Abstract
Phylogenetic studies of Trypanosoma cruzi have identified the existence of two groups: T. cruzi I and T. cruzi II. There are aspects that still remain unknown about the genetic variability within the T. cruzi I group. Given its epidemiological importance, it is necessary to have a better understanding of T. cruzi transmission cycles. Our purpose was to corroborate the existence of haplotypes within the T. cruzi I group and to describe the genetic variability and phylogenetic relationships, based on single nucleotide polymorphisms (SNPs) found in the miniexon gene intergenic region, for the isolates from different hosts and epidemiological transmission cycles in Colombian regions. 31 T. cruzi isolates were molecularly characterized. Phylogenetic relationships within T. cruzi I isolates showed four haplotype groups (Ia–Id), associated with their transmission cycle. In previous studies, we reported that haplotype Ia is mainly associated with the domestic cycle and domiciliated Rhodnius prolixus. Haplotype Ib is associated with the domestic cycle and peridomestic cycle, haplotype Ic is closely related with the peridomestic cycle, and haplotype Id is strongly associated with the sylvatic cycle. The phylogenetic methodologies applied in this study are tools that bolster the associations among isolates and thus shed light on Chagas disease epidemiology.
1. Introduction
Chagas disease is a very complex zoonosis and represents a major public health problem in Latin America, affecting about 15 million people, with 28 million people being at risk of acquiring the infection. It has been estimated that nearly 5% of the Colombian population is infected and that 20% are at risk of acquiring the disease [1]. The infection, caused by the protozoan Trypanosoma cruzi, affects people during acute and chronic phases with different degrees of severity. The acute stage appears shortly after the infection and the chronic stage may last several years. After a silent asymptomatic period lasting several years, about 25% of the patients develop cardiac symptoms that may lead to chronic heart failure and sudden death, 6% develop digestive lesions, mainly megacolon and megaesophagus, and 3% suffer peripheral nerve damage [2]. These manifestations are not equally distributed within countries [3].
As a result of their particular method of clonal propagation, T. cruzi isolates have been described as showing great phenotypical and genotypical heterogeneity [4, 5]. Despite their high genetic variability, T. cruzi isolates can be classified into two groups, T. cruzi I and T. cruzi II [6], the latter being divided into five subgroups IIa–e [7], whereas T cruzi I consists of a single, relatively homogeneous clade [8].
T. cruzi I, which has a very large geographical distribution from North to South America, predominates from the Amazonian basin northwards, where domestic and sylvatic triatomine species ensure the transmission of Chagas disease in various countries, including Venezuela, Colombia, Central America, and Mexico [9–13]. In the southern cone, countries such as Bolivia, Argentina, and Chile have reported the presence of T. cruzi I circulating in domestic and sylvatic transmission cycles [3, 14, 15].
Intraspecific variation within T. cruzi I has been extensively documented by molecular inferences and biological characterization. In recent years, O'Connor et al. have published studies about genetic variability in T.cruzi I [9] and our group proposed the existence of four haplotypes within T. cruzi I [16]. The inferences described in our study were based on polymorphisms found in the miniexon gene's intergenic partial region. This gene is involved in posttranscriptional events, such as mRNA processing, and it has been proposed as an important molecular marker due to its essential role as a control mechanism of differential protein expression and its high variability within T. cruzi populations [17–19].
The identification and characterization of T. cruzi I haplotypes would be a good epidemiological tool for the understanding of transmission dynamics in endemic areas of Latin America. With this study, we show how the previously proposed haplotypes, based on specific polymorphisms in the partial intergenic region of the miniexon gene that are related to the parasite's transmission cycles, are found in other Colombian isolates and can be usefull to identified other isolates from Latin America.
2. Materials and Methods
2.1. T. cruzi Isolates
A total of 31 T. cruzi isolates were evaluated from different hosts and Colombian regions. Characterized T. cruzi I and T. cruzi II isolates were used as controls (Table 1).
Table 1.
Geographical origin, host, reservoir, and cycle of the T. cruzi isolates analyzed from different parts of Colombia with T. cruzi II used as a control.
