Summary
Although autosomal forms of nonsyndromic mental retardation account for the majority of cases of mental retardation, the genes that are involved remain largely unknown. We sequenced the autosomal gene SYNGAP1, which encodes a ras GTPase-activating protein that is critical for cognition and synapse function, in 94 patients with nonsyndromic mental retardation. We identified de novo truncating mutations (K138X, R579X, and L813RfsX22) in three of these patients. In contrast, we observed no de novo or truncating mutations in SYNGAP1 in samples from 142 subjects with autism spectrum disorders, 143 subjects with schizophrenia, and 190 control subjects. These results indicate that SYNGAP1 disruption is a cause of autosomal dominant nonsyndromic mental retardation.
Mental retardation is the most prevalent severe handicap of children, affecting 1 to 3% of the population.1 Most patients have the nonsyndromic form of the disorder, which is characterized by the absence of associated morphologic, radiologic, and metabolic features.2 The genetic factors involved in nonsyndromic mental retardation remain poorly understood. Linkage and cytogenetic analyses have led to the identification of 29 X-linked and 5 autosomal recessive genes associated with nonsyndromic mental retardation, which together account for less than 10% of cases.1–6 Moreover, autosomal dominant genes have yet to be identified, mainly because mental retardation results in lower reproductive fitness, which in turn decreases the likelihood of identifying families that are amenable to linkage analysis. Nevertheless, the fact that de novo chromosomal rearrangements (usually involving a change in copy number of genomic regions) represent the most commonly recognized cause of mental retardation indicates that monoallelic lesions are sufficient to cause this disorder. This raises the possibility that smaller de novo genetic lesions, such as point mutations, also contribute to the pathogenesis of the disorder. Indeed, it has been estimated that one to three de novo mutations per zygote affect the amino acid sequence.7 Given the genetic heterogeneity of nonsyndromic mental retardation, de novo autosomal point mutations may thus explain a large fraction of cases. However, the identification of such mutations in the context of nonsyndromic mental retardation is not amenable to linkage or association approaches and relies on the sequencing of candidate genes.
The classic observation that various forms of mental retardation are associated with abnormalities in the morphology of dendritic spines suggests that disruption of pathways involved in synaptic plasticity may be a common mechanism of this disorder.8 Indeed, several X-linked nonsyndromic mental retardation genes regulate synaptic strength and spine morphogenesis.1,2,9
Nearly all presynaptic terminals that make synapses on dendritic spines release the neurotransmitter glutamate. Glutamate signaling through N-methyl-d-aspartate (NMDA) receptors on the surface of spines is necessary for the plasticity of excitatory synapses. The NMDA receptor is linked to multiple pathways through its association with a large complex of more than 185 proteins.9 Some forms of synaptic plasticity that are regulated by NMDA receptors require the insertion of the α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor at the postsynaptic membrane (i.e., at the dendritic spine).10 SYNGAP1, a GTPase-activating protein that is selectively expressed in the brain and is a component of the NMDA-receptor complex, acts downstream of the receptor, blocking the insertion of the AMPA receptor at the postsynaptic membrane11–14 by inhibition of the RAS–ERK pathway.12 Mice that are homozygous for null alleles of Syngap1 die shortly after birth, indicating an essential role for Syngap1 during early postnatal development, whereas Syngap1 heterozygous mice have impaired synaptic plasticity and learning, which is consistent with a role for SYNGAP1 as a component of the NMDA-receptor complex.15,16
We tested the hypothesis that de novo mutations in autosomal genes that are involved in synaptic plasticity are a common cause of nonsyndromic mental retardation. We first focused on SYNGAP1 because of its involvement in pathways regulated by NMDA receptors and because of the cognitive dysfunction seen in mice that have a heterozygous mutation in Syngap1. In this report, we describe the results of SYNGAP1 sequence analysis and identify three de novo protein-truncating mutations in 3 of 94 patients with nonsyndromic mental retardation.
