Summary
Human bitter taste is mediated by the hTAS2R family of G protein-coupled receptors [1-4]. The discovery of the hTAS2Rs enables the potential to develop specific bitter receptor antagonists that could be beneficial as chemical probes to examine the role of bitter receptor function in gustatory and non-gustatory tissues. In addition, they could have widespread utility in food and beverages fortified with vitamins, antioxidants and other nutraceuticals since many of these have unwanted bitter aftertastes. We employed a high-throughput screening approach to discover a novel bitter receptor antagonist (GIV3727) that inhibits activation of hTAS2R31 by saccharin and acesulfame K, two common artificial sweeteners. Pharmacological analyses revealed that GIV3727 likely acts as an orthosteric, insurmountable antagonist of hTAS2R31. Surprisingly, we also found that this compound could inhibit five additional hTAS2Rs, including the closely related receptor hTAS2R43. Molecular modeling and site-directed mutagenesis studies suggest that two residues in helix seven are important for antagonist activity in hTAS2R43/31. In human sensory trials, GIV3727 significantly reduced the bitterness associated with the two sulphonamide sweeteners, indicating that TAS2R antagonists are active in vivo. Our results demonstrate that small molecule bitter receptor antagonists can effectively reduce the bitter taste qualities of foods, beverages, and pharmaceuticals.
Keywords: taste, bitter, gustation, calcium imaging, heterologous expression
Results and Discussion
Identification and characterization of a small molecule bitter receptor antagonist
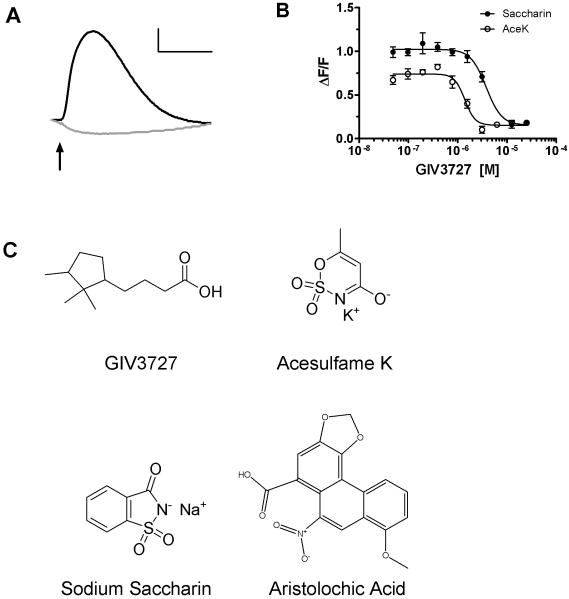
To identify putative antagonists of the human TAS2R31 bitter receptor, we constructed a double stable cell line expressing hTAS2R31 along with the chimeric G-protein G16gust44 in HEK293 cells. This cell line was used for high-throughput biomolecular screening via calcium imaging. After thorough data analysis of the screening set (17,854 compounds total) and hit selection, we identified 139 candidate antagonists. One compound in particular, GIV3727 [4-(2,2,3-trimethylcyclopentyl)butanoic acid], was selected as a robust inhibitor of hTAS2R31 (Figure 1A). Potency assessments in hTAS2R31 expressing cells revealed that the half-maximal inhibitory concentration (IC50) of GIV3727 for both agonists was 6.4 ± 2.4 μM (n=3) for acesulfame K and 7.9 ± 6.1 μM (n=3) for saccharin (Figure 1B). Inhibition of hTAS2R31 was reversible, since cells that were pre-exposed to GIV3727 responded fully to agonist upon washout of the antagonist (see Figure S1A). Furthermore, we found that GIV3727 similarly inhibited hTAS2R43 (see Figure S1B), which is 88% identical to hTAS2R31 and can also be activated by saccharin and acesulfame K [5], suggesting that this compound would be of interest for further study as a putative bitter taste inhibitor. The structural identity and purity of the original sample of GIV3727 (Figure 1C) were confirmed by GC-MS. GIV3727 was re-synthesized and hTAS2R31 inhibition by the new batch was confirmed before proceeding with further experiments.
Figure 1.