| Code Isolates | Abbreviated code | Host/Vector | Geographical origin | Cycle |
|---|---|---|---|---|
| MHOM/CO/03/CG | CGC | Human (acute phase,VIH) | Caqueta | Domestic |
| MHOM/CO/92/FCH | FChC | Human (acute phase) | Norte de Santander | Domestic |
| MHOM/CO/JEM | JEMC | Human (acute phase) | Putumayo | Domestic |
| MHOM/CO/92/JL | JLC | Human (acute phase) | Arauca | Domestic |
| MHOM/CO/SP | SPC | Human (acute phase) | Casanare | Domestic |
| MHOM/CO/07/EB [20, 21] | EBEBE | Human (Congenital) | Boyacá | Domestic |
| MHOM/CO/07/EMA [20, 21] | EMAMA | Human (acute phase) | Boyacá | Domestic |
| MCanis/CO/H135 | H135C | Canis familiaris | Boyacá | Domestic |
| MCanis/CO/H105 | H105C | Canis familiaris | Boyacá | Domestic |
| MDID/CO/28 | Dm28C | Didelphis marsupialis | Tolima | Sylvatic |
| MDID/CO/11 | Dm11C | Didelphis marsupialis | Tolima | Sylvatic |
| MDID/CO/38 | Dm38C | Didelphis marsupialis | Tolima | Sylvatic |
| IRHO/CO/SN6 | SN6C | Rhodnius prolixus | Magdalena | Domestic |
| IRHO/CO/SN8 | SN8C | Rhodnius prolixus | Magdalena | Domestic |
| IRHO/X/CO/380 | X380C | Rhodnius prolixus | Boyacá | Domestic |
| IRHO/CO/X236 | X236C | Rhodnius prolixus | Boyacá | Domestic |
| IRHO/CO/X150 | X150C | Rhodnius prolixus | Boyacá | Domestic |
| IRHO/CO/X1082 | X1082C | Rhodnius prolixus | Boyacá | Domestic |
| IRHO/CO/X1084 | X1084C | Rhodnius prolixus | Boyacá | Domestic |
| IRHO/CO/PAL | PALC | Rhodnius prolixus | Casanare | Sylvatic |
| IRHO/CO/JDI | JD1S | Rhodnius prolixus | Vichada | Sylvatic |
| IRHO/CO/03/PRC | PRC | Rhodnius prolixus | Caqueta | Sylvatic |
| IRPallescens/CO/Mg11 | Mg11C | Rhodnius pallescens | Magdalena | Peridomestic |
| ITr dimidiata/CO/Mg10 | Mg10C | Triatoma dimidiata | Magdalena | Peridomestic |
| ITr dimidiata/CO/Td11 | Td11C | Triatoma dimidiata | Boyacá | Peridomestic |
| ITr dimidiata/CO/Td3 | Td3C | Triatoma dimidiata | Boyacá | Peridomestic |
| ITri dimidiata /CO/HAT | HAT | Triatoma dimidiata | Boyacá | Peridomestic |
| ITri dimidiata /CO/EUR | EUR | Triatoma dimidiata | Boyacá | Peridomestic |
| ITri dimidiata /CO/EF | EFC | Triatoma dimidiata | Boyacá | Peridomestic |
| ITri dimidiata /CO/G11 | G11C | Triatoma dimidiata | Boyacá | Peridomestic |
| ITr venosa/CO/04/TV | TVC | Triatoma venosa | Boyacá | Peridomestic |
| MDID/CO/7 | Dm7(a) | Didelphis marsupialis | Tolima | Sylvatic |
| MHOM/Br/167 | Tc 167(b) | Human | Minas Gerais Brasil | Domestic |
a,b DNA obtained from previously characterized strains (as controls).
2.2. Sequencing of the Miniexon Gene Intergenic Region
DNA was obtained from parasites maintained in culture using the QIAmp DNA extraction kit (Qiagen) and subsequently stored at −4°C. Characterization of the miniexon was performed using previously described primers [22]. PCR was performed on a 20 μL reaction mixture containing 30 ng/μL DNA, 2.5 mM MgCl2, 0.2 μM of each primer, and 0.5 U/μL of BIOTOOLS DNA Polymerase. Amplification products were generated in a PT-150 Minicycler (MJ Research, Watertown, MA, USA) by 27 cycles of 30 seconds at 94°C, 30 seconds at 55°C, and 30 seconds at 72°C, followed by a final elongation of 5 minutes at 72°C. Primers and nucleotides were removed from PCR products by purification using the Ultra Clean PCR Clean-up DNA Purification System (MoBio, Solana Beach, CA, USA) according to the manufacturer's protocol and resuspended in 50 mL of 10 mM TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.6). DNA purity and concentration were determined using an Eppendorf Biophotometer 6131 at 260/280 nm wavelengths.
Sequencing was performed on both strands by the dideoxy chain-termination method and the Taq dye-terminator chemistry kit for the ABI 3730 and ABI 3700 capillary system (Perkin Elmer, Foster City, CA, USA). Multiple alignments were performed using the ClustalW application v.1.8 [23] with default parameters. The alignment was edited in MEGA Version 3.0 [24] and the Staden Package Version 1.5 [25].
2.2.1. Phylogenetic Inference
Phylogenies were inferred using Maximum Parsimony (MP) and Maximum-Likelihood (ML) estimated with PAUP v.4.0b10 [26]. MP analysis was performed using a heuristic algorithm. To assess the relative support of internal nodes, a bootstrap-resampling approach (with 1000 replicates) was performed.
The alignment of the 32 sequences corresponding to T. cruzi I (including the T. cruzi I sequence used as a control) was evaluated to obtain the best-fit model in MrModel Test 2.2 [27], a modified version of model test 3.6 [28] and by the Akaike Information Criterion (AIC) [29, 30]. An analysis of MP was performed using a heuristic search, adding 10 sequences randomly per cycle.
