VSP-II and one was positive for V. cholerae RC385 VSP-II (Table 2), which was negative for VSP-I and ctxA (Grim et al., 2010). A similar result was obtained for environmental V. cholerae O1 isolates, since these were all ctx and tcpA positive strains and, therefore likely related to the clinical strains. That is, all carried V. cholerae CIRS101 VSP-II, except one strain that did not have V. cholerae VSP-II or VSP-I (Table 2) (Grim et al., 2010). In contrast, all V. cholerae O139, both clinical and environmental, contained the canonical 7th pandemic VSP-II (Table 2), suggesting this serogroup is genetically isolated from the dominant V. cholerae O1 pandemic clones. Among V. cholerae non-O1/non-O139 isolates, 70% did not harbor VSP-II, 26% contained V. cholerae RC385 VSP-II and two the V. cholerae TMA21 VSP-II (Table 2), showing these are the most common variants in the non-epidemic V. cholerae population.
Discussion
Comparative genomic analysis of 23 V. cholerae strains belonging to different serotypes, widely distributed geographically and isolated over an extended period of time, has led to the discovery of three new variants of the VSP-II genomic island. This is remarkable, since VSP-I and VSP-II were originally considered to be conserved genetic markers of 7th pandemic V. cholerae (Dziejman et al., 2002; O’Shea et al., 2004). To date, two other examples of sequence variation whitin V. cholerae VSP-II were described (Dziejman et al., 2005; Nusrin et al., 2009). Our analysis adds further insight to the knowledge of this genomic cluster and its evolution in V. cholerae.
From the standpoint of genetic comparison, it is clear that the island has undergone significant genetic rearrangement. Two loci, at the 3′ end of the VC0498 and VC0511, may represent hot spots for recombination events within the conserved genomic backbone of the island. It appears that the VSP-II cluster has evolved into different variants by acquisition and loss of indels at specific loci within a conserved core.
The VSP-II variant found in V. cholerae O1 El Tor CIRS101 has a significant deletion compared to the other two variants presumably circulating among V. cholerae O1 El Tor strains: the 7th pandemic and the Peruvian VSP-II. Although its function remains to be elucidated, the CIRS101 VSP-II presence is clearly dominant in recent V. cholerae O1 isolates from two cholera endemic sites of Bangladesh, but not in V. cholerae O139 isolated from those sites, the latter possessing the prototypical 7th pandemic VSP-II. These data are surprising, given that in the endemic areas under study, V. cholerae O1 and O139 share the same environment and host population, but appear not to have exchanged this genomic island.
In Bangladesh, by tracking VSP-II variants, we were able to detect a shift between “old” and “new” pandemic clones of V. cholerae O1 El Tor, based on the fact that a 1994 strain (V. cholerae O1 MJ1236) carries the prototypical 7th pandemic VSP-II, while those isolated during 2004-2007, carry the new CIRS101 variant. It is of paramount importance to know whether the same shift occurred in clinical V. cholerae isolates from Africa or South America to be able to determine if this event is region-specific.
By not being present in non-epidemic isolates of V. cholerae non-O1/O139 suggests that the CIRS101 VSP-II confers a selective advantage when in the human host but not when in the aquatic environment. In this regard, it is noteworthy that V. cholerae O1 El Tor CIRS101 carries a variant of the CTX cluster found in a group of newly emerged 7th pandemic clones, referred to as El Tor/classical ‘hybrid’ or ‘altered’ strains (Nair et al., 2006) Therefore, the new V. cholerae CIRS101 VSP-II may have arisen in a genomic background positively selected in the human host (hybrid strains appear to produce more cholera toxin), likely becoming dominant among epidemic clones. A link between their evolutionary success of the two clusters (CTX and VSP-II) is not indicated, based on the presence of a canonical 7th pandemic VSP-II in two other hybrid strains, V. cholerae O1 MJ1236 (Bangladesh, 1994) and B33 (Mozambique, 2004).
The VSP-II circulating among V. cholerae non-O1/non-O139 and V. mimicus is the RC385 VSP-II. Despite different serotype and significant genetic diversity among the strains, this variant appears to be stable in isolates obtained at different times and geographical locations while TMA21 and MZO-3 VSP-II show only limited distribution. The presence of the new VSP-II variants was not correlated with the presence of a new VSP-I, indicating that the two gene clusters derive from a different history of genetic exchange among V. cholerae non-O1/non-O139 and V. mimicus.
References
- Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. DOI 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Chatterjee SN, Chaudhuri K. Lipopolysaccharides of Vibrio cholerae. I. Physical and chemical characterization. Biochim Biophys Acta. 2003;1639:65–79. doi: 10.1016/j.bbadis.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. PNAS. 2009;106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Kasif S, Fleischmann RD, Peterson J, White O, Salzberg SL. Alignment of whole genomes. Nucleic Acids Research. 1999;27(11):2369–2376. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, et al. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. PNAS. 2002;99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, et al. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. PNAS. 2005;102:3465–3. doi: 10.1073/pnas.0409918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim CJ, Choi J, Chun J, Jeon YS, Taviani E, Hasan NA, Haley B, Huq A, Colwell RR. Occurrence of the Vibrio cholerae Seventh Pandemic VSP-I Island and a New Variant. OMICS. 2010;14(1):1–7. doi: 10.1089/omi.2009.0087. doi:10.1089/omi.2009.0087. [DOI] [PubMed] [Google Scholar]
- Kalendar R, Lee D, Schulman AH. FastPCR Software for PCR Primer and Probe Design and Repeat Search. Genes, Genomes and Genomics. 2009;3:1. [Google Scholar]
- Kaper JB, Morris JG, Jr., Levine MM. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DKR, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, et al. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher A, Smoot M, Shumway M, et al. Versatile and open software for comparing large genomes. Genome Biology. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Murphy RA, Boyd EF. Three Pathogenicity Islands of Vibrio cholerae Can Excise from the Chromosome and Form Circular Intermediates. J Bacteriol. 2008;190:636–647. doi: 10.1128/JB.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair GB, Qadri F, Holmgren J, Svennerholm AM, Safa A, et al. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol. 2006;44:4211–4213. doi: 10.1128/JCM.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusrin S, Gil AI, Bhuiyan NA, Safa A, Asakura M, et al. Peruvian Vibrio cholerae O1 El Tor strains possess a distinct region in the Vibrio seventh pandemic island-II that differentiates them from the prototype seventh pandemic El Tor strains. J Med Microbiol. 2009;58:342–354. doi: 10.1099/jmm.0.005397-0. [DOI] [PubMed] [Google Scholar]
- O’Shea YA, Finnan S, Reen FJ, Morrissey JP, O’Gara F, et al. The Vibrio seventh pandemic island-II is a 26.9 kb genomic island present in Vibrio cholerae El Tor and O139 serogroup isolates that shows homology to a 43.4 kb genomic island in V. vulnificus. Microbiology. 2004;150:4053–4063. doi: 10.1099/mic.0.27172-0. [DOI] [PubMed] [Google Scholar]
- Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, et al. Emergence ofnovel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]