Inhibition of hTAS2R31 by GIV3727. (A) Representative FLIPR responses of cells expressing hTAS2R31 following stimulation with 500 μM sodium saccharin in the absence (black trace) or presence (gray trace) of 25 μM GIV3727. Arrows indicate agonist application and the data were corrected for and normalized to background fluorescence. Calibration: horizontal bar, 100 sec; vertical bar, ΔF/F = 0.2. (B) Dose-response profile of hTAS2R31 expressing cells stimulated with either 500 μM sodium saccharin (●) or 800 μM acesulfame K (○) in the presence of increasing concentrations of GIV3727. Data are presented as the mean ± s.e.m. of at least three separate experiments performed in duplicate and were fitted in GraphPad Prism using a 4-parameter logistic fit equation. (C) Chemical structures of compounds used in this study. See also Figures S1 and S3.
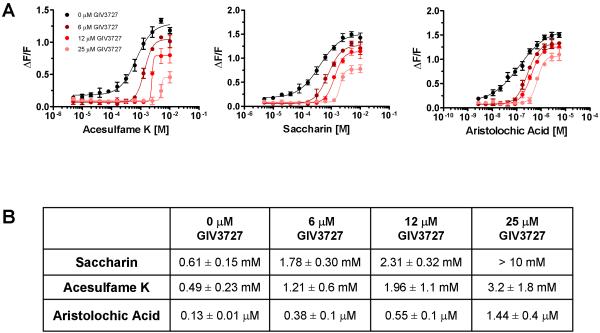
Given the structural dissimilarity between the antagonist and known agonists (Figure 1C), we examined whether GIV3727 was acting as a competitive or non-competitive receptor antagonist. Concentration curves of hTAS2R31 agonists were constructed in the presence of fixed concentrations of the inhibitor. Increasing concentrations of GIV3727 resulted in a rightward shift of the dose-response curves for all three cognate bitter agonists (Figure 2), suggesting that it might act at an orthosteric position and competitively inhibit receptor activation [6, 7]. However, agonist efficacy was also diminished by GIV3727 as evidenced by the lower maximal signal amplitudes (Figure 2A), which is typically indicative of allosteric antagonism or insurmountable inhibition [6, 7]. We observed that the EC50 increased 3- to 10-fold for all three agonists while the signal amplitude decreased by 35-70 % upon inclusion of 25 μM GIV3727 (Figure 2), and we conclude that GIV3727 is likely an orthosteric, insurmountable bitter receptor antagonist [6, 7].
Figure 2.
GIV3727 is an insurmountable antagonist of hTAS2R31. (A) Dose-response curves for acesulfame K, sodium saccharin and aristolochic acid were prepared and the effects of 6, 12 or 25 μM GIV3727 on agonist activation of hTAS2R31 expressing cells were assessed via calcium imaging in the FLIPR. Data are presented as the mean ± s.e.m. of at least three separate experiments performed in quadruplicate and fitted in GraphPad Prism using a 4-parameter logistic fit equation. (B) Average EC50 values obtained for acesulfame K, saccharin and aristolochic acid in the absence or presence of GIV3727, which were derived from each experiment used to generate the data in (A). There was a significant effect (p < 0.0001) of antagonist concentration on each agonist as assessed by two-way ANOVA. Post-hoc tests (Bonferroni) showed that all three concentrations of the antagonist significantly (p < 0.0001) reduced receptor activation across all concentrations of agonist.
GIV3727 inhibits multiple hTAS2Rs
Since GIV3727 inhibited both hTAS2R43 and hTAS2R31, we were interested to determine whether it can inhibit other bitter receptors. Therefore, we examined the effects of 25 μM GIV3727 via calcium imaging using a panel of transfected hTAS2Rs with known bitter agonists (18 of the 25 hTAS2Rs). The receptors and the corresponding bitter agonists [8] tested are shown in Table 1 along with the effects of GIV3727 on receptor activation. When possible we utilized an EC90 concentration of bitter agonist, with the exception of four hTAS2Rs. The low potency and the limited number of available agonists for these four receptors (hTAS2R3/5/7/13) did not allow for an accurate EC90 value to be determined prior to testing of GIV3727. Therefore, we used the maximal concentration of agonist that activated the receptor without eliciting an artifactual calcium transient. We found that GIV3727 could significantly inhibit a total of six bitter taste receptors (hTAS2R4/7/40/43/31/20) amongst those tested (Table 1) and four receptors exhibited dose-dependent inhibition by GIV3727 with varying potencies (Figure S2A). hTAS2R7 was among the receptors inhibited by GIV3727 but it is not clear whether inhibition occurs at an equivalent EC90 concentration of agonist due to the low potency of the bitter agonist. Inhibition was reversible in hTAS2R40 and hTAS2R43, similar to hTAS2R31 (Figure S2B). Finally, we did not observe inhibition of the remaining hTAS2Rs by the antagonist. Together, these studies suggest that GIV3727 acts selectively in a subset of hTAS2Rs.