CASE REPORT
PATIENTS AND CONTROL SUBJECTS
We recruited a series of 45 male patients and 49 female patients with sporadic nonsyndromic mental retardation, including 63 French Canadians, 6 European whites, 9 non-European whites, 2 South Americans, 6 nonwhites, and 8 patients of mixed race or ethnic background. We used a series of open-ended questions to determine race or ethnic background by asking the parents of the patients about the origin of each of their parents. The diagnosis of mental retardation was made on a clinical basis with the use of standardized developmental or IQ tests.
All patients were examined by at least one experienced clinical geneticist, who ruled out the presence of specific dysmorphic features. For all patients, the birth weights and history of postnatal growth were within the normal range, and the head circumference was normal at birth. Mental retardation was unexplained in these patients despite standard investigations, including karyotyping, subtelomeric fluorescence in situ hybridization analysis, or comparative genomic hybridization targeting regions associated with known syndromes, molecular testing for the common expansion mutation in FMR1, and computed tomography or magnetic resonance imaging of the brain.
We also studied three series of other subjects, including 142 subjects with nonsyndromic autism spectrum disorders, 143 subjects with schizophrenia, and 190 healthy control subjects matched for ancestral origin. The subjects in all three series were mainly of French Canadian or European origin (for details regarding case definitions, see the Supplementary Appendix, available with the full text of this article at NEJM.org). Blood samples were obtained from all subjects and from each set of parents, after we had obtained written informed consent and approval from the ethics committee at each institution.
Genomic DNA was extracted from blood samples with the use of the Puregene DNA kit (Gentra Systems). Paternity and maternity of each patient were confirmed with the use of six highly informative unlinked microsatellite markers (D2S1327, D3S1043, D4S3351, D6S1043, D8S1179, and D10S677).
GENETIC SCREENING, VALIDATION ANALYSES, AND BIOINFORMATICS
We amplified coding regions in SYNGAP1 (National Center for Biotechnology Information build number, 36.1) and their intronic flanking regions using a polymerase-chain-reaction (PCR) assay of genomic DNA and then sequenced the resulting products. PCR primers targeting the 19 exons of SYNGAP1 were designed with the use of Exon-Primer from the University of California, Santa Cruz, Genome Browser (Table 1 in the Supplementary Appendix). PCR assays were performed in 384 well plates with the use of 5 ng of genomic DNA, according to standard procedures. PCR products were sequenced at the McGill University and Genome Quebec Innovation Centre in Montreal (www.genomequebecplatforms.com/mcgill) on a 3730XL DNA analyzer. In each case, unique mutations were confirmed by reamplification of the fragment and resequencing of the proband and both parents with the use of reverse and forward primers. PolyPhred (version 5.04), PolyScan (version 3.0), and Mutation Surveyor (version 3.10) were used for mutation-detection analyses.
DISCOVERY OF TRUNCATING MUTATIONS
We identified two patients who were heterozygous for nonsense mutations, K138X in Patient 1 and R579X in Patient 2. In addition, we identified a patient (Patient 3) who was heterozygous for the mutation c.2438delT, which was predicted to cause a frame shift starting at codon 813, producing a premature stop codon at position 835 (L813RfsX22) (Fig. 1). Each mutation was absent in the DNA samples we obtained from the parents of the affected patients (indicating that the mutations were de novo) and in a control series of 190 healthy subjects, in which all SYNGAP1 exons and intronic junctions were sequenced. One heterozygous missense variant (I1115T), probably a benign polymorphism, was present in patients with nonsyndromic mental retardation and control subjects alike (Table 1).