The ML method was performed based on the maximum parsimony results, and according to these data, maximum-likelihood trees were reconstructed by TreeView (Win32) v1.6.6. The robustness of the nodes was evaluated by bootstrap on 1000 replications with heuristic research [31]. Bayesian inference of phylogeny was made using MrBayes v 3.1 [32] (settings according to MrModel test 2.2), 10 million Monte Carlo Markov chain generations (Bayesian-Monte Carlo simulation by MrBayes sampling every 100 simulations, burn-in 10000, number of chains 4).
3. Results
3.1. Miniexon Gene Intergenic Region Sequences
A length of 319 bp was obtained in all isolates for the miniexon SL region gene (spliced leader region). In the alignment of sequences, there were 263 constant sites and 56 parsimony informative sites. A total of 19.43% polymorphic sites were observed, which corresponded to 23 transitions, 14 transversions, and 19 insertion-deletions. The average nucleotide composition (58.4%) was based on GC content (Table 2). The sequences obtained were deposited in GenBank-EMBL-DDJB databases (accession numbers AM259467-AM259480, EU127299-EU127315, EU344771, and EU344772).
Table 2.
Length, GC content (%), and GenBank accession numbers for reported sequences of T. cruzi I and T. cruzi II isolates.
| TCI isolates | % GC | Bp length | GB accession No. |
|---|---|---|---|
| CGC | 58.7 | 313 | AM259467 |
| JLC | 58.9 | 314 | AM259468 |
| FChC | 58.8 | 314 | AM259469 |
| JEMC | 58.7 | 313 | EU127299 |
| Dm28colC | 57.7 | 303 | AM259470 |
| EBEBE | 58.7 | 314 | EU344771 |
| EMAMA | 58.7 | 314 | EU344772 |
| SN6C | 58.9 | 312 | AM259471 |
| X380C | 58.3 | 310 | AM259472 |
| PAL C | 59 | 303 | AM259473 |
| EFC | 57.1 | 313 | AM259474 |
| Td 11C | 58.6 | 312 | AM259475 |
| TVC | 58.5 | 314 | AM259476 |
| Mg10C | 57.8 | 311 | AM259477 |
| JD1S | 58 | 305 | AM259478 |
| SPC | 58 | 314 | EU127300 |
| H135C | 58.2 | 316 | EU127301 |
| H105C | 58.2 | 316 | EU127302 |
| Dm38C | 58 | 305 | EU127303 |
| Dm11C | 58 | 305 | EU127304 |
| SN8C | 58.7 | 313 | EU127305 |
| X236C | 58.9 | 314 | EU127306 |
| X150C | 58.9 | 314 | EU127307 |
| X1082C | 58.2 | 314 | EU127308 |
| X1084C | 58.2 | 314 | EU127309 |
| PRCC | 58.6 | 314 | EU127310 |
| Mg11C | 58.9 | 314 | EU127311 |
| Td3C | 58.6 | 314 | EU127312 |
| HATC | 58 | 314 | EU127312 |
| EURC | 58 | 322 | EU127314 |
| G11C | 58.6 | 314 | EU127315 |
| TCIC | 58.2 | 304 | AM259479 |
| TCIIC | 58 | 298 | AM259480 |
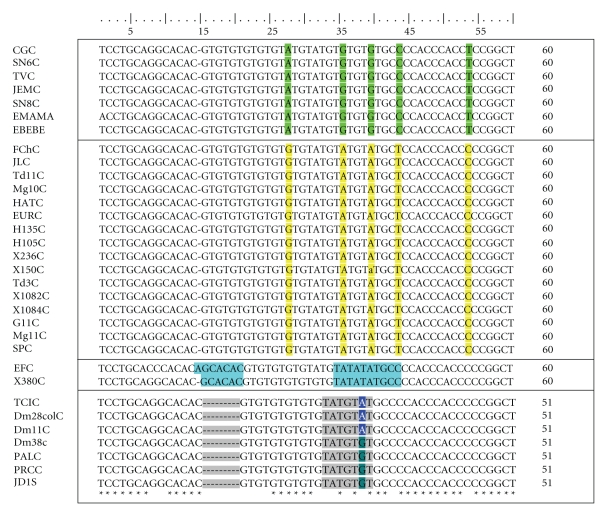
The isolates were clustered into four haplotype groups based on the polymorphic sites in the initial region composed of 56 nucleotides (nt) of the no transcriptional miniexon gene, which showed a continuous microsatellite region (TG)n (TATG)m (TG)x. The number of different repetitions varied between 4 and 7 (n), 2 and 4 (m) and 0 and 3 (x) (positions 15–40) (Figure 1).
Figure 1.
A microsatellite region with an average of 56 nt in which the variability between the 4 proposed haplotypes can be clearly observed. Alignment of the miniexon gene from T. cruzi strains. *Differences are not observed in the alignment. The boxed area corresponds to the nucleotide polymorphisms (SNPs) found between the four proposed haplotypes.