Table 1. Inhibition of multiple hTAS2Rs by GIV3727.
| Receptor | Bitter Agonist | % activation in presence of GIV3727 |
p-value |
|---|---|---|---|
| hTAS2R1 | 30 μM trans-isohumulone | 114.3 ± 6.2% | 0.2268 |
| hTAS2R3 | 3 mM chloroquine | 76.7 ± 21.5% | 0.2762 |
| hTAS2R4 | 10 mM colchicine | 45.8 ± 17.4% | 0.0067 |
| hTAS2R5 | 300 μM phenanthrolin | 100.2 ± 56.9% | 0.9954 |
| hTAS2R7 | 10 mM cromolyn | 47.8 ± 17.0% | 0.0004 |
| hTAS2R8 | 100 μM chloramphenicol | 95.7 ± 30.6% | 0.8422 |
| hTAS2R10 | 100 μM strychnine | 93.9 ± 16.4% | 0.4598 |
| hTAS2R13 | 3 mM denatonium benzoate | 97.5 ± 15.1% | 0.7814 |
| hTAS2R14 | 3 μM aristolochic acid | 94.3 ± 14.8% | 0.618 |
| hTAS2R16 | 10 mM salicin | 101.1 ± 13.5% | 0.8977 |
| hTAS2R38 | 10 μM PTC | 104.0 ± 14.6% | 0.5599 |
| hTAS2R39 | 100 μM EGCG | 93.7% ± 12.8% | 0.6043 |
| hTAS2R40 | 0.3 μM cohumulone | 3.2 ± 6.5% | 9.1×10−6 |
| hTAS2R43 | 0.3 μM aristolochic acid | 2.5 ± 4.9% | 1.4×10−7 |
| hTAS2R31 | 3 μM aristolochic acid | 61.9 ± 9.3% | 0.0013 |
| hTAS2R46 | 3 μM strychnine | 92.6 ± 8.9% | 0.1571 |
| hTAS2R20 | 100 μM cromolyn | 62.9 ± 31.8% | 0.034 |
| hTAS2R50 | 300 μM andrographolide | 119.0 ± 14.2% | 0.0263 |
Data are presented as the mean ± s.e.m. of n=3 experiments and have been expressed as a percent of the signal obtained with agonist in the absence of inhibitor. Statistical significance was assessed by student’s t-test and p < 0.05 is considered statistically significant. Abbreviations; phenythiocarbamide (PTC), epigallocatechin gallate (EGCG). See also Figure S2.
Inhibition of several hTAS2Rs by GIV3727 was somewhat surprising given the low sequence homology in this receptor family. Overall homology among the hTAS2Rs is only 30-70 % [1] and we initially expected that the antagonist might interact with only hTAS2R43/31. However, our observation that GIV3727 modulates several receptors seems to parallel emerging trends for bitter receptor agonists. For example, aristolochic acid actually activates three different bitter receptors: hTAS2R14, hTAS2R43 and hTAS2R31 [5, 9]. Similarly, we found that chlorpheniramine activates eight different hTAS2Rs, while diphenidol appears to activate 15 hTAS2Rs [8]. This suggests that antagonists, like agonists, have the capacity to interact with several bitter receptors, which raises interesting questions regarding possible mechanisms. One possible interpretation is that the three-dimensional architecture is still maintained among the hTAS2Rs despite the low sequence homology and GIV3727 is capable of binding to multiple receptors. An alternate explanation is that GIV3727 is disrupting G-protein coupling, which could also explain inhibition of several relatively unrelated receptors. However, a majority of receptors were not inhibited by GIV3727 despite signaling through the same chimeric G-protein (Table 1), suggesting that inhibition of agonist-evoked signaling is specific to the hTAS2R subset. It will be of interest to determine whether the promiscuous activity of GIV3727 is a property shared by other chemical classes of bitter receptor antagonists.