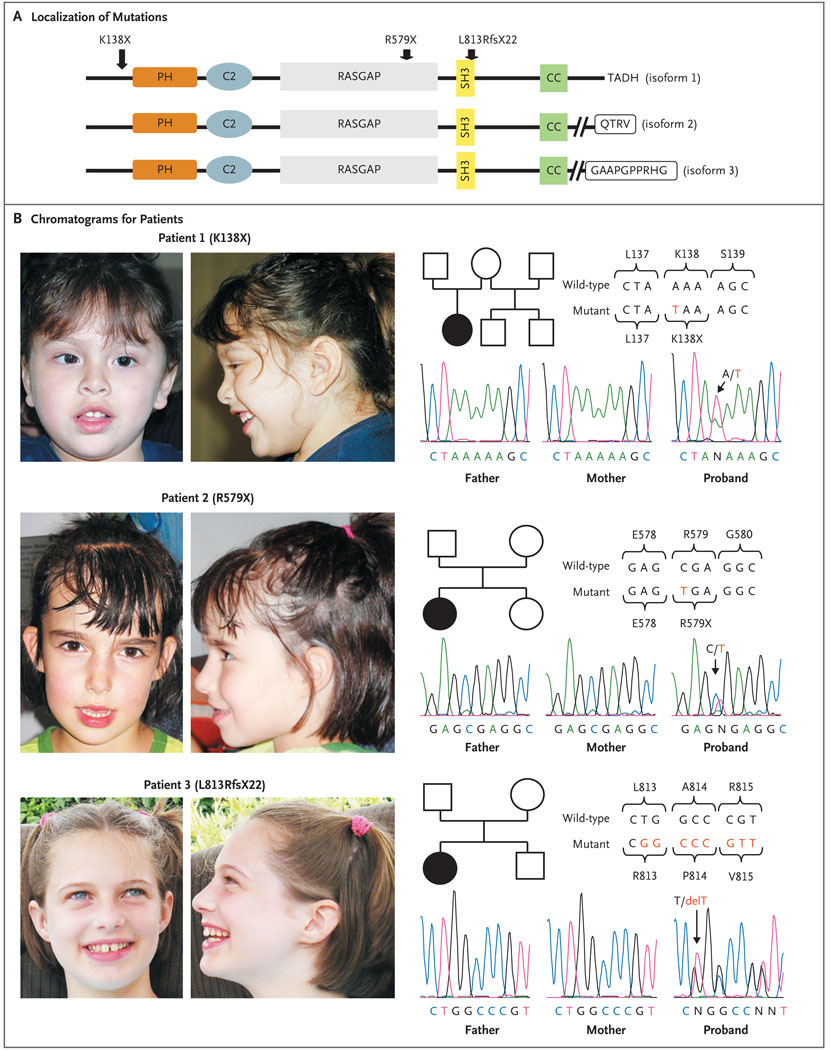
Figure 1. De Novo SYNGAP1 Mutations in Three Patients with Nonsyndromic Mental Retardation.
Panel A shows the localization of de novo SYNGAP1 mutations identified in patients with nonsyndromic mental retardation. Amino acid positions are based on the NCBI Reference Sequence project number NP_006763 (from NM_006772) (for isoform 1, which consists of 1343 amino acids). The various predicted functional domains are highlighted: pleckstrin homology domain (PH; positions 150 to 251), C2 domain (positions 263 to 362), RASGAP (positions 392 to 729), SH3 (positions 785 to 815), CC domain (positions 1189 to 1262), T/SXV type 1 PDZ-binding motif (QTRV) (isoform 2), and CamKII binding (GAAPGPPRHG) (isoform 3). In addition, the last amino acids of isoform 1 (TADH) are shown. The variable C-terminals of the three SYNGAP1 isoforms that are shown here correspond to GenBank complementary DNA accession numbers AB067525 (for isoform 1), AK307888 (for isoform 2), and AL713634 (for isoform 3). The hatched lines for isoforms 2 and 3 indicate that the amino acid sequence has been abbreviated. Panel B shows chromatograms corresponding to the SYNGAP1 sequence for each of the three patients with de novo mutations and for their parents. Wild-type and mutant SYNGAP1 DNA sequences are shown, along with the corresponding amino acids.
Table 1.
SYNGAP1 Mutations in the Study Subjects.*
| Series of Subjects and Coding Sequence of Mutation |
Change in Amino Acid |
Occurrence no./total no. |
Inheritance |
|---|---|---|---|
| Nonsyndromic mental retardation | |||
| c.412A→T | K138X | 1/94 | De novo |
| c.1735C→T | R579X | 1/94 | De novo |
| c.2438delT | L813RfsX22 | 1/94 | De novo |
| c.3344T→C | I1115T | 2/94 | ND |
| Autism spectrum disorders | |||
| c.3344T→C | I1115T | 8/142 | ND |
| c.3848C→T | P1283L | 1/142 | Mother† |
| Schizophrenia | |||
| c.3344T→C | I1115T | 4/143 | ND |
| c.3929C→T | T1310M | 1/143 | Mother† |
| c.2369C→A | T790N | 1/143 | Father† |
| Control subjects | |||
| c.603T→G | D201E | 1/190 | Father† |
| c.2246G→A | R749Q | 1/190 | Father† |
| c.3344T→C | I1115T | 4/190 | ND |
All reported mutations are heterozygous. Mutation positions are listed according to the coding sequence of SYNGAP1, as listed in NCBI Reference Sequence number NM_006772. ND denotes not determined.