3.2. Phylogenetic Analyses Inferred from Partial Miniexon Gene Intergenic Region Sequences
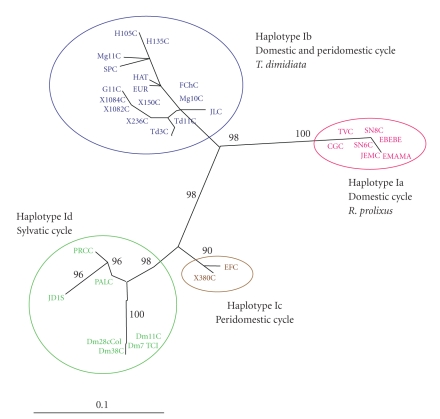
The best ML model fitting the intergenic region data was GTR+G+I. The ML tree obtained (-lnL = 900.8816; proportion of invariable sites = 0.560; gamma distribution shape parameter = 0.893) clearly corroborated the existence of four well-supported haplotypes within the T. cruzi I isolates analyzed and their relation with the transmission cycle (Figure 2).
Figure 2.
Unrooted phylogenetic tree depicting the evolutionary relationships among T. cruzi I isolates. The tree was constructed by the maximum likelihood method. Numbers in larger fonts represent the bootstrap values for the main cluster of the haplotypes. The topology obtained for this method represents the consensus from 236 trees (100 replicates each producing, on average, 4 most parsimonious trees), bootstrap 50% majority rule consensus.
The ML analyses showed four clusters supported by significant bootstrap values (>95%). In Figures 2 and 3, the phylogenetic analyses showed haplotype Id, corresponding to isolates from Didelphis marsupialis and wild triatomines, with 96% bootstrap support and 100% Bayesian probabilities. The haplotypes Ia, Ib, and Ic were supported with 98% bootstrap. Haplotypes Ia and Ib corresponded to the domestic and peridomestic cycles, respectively, showing a bootstrap value of 98%, and they were more related in comparison with haplotype Ic. Haplotype Ic was formed by isolates from the peridomestic cycle; however, although they belonged to this cycle, they were closely related to haplotype Id (sylvatic cycle), with a bootstrap value of 98%.
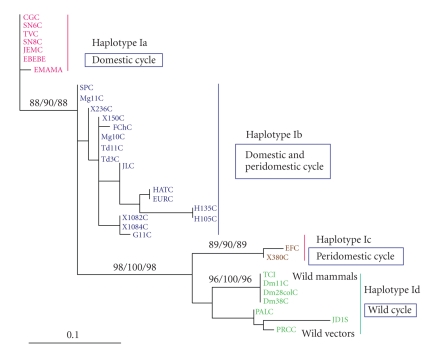
Figure 3.
Unrooted phylogenetic tree depicting the evolutionary relationships between T. cruzi I isolates. Phylogram showing support nodes from Bayesian-estimated likelihood with 10 million Monte-Carlo Markovian chain generation node support from the >50%-majority rule consensus GTR+I+G model. Invariant sites and gamma distribution values are at the right side of the slash (MP bootstrap values/Bayesian probabilities × 100/ML bootstrap values).
Haplotype Id was mainly related with the sylvatic environment (isolates obtained from D. marsupialis and Rhodnius prolixus sylvatic population) supported by a significant bootstrap value of 96% (Figure 3).
A MrBayes credibility method was used to give statistical validity. The Bayesian probabilities (>95%) showed three significant clades: one formed by the domestic isolates, haplotype Ia, which showed a strong polytomy, a second clade (Ib) with a Bayesian probability of 100% formed by isolates from domestic and peridomestic cycles, and a third main clade formed in which haplotypes Ic and Id were closely related. However, these isolates presented punctual mutations that allowed us to group them in different haplotype groups.
4. Discussion
T. cruzi I isolates have been found mainly associated with the sylvatic transmission cycle [33, 34] and parasites isolated from marsupials [35, 36]. T. cruzi II has been mainly associated with the domestic transmission cycle and infection in primates [33]. This has led to the misconception that T. cuzi I is always associated with the sylvatic cycle of transmission [8]. Recent epidemiological studies performed in the northern regions of South America, in Central America, and Mexico have demonstrated a predominance of T. cruzi I in the sylvatic cycle of transmission, as well as in the domestic and peridomestic cycles [11, 12, 37].
The miniexon gene has been widely used as a taxonomic marker due to its high heterogeneity and the presence of large multicopy genomic arrays in the parasite genome [16, 17]. The haplotypes within the T. cruzi I group of Colombia have been identified from phylogenetic analyses such as MP, ML, and BI as follows: haplotype Ia, associated with the domestic cycle and the vector R. prolixus; haplotype Ib, associated with the domestic and peridomestic cycles and the vector Triatoma dimidiata; haplotype Ic, associated with the peridomestic cycle; and haplotype Id, associated with the sylvatic cycle of transmission.
Haplotype Ib is associated with humans, peridomestic vectors (T. dimidiata), and isolates from reservoirs associated with the peridomicile (Canis familiaris). Haplotype Ic is not clearly defined within a specific transmission cycle and seems to play an intermediary role between the domestic and sylvatic cycles. Furthermore, there is a phylogenetic association with haplotype Id that is supported by a bootstrap of 98%, grouping the isolates in a monophyletic clade (Figures 2 and 3). To confirm this observation requires a larger sample of isolates for either defining haplotype Ic as an independent group or classifying it into sylvatic haplotype Id [16, 18].