Molecular Determinants of Antagonist Binding
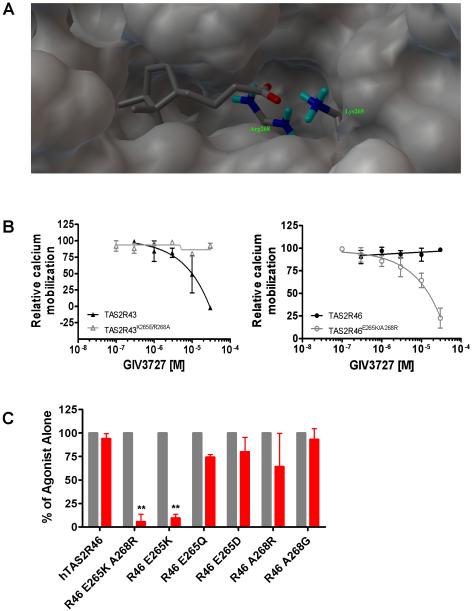
Since GIV3727 inhibited several hTAS2Rs, it was of interest to identify key residues for recognition of this compound in the targeted receptors, hTAS2R31 and hTAS2R43. Such information could be useful for improving potency and selectivity of this antagonist series. During previous studies in our laboratory examining agonist specificity of the hTAS2Rs, we identified residues important for activation of the bitter taste receptors hTAS2R43/31/46 [10]. We focused on these receptors because they are closely related, possess highly distinct agonist activation patterns, and the relatively few amino acid sequence differences should account for the pronounced differences in bitter agonist selectivities. The most obvious amino acid differences between these receptors are in transmembrane helix seven where hTAS2R46 contains an acidic Glu265 and a hydrophobic Ala268, while hTAS2R43/31 both contain basic residues instead (Lys265 and Arg268). Point-mutagenesis of the corresponding positions in all 3 receptors revealed that agonist activation and receptor selectivity is largely determined by these two residues [10]. During the course of the current study, we constructed homology models for hTAS2R43 and hTAS2R31 using the crystal structure reported for the β-adrenergic receptor [11, 12] to examine these receptors for residues that may interact with GIV3727. Chemical optimization to improve potency of this scaffold indicated that the -COOH moiety is essential for antagonist activity since replacement of this group with an ester or corresponding alcohol abolished activity (Figure S3). Therefore, we focused on possible key interaction sites for this moiety in the homology models. Docking of GIV3727 in the putative binding pocket indicated that Lys265 and Arg268 were also likely interaction sites for the –COOH group of the antagonist (Figure 3A). In Ballesteros-Weinstein nomenclature [13], these residues occupy positions 7.39 and 7.42, respectively, and have been previously shown to participate in ligand binding in other GPCRs [14-19]. Taken together, we reasoned that interaction of GIV3727 may also depend on the same two residues that we previously found to be important for agonist selectivity and activation.
Figure 3.
Residues in helix seven of hTAS2R43/31 are important for antagonist activity. (A) Three-dimensional view of the hTAS2R31 domain binding pocket for GIV3727 showing the interaction of the antagonist with Lys265/Arg268. (B) Site-directed mutagenesis of hTAS2R43 and hTAS2R46 reveals that Lys265/Arg268 are important contact sites for GIV3727. Receptors were stimulated with a fixed agonist in the absence or presence of increasing concentrations GIV3727. The following agonists and concentrations: TAS2R43: 0.3 μM aristolochic acid; hTAS2R43K265E/R268A: 10 μM strychnine; TAS2R46: 3 μM strychnine; hTAS2R46E265K/A268R: 1 μM aristolochic acid. An n=2-3 is shown for each dose-response curve. The data are normalized in % to the maximal signal obtained with the agonist in the absence of the inhibitor and were fitted in GraphPad Prism using a 4-parameter logistic fit equation. (B) Site-directed mutagenesis of hTAS2R46 reveals that Lys265 is sufficient for activity of GIV3727. Receptors were stimulated with an EC90 concentration of agonist in the absence (black bars) or presence (red bars) of 25 μM GIV3727. The data are normalized in % to the maximal signal obtained with the agonist in the absence of the inhibitor. The specific ligands and concentrations used for each construct can be found in the Supplemental Experimental Procedures. An n=2-3 experiments were conducted for each mutant receptor. Statistical significance is indicated by ** where p < 0.005 based on a student’s t-test. See also Figure S4.