This relative of the subject with the mutation had no symptoms suggestive of a neurodevelopmental disorder.
The three patients with the de novo mutations, whose ages ranged from 4 to 11 years, had similar clinical profiles (Table 2, and the Supplementary Appendix). They were all born to nonconsanguineous parents after uneventful pregnancies and deliveries. Their early development was characterized by global delay and hypotonia, with the onset of walking at the age of 2 years. Scores on the Mullen Scales of Early Learning and the Vineland Adaptive Behavior Scales showed profiles that were consistent with moderate-to-severe mental retardation in all patients. Nonverbal social interactions were within the normal range. In particular, evaluation of Patient 3 with the Autism Diagnostic Observation Schedule was negative. Ophthalmologic assessment revealed a strabismus in Patient 1. Two of the patients were mildly epileptic. Patient 1 had brief, generalized tonic–clonic seizures and was seizure-free while taking topiramate, whereas Patient 2 had some myoclonic and absence seizures, which were well controlled with valproate. In both patients, electroencephalography revealed bioccipital spikes during intermittent light stimulation.
Table 2.
Clinical Features of Three Patients with SYNGAP1 De Novo Mutations.*
| Variable | Patient 1 | Patient 2 | Patient 3 |
|---|---|---|---|
| Mutation | K138X | R579X | L813RfsX22 |
| Age | 4 yr 5 mo | 5 yr 10 mo | 12 yr 2 mo |
| Sex | Female | Female | Female |
| Ethnic background | South American | French Canadian | French Canadian |
| Weight — kg (percentile rank) | 21.9 (95th) | 18.0 (50th) | 39.1 (25th to 50th) |
| Height — cm (percentile rank) | 104 (50th) | 108.7 (50th) | 141.5 (10th) |
| Head circumference — cm (percentile rank) | 48.3 (3rd to 10th) | 52 (75th) | 52 (25th) |
| Diagnosis of epilepsy | Yes | Yes | No |
| Mullen Scales of Early Learning — percentile rank (age equivalent) |
|||
| Fine motor skills | <1st (17 mo) | <1st (27 mo) | <1st (31 mo) |
| Visual reception | <1st (25 mo) | <1st (27 mo) | <1st (34 mo) |
| Receptive language | <1st (14 mo) | <1st (28 mo) | <1st (36 mo) |
| Expressive language | <1st (10 mo) | <1st (26 mo) | <1st (23 mo) |
| Vineland Adaptive Behavior Scales — percen- tile rank |
|||
| Communication | <1st | 1st | <1st |
| Daily living skills | <1st | 6th | <1st |
| Socializing | <1st | 2nd | <1st |
| Motor skills | <1st | 1st | <1st |
| Composite of skills | <1st | 1st | 1st |
| Results of brain imaging | |||
| Magnetic resonance imaging | Normal | Normal | ND |
| Computed tomography | ND | ND | Normal |
ND denotes not determined.