The isolates from wild R. prolixus captured in palms (Attalea butyracea) and the wild reservoir D. marsupialis have been grouped in haplotype Id. Phylogenetic analysis shows a strong group (haplotype Id), which includes the isolates with sylvatic origin, and the bootstrap value (96%–100%) statistically supports this group. This result also corroborates the observation by O'Connor et al. [9] in which the “Didelphis-group” was proposed, adding in from the present study the isolates from sylvatic vectors. We found a subdivision, based on nucleotide polymorphisms at position 39, in haplotype Id. This small difference linked to the wild origin of the vectors found in this group and was not considered as a fifth haplotype (Figure 1).
The proposed haplotypes have a direct association with the epidemiologic cycle from which they come, but there was not a direct relationship with the geographical isolation. The variability found between the haplotypes can be explained by the diverse fauna in different regions and by clearance processes that are occurring as environmental changes are occurring. Unfavorable changes and the subsequent rarefaction that occurs in wild fauna favor the overlapping of transmission cycles and the consequent molecular diversification of parasite strains [38].
This phenomenon is influenced by environmental and immunological factors, including virulence, pathogenicity, and possible selection of strains and clones after interacting with vectors and vertebrate hosts. The combination of several of these factors might explain the variability in the biological behavior of the parasite [39].
The primitive transmission was restricted to tropical forest environments, where the parasite spread initially by anal gland secretions and urine from the didelphides [40, 41]. Different genetic characterizations performed on T. cruzi I isolates have suggested a strong association with Didelphis spp., while at the same time, there is an association of T. cruzi II with armadillos [8]. Interestingly, R. prolixus has been related to mammals in palms within wild ecotopes. In addition, these relationships were observed throughout North and South America [42].
The opossum genus presents the highest distribution in the world and ranges found from southern Canada to southern Argentina. D. albiventris lives preferentially in the cold climates of South America. D. marsupialis inhabits warm forest areas, mainly in the northern part of South America and Central America, where they maintain the same distribution as T. cruzi I [9].
The association of T. cruzi I with Didelphis spp. has also been observed in the United States, where D. virginiana migrated to North America during the Pleistocene approximately 40 million years ago [9, 43, 44]. The molecular characterization of parasites isolated from D. albiventris has also demonstrated the presence of T. cruzi I. It has been demonstrated that phylogenetic groups of T. cruzi infect certain hosts in a preferential manner. Furthermore, there is evidence that T. cruzi II produces high parasitemia in rodents, while in D. marsupialis, T. cruzi II is eliminated and T. cruzi I is withheld [45].
The relationship between the sylvatic and domestic cycle is extremely complex; however, a common feature in the two cycles is the presence of didelphydes, which allegedly play an important role in the dynamics of transmission [46]. The coevolution of T. cruzi I and D. marsupialis suggests an ancestral and parental relation that has produced all genetic subdivisions of the parasite identified in this study. This reported variability has favored the parasite's dispersion from the insect to wild mammals to domestic mammals [8, 16, 33, 34, 47].
Based on these observations, we suggest that T. cruzi I isolates are part of an ancestral group in America beginning with the sylvatic cycle of transmission between vectors and wild reservoirs, in which D. marsupialis acted as a transmission bridge between the domiciliary and peridomiciliary cycles, given its ability to shift from domestic to wild environments where this marsupial sometimes goes in search of food [33, 48].
Different studies have reported the vector R. prolixus as a factor for the final selective transmission of T. cruzi I for humans and reservoirs. This suggests that these sylvatic R. prolixus, widely distributed in palm trees [49, 50], may have begun the process of the passage of T. cruzi I from the wild cycle to a domestic cycle at the time they began their domiciliation process [49, 51].
These results support, for the first time, a very close relationship between T. cruzi I haplotypes Ib and Id and vectors R. prolixus and T. dimidiata, respectively. Previous studies, with an emphasis on the role of vectors in the transmission of trypanosome subpopulation, have shown a close correlation between genotypes of T. rangeli and Rhodnius species [51–54].
The trypanosome interaction with different triatomine species and populations is apparently an important factor determining the trypanosome epidemiology that infects humans in Latin America. Frequently, specific subpopulations of trypanosomes are transmitted by specific vectors in a particular geographic area.
Future studies focused on the trypanosome-vector interaction will allow the identification of coevolutionary processes, which in turn could support the evolutionary hypothesis and the distribution of T. cruzi and T. rangeli vectors in America. Additionally, these studies will help to identify the mechanisms that facilitate or impede the transmission of parasites by different vector species [43]. However, to establish a relationship between the different T. cruzi I haplotypes, further analyses using additional suitable molecular markers, such as mitochondrial and flagellar genes, are required.