To address this question, the existing mutants from our prior agonist specificity studies were tested for sensitivity to GIV3727. Consistent with our hypothesis, GIV3727 was more active in receptors harboring basic residues (native hTAS2R43 or hTAS2R46E265K/A268R), while introduction of the hTAS2R46-specific residues (E265/A268) into hTAS2R43 abolished activity (Figure 3B). Our data indicate that these two residues in helix seven are important for antagonist activity in TAS2R43/31 and are consistent with the molecular modeling predictions. This also suggests that GIV3727 is likely binding in proximity to bitter agonists since these same two residues also affect agonist specificity [10]. For example, introduction of K265/R268 into hTAS2R46 confers sensitivity to aristolochic acid, which activates hTAS2R43 but is inactive in the native hTAS2R46 (Figure S4). Conversely, the hTAS2R43K265E/R268A mutant can be activated by the hTAS2R46-agonist strychnine, while the native hTAS2R43 is insensitive to this compound (Figure S4). Thus, these residues appear important for both agonist and antagonist activity. The importance of helix seven in agonist and antagonist binding has been observed for other GPCRs including hTAS1R3, a component of the sweet and umami taste receptor heterodimers [15, 20]. Elucidation of other residues within hTAS2R43/31 should further define the degree of overlap between the agonist and antagonist binding sites.
To determine the individual contributions of each residue, we generated mutants of hTAS2R46 in which Lys265 or Arg268 were introduced individually and examined them for sensitivity to GIV3727. We found that introduction of Lys265 into hTAS2R46 was sufficient to confer sensitivity to the antagonist that was indistinguishable from the double mutant, while we did not observe inhibition in the A268R single mutant (Figure 3C). We also did not observe inhibition by GIV3727 in other hTAS2R46 mutants harboring non-basic amino acid substitutions such as E265D, E265Q or A268G (Figure 3C). Together, these results suggest that a basic residue at position 7.39 is a key contact point for GIV3727, at least within the mutated hTAS2R46 backbone. However, since the GIV3727 is active in other hTAS2Rs lacking these two basic residues, we do not rule out the existence of other residues in determining antagonist binding or specificity for other hTAS2Rs. Furthermore, presence of a lysine in this position is not sufficient to confer sensitivity to GIV3727 in other hTAS2Rs since hTAS2R50 also has a lysine at this site but is insensitive to the antagonist. This implies that residues from other helices or extracellular loops are more likely to determine sensitivity to GIV3727 in receptors other than hTAS2R43/31.