The K138X mutation is predicted to result in a truncated SYNGAP1 protein that lacks the RASGAP domain (which activates ras GTPases) in addition to other functional domains (Fig. 1).11,17 The R579X and c.2438delT mutations are predicted to truncate SYNGAP1 in the middle and just after the RASGAP domain, respectively. These three mutations occur upstream of the 3′ terminus of the gene (encoding the C-terminal part of the protein) that can be alternatively spliced. This splicing process produces one of at least three possible isoforms, which differentially bind to other components of the NMDA-receptor complex, such as PSD95 and DLG3 (through the PDZ-binding motif, QTRV, in isoform 2) or CamKII (through GAAPGPPRHG in isoform 3).11,18 The importance of the QTRV motif is highlighted by the observation that its deletion impairs the ability of SYNGAP1 to regulate dendritic-spine formation.19
Mental retardation and autism spectrum disorders tend to be concurrent. Because of the phenotypic overlap between these conditions, we sequenced SYNGAP1 in a series of 142 patients with sporadic autism spectrum disorders. Because SYNGAP1 interacts with NMDA receptor, which is thought to be involved in schizophrenia,20 we also sequenced SYNGAP1 in a series of 143 patients with schizophrenia. We observed no de novo, splicing, or truncating mutations in these series of subjects (Table 1). The number of de novo mutations in the SYNGAP1 coding sequence in the patients with nonsyndromic mental retardation was significantly higher than that in the rest of the tested series consisting of patients with autism spectrum disorders or schizophrenia and control subjects (P = 0.004 by Fisher’s exact test). In addition to the common I1115T variant, three unique heterozygous missense variants were found in subjects with autism spectrum disorders (P1238L) and schizophrenia (T1310M and T790N) (Table 1). These variants are not present in any SYNGAP1 functional domains, are unlikely to be pathogenic because they were transmitted from an unaffected parent, and are predicted to have no effect on protein function (according to several programs analyzing amino acid substitutions) (Table 2 in the Supplementary Appendix).
STATISTICAL ANALYSIS
We used Fisher’s exact test to compare the rates of de novo mutations in the coding sequence of SYNGAP1 in samples from 94 patients with nonsyndromic mental retardation (188 chromosomes) with those of samples from 475 subjects in the other three series in the study (950 chromosomes). This comparison was performed with the use of the 2BY2 program (version 1.50 by Jurg Ott).
DISCUSSION
We identified protein-truncating de novo mutations in the autosomal gene SYNGAP1 in approximately 3% of our series of patients with nonsyndromic mental retardation. These mutations are probably pathogenic for several reasons. First, they result in the production of proteins that lack domains, such RASGAP and QTRV, that have been shown to be important for the synaptic plasticity and spine morphogenesis that are required for learning and memory.16,19 In addition, each of the resulting premature stop codons could also destabilize the SYNGAP1 messenger RNA (mRNA) transcript through the nonsense-mediated mRNA decay mechanism.21 Second, mice that are heterozygous for null alleles of Syngap1 have impaired synaptic plasticity and learning, which suggests that the disruption of a single SYNGAP1 allele is sufficient to cause cognitive dysfunction in humans.15,16 Third, extensive screening of 475 subjects without nonsyndromic mental retardation, including a subgroup with autism spectrum disorders and another with schizophrenia, did not identify any truncating, splicing, or de novo amino acid–altering variants in SYNGAP1. This finding reinforces the idea that the disruption of this gene is specifically associated with nonsyndromic mental retardation.
The disruption of SYNGAP1 appears to be associated with a homogeneous clinical phenotype that is characterized by moderate-to-severe mental retardation accompanied by severe language impairment. The absence of specific dysmorphic features and growth abnormalities in these patients is consistent with the fact that SYNGAP1 is exclusively expressed in the brain. The behavioral profile of the three patients and the absence of SYNGAP1 deleterious mutations in the series of patients with autism spectrum disorders indicate that SYNGAP1 disruption is unlikely to be associated with autism spectrum disorders. Two of the patients in our study had been treated for generalized forms of mild epilepsy. The disruption of SYNGAP1 could predispose them to seizures by increasing the recruitment of AMPA receptor at postsynaptic glutamatergic synapses, resulting in increased excitatory synaptic transmission, as has been described in mice with a Syngap1 mutation.12,14 The fact that epilepsy was well controlled in both patients by topiramate or valproate is consistent with this hypothesis. Indeed, topiramate directly inhibits AMPA-receptor activity, whereas valproate reduces the level of GluR1 AMPA-receptor subunit at hippocampal synapses and therefore indirectly reduces AMPA-receptor activity.22,23
The identification of genes associated with nonsyndromic mental retardation that encode proteins in well-characterized synaptic pathways offers the possibility of developing pharmacologic treatments to target associated complications, such as epilepsy, in addition to improving cognitive processes. Moreover, current therapeutic approaches that are aimed at allowing the translation and production of a normal protein in a fraction of mRNAs bearing nonsense mutations24 would be relevant for at least two of the patients in our study.