Acknowledgments
Financial support was obtained from the Projects: The European Union Seventh Framework Programme (ChagasEpinet) Contract number 223034 and the “Red de Investigación de Centros de Enfermedades Tropicales” (RICET, Projects no. C03/04, no. ISCIII2005-PI050574, and no. ISCIII-RETIC RD06/0021/0017) of the Program of Redes Temáticas de Investigación Cooperativa, FIS, Ministry of Health, Madrid. Colciencias no 7155-CO, “Support to the Scientific Community, National Doctorates Programme,” Colciencias no. 1204-343-19188, Faculty of Sciences, Universidad de los Andes, Bogotá, Colombia, and Servicio Central de Soporte a la Investigación SCSIE, Universidad de Valencia, Spain. The authors wish to thank Omar Triana and the Congenital Chagas Disease Study group (ColcienciasProject no. 1109-04-18231) for their contribution with some isolates involved in this study and Juan A. Sánchez for their consultantship in phylogenetics methods.
References
- 1.WHO. Report of Scientific Group in Chagas Disease. TDR/SWG/09. Buenos Aires, Argentina: World Health Organization; 2007. Special programme for research and training in tropical diseases (TDR) [Google Scholar]
- 2.Moncayo A, Ortiz M. An update on Chagas disease (human American trypanosomiasis) Annals of Tropical Medicine and Parasitology. 2006;100:663–677. doi: 10.1179/136485906X112248. [DOI] [PubMed] [Google Scholar]
- 3.Brenière SF, Carrasco R, Revollo S, Aparicio G, Desjeux P, Tibayrenc M. Chagas’ disease in Bolivia: clinical and epidemiological features and zymodeme variability of Trypanosoma cruzi strains isolated from patients. American Journal of Tropical Medicine and Hygiene. 1989;41(5):521–529. doi: 10.4269/ajtmh.1989.41.521. [DOI] [PubMed] [Google Scholar]
- 4.Tibayrenc M, Ward P, Moya A, Ayala FJ. Natural populations of Trypanosoma cruzi, the agent of Chagas disease, have a complex multiclonal structure. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(1):115–119. doi: 10.1073/pnas.83.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tibayrenc M, Ayala FJ. The clonal theory of parasitic protozoa: 12 years on. Trends in Parasitology. 2002;18(9):405–410. doi: 10.1016/s1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- 6.Recommendations from a satellite meeting. Memorias do Instituto Oswaldo Cruz. 1999;194(supplement 1):429–432. doi: 10.1590/s0074-02761999000700085. [DOI] [PubMed] [Google Scholar]
- 7.Brisse S, Dujardin J-C, Tibayrenc M. Identification of six Trypanosoma cruzi lineages by sequence-characterised amplified region markers. Molecular and Biochemical Parasitology. 2000;111(1):95–105. doi: 10.1016/s0166-6851(00)00302-9. [DOI] [PubMed] [Google Scholar]
- 8.Yeo M, Acosta N, Llewellyn M, et al. Origins of Chagas disease: didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. International Journal for Parasitology. 2005;35(2):225–233. doi: 10.1016/j.ijpara.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 9.O’Connor O, Bosseno M-F, Barnabé C, Douzery EJP, Breniére SF. Genetic clustering of Trypanosoma cruzi I lineage evidenced by intergenic miniexon gene sequencing. Infection, Genetics and Evolution. 2007;7(5):587–593. doi: 10.1016/j.meegid.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Bosseno M-F, Barnabé C, Magallón E, et al. Predominance of Trypanosoma cruzi lineage I in Mexico. Journal of Clinical Microbiology. 2002;40(2):627–632. doi: 10.1128/JCM.40.2.627-632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Añez N, Crisante G, da Silva FM, et al. Predominance of lineage I among Trypanosoma cruzi isolates from Venezuelan patients with different clinical profiles of acute Chagas’ disease. Tropical Medicine and International Health. 2004;9(12):1319–1326. doi: 10.1111/j.1365-3156.2004.01333.x. [DOI] [PubMed] [Google Scholar]
- 12.Crisante G, Rojas A, Teixeira MMG, Añez N. Infected dogs as a risk factor in the transmission of human Trypanosoma cruzi infection in western Venezuela. Acta Tropica. 2006;98(3):247–254. doi: 10.1016/j.actatropica.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Sousa OE, Samudio F, Junca C, Calzada JE. Molecular characterization of human Trypanosoma cruzi isolates from endemic areas in Panama. Memorias do Instituto Oswaldo Cruz. 2006;101(4):455–457. doi: 10.1590/s0074-02762006000400018. [DOI] [PubMed] [Google Scholar]
- 14.Diosque P, Padilla AM, Cimino RO, et al. Chagas disease in rural areas of Chaco Province, Argentina: epidemiologic survey in humans, reservoirs, and vectors. American Journal of Tropical Medicine and Hygiene. 2004;71(5):590–593. [PubMed] [Google Scholar]