GIV3727 decreases bitter taste intensity in humans
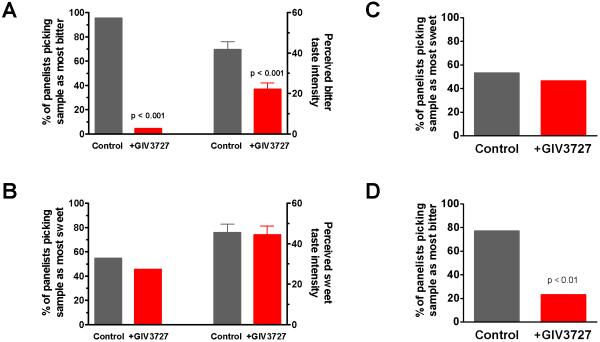
Acesulfame K and saccharin, to a lesser extent, are commonly used as sugar substitutes for low calorie food products and their acceptance could be significantly increased by blocking the undesirable bitter tastes associated with them [21]. Therefore, we wanted to determine whether GIV3727 could reduce the bitter taste of acesulfame K in vivo using structured human taste trials. We employed a 2-alternative forced choice paradigm (2-AFC) [22] coupled with anchored taste intensity ratings in order to quantify the reduction in bitterness. Subjects were presented with two aqueous solutions in random order: a 2 mM acesulfame K control solution and the same acesulfame K solution containing 30 ppm (150 μM) of GIV3727. Panelists were asked to identify the solution having the strongest bitter aftertaste and then instructed to rate the bitter taste intensity of the solutions using an anchored line scale. In a separate experiment with the same cohort, subjects identified the sweetest solution and then rated the sweet taste intensity. We found that nearly all of the panelists (95% of cohort) selected the GIV3727 solution as the least bitter, indicating that inclusion of GIV3727 led to a significant (p < 0.001) and perceivable decrease in the bitter taste associated with acesulfame K (Figure 4A). Furthermore, taste intensity ratings of the solutions revealed that GIV3727 significantly (p<0.001) decreased the bitterness associated with the acesulfame K from moderate to merely recognizable (Figure 4A). In contrast to the effects on bitter perception, GIV3727 had no effect on the perceived sweetness of acesulfame K since only 55% of the panelists selected the control solution as more sweet (Figure 4B). Similarly, the perceived sweet intensity ratings obtained for the two solutions were not significantly different (Figure 4B). The lack of an effect on sweet taste perception was further confirmed in 2-AFC studies using sucrose where the sample containing GIV3727 could not be discriminated from the 7% sucrose reference (Figure 4C), further confirming that GIV3727 specifically affects only bitter taste perception. Furthermore, we found that GIV3727 has no discernable taste or aroma of its own since an aqueous solution containing 30 ppm GIV3727 could not be discriminated from a water reference in a triangle test (n=50, p=0.196) [22]. Finally, similar results were also obtained in 2-AFC studies using 3 mM saccharin as the taste stimulus in which the reference was also identified as more bitter than the saccharin solution containing GIV3727 (Figure 4D). Together, our findings indicate that GIV3727 effectively reduces only the bitter taste associated with these two sweeteners.
Figure 4.
Bitter taste perception is decreased by GIV3727. The effects of GIV3727 on the bitterness (A) or sweetness (B) of 2 mM acesulfame K were determined by 2-AFC paradigm paired with anchored intensity ratings. Panelists were presented with sweetener alone or plus 30 ppm GIV3727 in random order and asked to select the solution that was most bitter (A) or most sweet (B). The data on the left panels of each graph are the fraction of panelists (out of 22 total) that chose a given solution as the most bitter or sweet. We found that nearly all of the panelists (95% of cohort) selected the GIV3727 solution as less bitter but they were unable to detect any effect on sweetness [55% (control) vs. 45% (+GIV3727)]. The right portions of the graphs show the effects of 30 ppm GIV3727 on the perceived bitter and sweet taste intensities, which were generated using a 0-100 line scale with anchors at the following points: 0 = no bitterness, 25 = recognizable, 50 = moderate, 75 = strong, 100 = extreme. The taste intensity data are plotted as the mean ± s.e.m. and ANOVA was used to assess differences (samples and judges as main effects). We found that sample intensity ratings for bitterness were significantly different (F1,21=83.0; p<0.001) but there was no effect on perceived sweet taste intensity of the GIV3727 solution (F1,21=0.10; p=0.754). (C) Lack of an effect of GIV3727 on sweet taste of sucrose, as assessed using the 2-AFC method. 30 panelists were presented with 7% sucrose alone or plus 36 ppm GIV3727 in random order and asked to select the solution that was most sweet. Data are presented as the fraction of panelists that chose a given solution as the most sweet. Inclusion of GIV3727 had no significant effect on sweet taste perception of the sucrose solution (p = 0.856). (D) Effects of GIV3727 on bitterness of saccharin as determined by 2-AFC. Statistical significance is indicated above the bar and was determined by a beta-binomial analysis.