- 15.Solari A, Wallace A, Ortiz S, Venegas J, Sanchez G. Biological characterization of Trypanosoma cruzi stocks from Chilean insect vectors. Experimental Parasitology. 1998;89(3):312–322. doi: 10.1006/expr.1998.4289. [DOI] [PubMed] [Google Scholar]
- 16.Herrera C, Bargues MD, Fajardo A, et al. Identifying four Trypanosoma cruzi I isolate haplotypes from different geographic regions in Colombia. Infection, Genetics and Evolution. 2007;7(4):535–539. doi: 10.1016/j.meegid.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Thomas S, Westenberger SJ, Campbell DA, Sturm NR. Intragenomic spliced leader RNA array analysis of kinetoplastids reveals unexpected transcribed region diversity in Trypanosoma cruzi . Gene. 2005;352(1-2):100–108. doi: 10.1016/j.gene.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Falla A, Herrera C, Fajardo A, Montilla M, Vallejo GA, Guhl F. Haplotype identification within Trypanosoma cruzi I in Colombian isolates from several reservoirs, vectors and humans. Acta Tropica. 2009;110(1):15–21. doi: 10.1016/j.actatropica.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Fernandas O, Santos SS, Cupolillo E, et al. A mini-exon multiplex polymerase chain reaction to distinguish the major groups of Tryopanosoma cruzi and T. rangeli in the Brazilian Amazon. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2001;95(1):97–99. doi: 10.1016/s0035-9203(01)90350-5. [DOI] [PubMed] [Google Scholar]
- 20.Manrique FG, Ospina JM, Herrera GM, et al. Enfermedad de Chagas transplacentaria en Miraflores y Moniquirá, Boyacá. Biomédica. 2007;27(supplement 1):p. 172. [Google Scholar]
- 21.Pavía PJ, Montilla M, Nicholls RS, Manrique F, Herrera G, Puerta CJ. Análisis de un caso de enfermedad de Chagas transplacentario en Moniquirá (Boyacá) mediante AP-PCR. Biomédica. 2007;27(supplement 1):p. 238. [PubMed] [Google Scholar]
- 22.Souto RP, Fernandes O, Macedo AM, Campbell DA, Zingales B. DNA markers define two major phylogenetic lineages of Trypanosoma cruzi . Molecular and Biochemical Parasitology. 1996;83(2):141–152. doi: 10.1016/s0166-6851(96)02755-7. [DOI] [PubMed] [Google Scholar]
- 23.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 25.Staden R, Judge DP, Bonfield JK. Sequence assembly and finishing methods. Methods of Biochemical Analysis. 2001;43:303–322. doi: 10.1002/0471223921.ch13. [DOI] [PubMed] [Google Scholar]
- 26.Swofford DL. PAUP∗ phylogenetic analysis using parsimony (and other methods) Version 40b10. Sunderland, Sinauer Associates, 2002.
- 27.Nylander JAA. MrModeltest v2.2. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- 28.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 29.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19(6):716–723. [Google Scholar]
- 30.Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Systematic Biology. 2004;53(5):793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Ronquist F, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 33.Fernandes O, Sturm NR, Derré R, Campbell DA. The mini-exon gene: a genetic marker for zymodeme III of Trypanosoma cruzi . Molecular and Biochemical Parasitology. 1998;95(1):129–133. doi: 10.1016/s0166-6851(98)00073-5. [DOI] [PubMed] [Google Scholar]
- 34.Fernandes O, Mangia RH, Lisboa CV, Pinho AP, Morel CM, Zingales B. The complexity of the sylvatic cycle of Trypanosoma cruzi in Rio de Janeiro state (Brazil) revealed by the non-transcribed spacer of the mini-exon gene. Parasitology. 1999;118(2):161–166. doi: 10.1017/s0031182098003709. [DOI] [PubMed] [Google Scholar]
- 35.Kawashita SY, Sanson GFO, Fernandes O, Zingales B, Briones MRS. Maximum-likelihood divergence date estimates based on rRNA gene sequences suggest two scenarios of Trypanosoma cruzi intraspecific evolution. Molecular Biology and Evolution. 2001;18(12):2250–2259. doi: 10.1093/oxfordjournals.molbev.a003771. [DOI] [PubMed] [Google Scholar]
- 36.WHO. Tech. Rep. 905. Geneva, Switzerland: World Health Organization; 2002. Second report from the Committee of Experts Series of technical reports. [Google Scholar]
- 37.Carrasco HJ, Torrellas A, Garcıa C, Segovia M, Feliciangeli MD. Risk of Trypanosoma cruzi I (Kinetoplastida: Trypanosomatidae) transmission by Panstrongylus geniculatus (Hemiptera: Reduviidae) in Caracas (Metropolitan District) and neighboring States, Venezuela. International Journal for Parasitology. 2005;35(13):1379–1384. doi: 10.1016/j.ijpara.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Noireau F, Diosque P, Jansen AM. Trypanosoma cruzi: adaptation to its vectors and its hosts. Veterinary Research. 2009;40(2):p. 26. doi: 10.1051/vetres/2009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Devera R, Fernandes O, Coura JR. Should Trypanosoma cruzi be called “cruzi” complex? A review of the parasite diversity and the potential of selecting population after in vitro culturing and mice infection. Memorias do Instituto Oswaldo Cruz. 2003;98(1):1–12. doi: 10.1590/s0074-02762003000100001. [DOI] [PubMed] [Google Scholar]