It is notable that while GIV3727 abolished hTAS2R43/31 receptor activation in vitro (Figure 1), not all panelists detected a complete reduction in bitterness in vivo (Figure 4A). The taste intensity ratings revealed that GIV3727 reduced the bitterness of acesulfame K from moderate to recognizable, thus it did not fully abolish the bitterness. Several factors may contribute to this apparent discrepancy between in vitro and in vivo results. First, in cell-based assays receptor access is relatively unimpeded since the cells are bathed in the solution containing the antagonist whereas in vivo, anatomical constraints may limit compound access leading to reduced efficacy. Single nucleotide polymorphisms (SNPs) have been identified in the hTAS2R gene family that affect bitter agonist activity in vitro and in vivo [23-25]. It is possible that non-synonymous SNPs or even genomic deletions within the TAS2R43/31 genes could diminish the effects of GIV3727 in a small number of individuals. Finally, the artificial sweeteners used in the current study elicit both sweet and bitter sensations, which may hinder discrimination between different taste solutions or taste attributes [26]. Nevertheless, our studies revealed that GIV3727 has efficacy in vivo in reducing the bitterness associated with the sulfonamide sweeteners, consistent with our in vitro findings.
Concluding Remarks
In this paper we describe the identification and characterization of GIV3727, which to our knowledge is the first reported small molecule bitter taste receptor antagonist with in vivo efficacy. While lactisole or 2-(4-methoxyphenoxy)-propanoic acid is widely known as an inhibitor of the human sweet taste receptor [15, 27, 28], this is the first antagonist targeting the hTAS2R bitter receptors. These results are an important step toward understanding how bitter taste perception and/or signaling can be modulated via small molecules. Furthermore, hTAS2R inhibitors could help determine the contributions of bitter taste perception during the ingestion of a complex meal. Moreover, recent evidence indicates that bitter receptors are also expressed in other non-gustatory tissues with proposed roles in the detection of noxious airborne chemicals [29] or regulation of glucose homeostasis via the gastrointestinal tract [30]. Bitter receptor antagonists such as GIV3727 could potentially be very useful as chemical probes to examine the precise contributions of bitter receptor signaling in non-gustatory systems. Finally, this approach also has the potential to improve the taste profile of packaged foods and beverages, and to improve oral administration of bitter-tasting medicines.
Highlights
Discovery and characterization of GIV3727, the first specific bitter receptor antagonist
GIV3727 selectively inhibits six different hTAS2Rs including hTAS2R31 and 43
Lys265 in helix 7 of hTAS2R43/31 is important for inhibition by GIV3727
GIV3727 significantly inhibits the bitterness of acesulfame K and saccharin in vivo
Supplementary Material
Acknowledgments
We thank the research volunteers for their participation and Nicole Brune for managing the high-throughput screening program. This work was supported by a grant of the German Research Foundation to W.M. (Me 1024/2-3) and by NIH and NCRR grants to C.G.B. (1U54MH084690 and P20 RR016480). J.P.S., C.T.S, K.D., S.F. and I.U. note that they are all employees of Givaudan Flavors Corporation, where some of the studies were conducted. J.P.S., C.T.S. and I.U. are inventors on provisional patents related to aspects of this work (WO 2008119197, WO 2008119196, WO 2009015504).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Data
Supplemental Data include five Supplemental Figures, Supplemental Experimental Procedures, and Supplemental References.
References
- 1.Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJP, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 2.Bufe B, Hofmann T, Krautwurst D, Raguse J-D, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to β-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- 3.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- 4.Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature. 2000;404:601–604. doi: 10.1038/35007072. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaddum JH. Theories of drug antagonism. Pharmacol Rev. 1957;9:211–218. [PubMed] [Google Scholar]
- 7.Kenakin T, Jenkinson S, Watson C. Determining the potency and molecular mechanism of action of insurmountable antagonists. J Pharmacol Exp Ther. 2006;319:710–723. doi: 10.1124/jpet.106.107375. [DOI] [PubMed] [Google Scholar]
- 8.Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. The Molecular Receptive Ranges of Human TAS2R Bitter Taste Receptors. Chem. Senses. 2009;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- 9.Sainz E, Cavenagh MM, Gutierrez J, Battey JF, Northup JK, Sullivan SL. Functional characterization of human bitter taste receptors. Biochem J. 2007;403:537–543. doi: 10.1042/BJ20061744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockhoff A, Behrens M, Niv MY, Meyerhof W. Proc Natl Acad Sci U S A. 2010;107:11110–5. doi: 10.1073/pnas.0913862107. PMID: 20534469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Kuhn P, Weis WI, Kobilka BK, et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasmussen SGF, Choi H-J, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, et al. Crystal structure of the human β2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 13.Ballesteros JA, Weinstein HW. Integrated Methods for the Construction of Three-Dimensional Models and Computational Probing of Structure-Function Relations in G-Protein-Coupled Receptors. Methods in Neurosci. 1995;25:366–428. [Google Scholar]
- 14.Jaakola V-P, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, IJzerman AP, Stevens RC. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang P, Cui M, Zhao B, Liu Z, Snyder LA, Benard LMJ, Osman R, Margolskee RF, Max M. Lactisole interacts with the transmembrane domains of human T1R3 to inhibit sweet taste. J Biol Chem. 2005;280:15238–15246. doi: 10.1074/jbc.M414287200. [DOI] [PubMed] [Google Scholar]
- 16.Malherbe P, Kratochwil N, Knoflach F, Zenner M-T, Kew JNC, Kratzeisen C, Maerki HP, Adam G, Mutel V. Mutational analysis and molecular modeling of the allosteric binding site of a novel, selective, noncompetitive antagonist of the metabotropic glutamate 1 receptor. J Biol Chem. 2003;278:8340–8347. doi: 10.1074/jbc.M211759200. [DOI] [PubMed] [Google Scholar]
- 17.Petrel C, Kessler A, Maslah F, Dauban P, Dodd RH, Rognan D, Ruat M. Modeling and mutagenesis of the binding site of Calhex 231, a novel negative allosteric modulator of the extracellular Ca2+-sensing receptor. J Biol Chem. 2003;278:49487–49494. doi: 10.1074/jbc.M308010200. [DOI] [PubMed] [Google Scholar]
- 18.Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klabunde T, Giegerich C, Evers A. Sequence-derived three-dimensional pharmacophore models for G-protein-coupled receptors and their application in virtual screening. J Med Chem. 2009;52:2923–2932. doi: 10.1021/jm9001346. [DOI] [PubMed] [Google Scholar]
- 20.Winnig M, Bufe B, Kratochwil N, Slack J, Meyerhof W. The binding site for neohesperidin dihydrochalcone at the human sweet taste receptor. BMC Struct Biol. 2007;7:66. doi: 10.1186/1472-6807-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiffman SS, Booth BJ, Losee ML, Pecore SD, Warwick ZS. Bitterness of sweeteners as a function of concentration. Brain Res Bull. 1995;36:505–513. doi: 10.1016/0361-9230(94)00225-p. [DOI] [PubMed] [Google Scholar]
- 22.Lawless HT, Heymann H. Sensory evaluation of food: principles and practices. Chapman & Hall; New York: 1998. [Google Scholar]
- 23.Bufe B, Breslin PAS, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim U-K, Drayna D, Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim U, Wooding S, Ricci D, Jorde LB, Drayna D. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci. Human Mutation. 2005;26:199–204. doi: 10.1002/humu.20203. [DOI] [PubMed] [Google Scholar]
- 25.Pronin AN, Xu H, Tang H, Zhang L, Li Q, Li X. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Curr Biol. 2007;17:1403–1408. doi: 10.1016/j.cub.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 26.Keast RSJ, Breslin PAS. An overview of binary taste-taste interactions. Food Qual Pref. 2003;14:111–124. [Google Scholar]
- 27.Schiffman SS, Booth BJ, Sattely-Miller EA, Graham BG, Gibes KM. Selective inhibition of sweetness by the sodium salt of {+/−}2-(4-methoxyphenoxy)propanoic acid. Chem Senses. 1999;24:439–447. doi: 10.1093/chemse/24.4.439. [DOI] [PubMed] [Google Scholar]
- 28.Xu H, Staszewski L, Tang H, Adler E, Zoller M, Li X. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc Natl Acad Sci USA. 2004;101:14258–14263. doi: 10.1073/pnas.0404384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dotson CD, Zhang L, Xu H, Shin Y-K, Vigues S, Ott SH, Elson AET, Choi HJ, Shaw H, Egan JM, et al. Bitter taste receptors influence glucose homeostasis. PLoS ONE. 2008;3:e3974. doi: 10.1371/journal.pone.0003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.