- 40.Deane MP, Jansen A, Lenzi HL. Trypanosoma cruzi: vertebrate and invertebrate cycles in the same mammal host, the opossum Didelphis marsupialis . Memorias do Instituto Oswaldo Cruz. 1984;79(4):513–515. doi: 10.1590/s0074-02761984000400021. [DOI] [PubMed] [Google Scholar]
- 41.Guhl F, Jaramillo C, Vallejo GA, Cárdenas F, Aufderheide A. Chagas disease and human migration. Memorias do Instituto Oswaldo Cruz. 2000;95(4):553–555. doi: 10.1590/s0074-02762000000400018. [DOI] [PubMed] [Google Scholar]
- 42.Gaunt M, Miles M. The ecotopes and evolution of triatomine bugs (Triatominae) and their associated trypanosomes. Memorias do Instituto Oswaldo Cruz. 2000;95(4):557–565. doi: 10.1590/s0074-02762000000400019. [DOI] [PubMed] [Google Scholar]
- 43.Barnabé C, Yaeger R, Pung O, Tibayrenc M. Trypanosoma cruzi: a considerable phylogenetic divergence indicates that the agent of chagas disease is indigenous to the native fauna of the United States. Experimental Parasitology. 2001;99(2):73–79. doi: 10.1006/expr.2001.4651. [DOI] [PubMed] [Google Scholar]
- 44.Diosque P, Padilla AM, Cimino RO, et al. Chagas disease in rural areas of Chaco Province, Argentina: epidemiologic survey in humans, reservoirs, and vectors. American Journal of Tropical Medicine and Hygiene. 2004;71(5):590–593. [PubMed] [Google Scholar]
- 45.Jansen AM, Madeira F, Carreira JC, Medina-Acosta E, Deane MP. Trypanosoma cruzi in the opossum Didelphis marsupialis: a study of the correlations and kinetics of the systemic and scent gland infections in naturally and experimentally infected animals. Experimental Parasitology. 1997;86(1):37–44. doi: 10.1006/expr.1997.4144. [DOI] [PubMed] [Google Scholar]
- 46.Gurgel-Gonçalves R, Ramalho ED, Duarte MA, Palma AR. Enzootic transmission of Trypanosoma cruzi and T. rangeli in the Federal District of Brazil. Revista do Instituto de Medicina Tropical de Sao Paulo. 2004;46(6):323–330. doi: 10.1590/s0036-46652004000600005. [DOI] [PubMed] [Google Scholar]
- 47.Carreira JCA, Jansen AM, de Nazareth Meirelles M, Costa e Silva F, Lenzi HL. Trypanosoma cruzi in the scent glands of Didelphis marsupialis: the kinetics of colonization. Experimental Parasitology. 2001;97(3):129–140. doi: 10.1006/expr.2001.4603. [DOI] [PubMed] [Google Scholar]
- 48.Fernandas O, Santos SS, Cupolillo E, et al. A mini-exon multiplex polymerase chain reaction to distinguish the major groups of Tryopanosoma cruzi and T. rangeli in the Brazilian Amazon. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2001;95(1):97–99. doi: 10.1016/s0035-9203(01)90350-5. [DOI] [PubMed] [Google Scholar]
- 49.Miles M, Feliciangeli MD, Rojas A. American trypanosomiasis (Chagas disease) and the role of molecular epidemiology in guiding control strategies. BMJ. 2003;28:p. 144. doi: 10.1136/bmj.326.7404.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fitzpatrick S, Feliciangeli MD, Sanchez-Martin MJ, Monteiro FA, Miles MA. Molecular genetics reveal that silvatic Rhodnius prolixus do colonise rural houses. PLoS Neglected Tropical Diseases. 2008;2(4, article e210) doi: 10.1371/journal.pntd.0000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vallejo GA, Guhl F, Carranza JC, Moreno J, Triana O, Grisard EC. Parity between kinetoplast DNA and mini-exon gene sequences supports either clonal evolution or speciation in Trypanosoma rangeli strains isolated from Rhodnius colombiensis, R. pallescens and R. prolixus in Colombia. Infection, Genetics and Evolution. 2003;3(1):39–45. doi: 10.1016/s1567-1348(02)00150-8. [DOI] [PubMed] [Google Scholar]
- 52.Maia da Silva F, Junqueira AC, Campaner M, et al. Comparative phylogeography of Trypanosoma rangeli and Rhodnius (Hemiptera: Reduviidae) supports a long coexistence of parasite lineages and their sympatric vectors. Molecular Ecology. 2007;16(16):3361–3373. doi: 10.1111/j.1365-294X.2007.03371.x. [DOI] [PubMed] [Google Scholar]
- 53.Urrea DA, Carranza JC, Cuba CAC, et al. Molecular characterisation of Trypanosoma rangeli strains isolated from Rhodnius ecuadoriensis in Peru, R. colombiensis in Colombia and R. pallescens in Panama, supports a co-evolutionary association between parasites and vectors. Infection, Genetics and Evolution. 2005;5(2):123–129. doi: 10.1016/j.meegid.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 54.Vallejo GA, Guhl F, Schaub GA. Triatominae-Trypanosoma cruzi/T. rangeli: vector-parasite interactions. Acta Tropica. 2008;110(2-3):137–147. doi: 10.1016/j.actatropica.2008.10.001. [DOI] [PubMed] [Google Scholar]