Supplementary Material
Acknowledgments
Supported by grants from the Canadian Institute of Health Research (CIHR) (to Drs. Lacaille, Rouleau, and Michaud), from Fonds de la Recherche en Santé (to Dr. Michaud), and from Genome Canada and Genome Quebec and cofunding by Université de Montréal for the Synapse to Diseases project (to Drs. Drapeau and Rouleau). Dr. Michaud is the recipient of a Clinical Investigatorship Award of the CIHR (Institute of Genetics). The schizophrenia series was collected partly through the Collaborative Network for Family Study in Psychiatry, which is supported by the Fondation Pierre Deniker.
We thank the patients and their parents for participating in this study; the members of the Synapse to Disease Group, including Edouard Henrion and Ousmane Diallo of the bioinformatics division and Sandra Laurent, Frederic Kuku, Joannie Duguay, Laurie Destroismaisons, Karine Lachapelle, Philippe Jolivet, Pascale Thibodeau, and Annie Raymond of the genetic screening division; Annie Levert and Judith St.-Onge for performing DNA extraction and paternity testing, respectively; and Pierre Lepage, Sébastien Brunet, and Hao Fan Yam of McGill University and Génome Québec Innovation Centre sequencing group and Louis Létourneau and Louis Dumond Joseph of the bioinformatics group.
Footnotes
No potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Chelly J, Khelfaoui M, Francis F, Chérif B, Bienvenu T. Genetics and pathophysiology of mental retardation. Eur J Hum Genet. 2006;14:701–713. doi: 10.1038/sj.ejhg.5201595. [DOI] [PubMed] [Google Scholar]
- 2.Ropers HH, Hamel BC. X-linked mental retardation. Nat Rev Genet. 2005;6:46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- 3.Basel-Vanagaite L. Genetics of autosomal recessive non-syndromic mental retardation: recent advances. Clin Genet. 2007;72:167–174. doi: 10.1111/j.1399-0004.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- 4.Garshasbi M, Hadavi V, Habibi H, et al. A defect in the TUSC3 gene is associated with autosomal recessive mental retardation. Am J Hum Genet. 2008;82:1158–1164. doi: 10.1016/j.ajhg.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molinari F, Foulquier F, Tarpey PS, et al. Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation. Am J Hum Genet. 2008;82:1150–1157. doi: 10.1016/j.ajhg.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motazacker MM, Rost BR, Hucho T, et al. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am J Hum Genet. 2007;81:792–798. doi: 10.1086/521275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat Rev Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 8.Purpura DP. Dendritic spine “dysgenesis” and mental retardation. Science. 1974;186:1126–1128. doi: 10.1126/science.186.4169.1126. [DOI] [PubMed] [Google Scholar]
- 9.Laumonnier F, Cuthbert PC, Grant SG. The role of neuronal complexes in human X-linked brain diseases. Am J Hum Genet. 2007;80:205–220. doi: 10.1086/511441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol. 2007;23:613–643. doi: 10.1146/annurev.cellbio.23.090506.123516. [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, Liao D, Lau LF, Huganir RL. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 12.Kim MJ, Dunah AW, Wang YT, Sheng M. Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron. 2005;46:745–760. doi: 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 13.Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham DE. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 2004;43:563–574. doi: 10.1016/j.neuron.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Rumbaugh G, Adams JP, Kim JH, Huganir RL. SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons. Proc Natl Acad Sci U S A. 2006;103:4344–4351. doi: 10.1073/pnas.0600084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JH, Lee HK, Takamiya K, Huganir RL. The role of synaptic GTPase-activating protein in neuronal development and synaptic plasticity. J Neurosci. 2003;23:1119–1124. doi: 10.1523/JNEUROSCI.23-04-01119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komiyama NH, Watabe AM, Carlisle HJ, et al. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci. 2002;22:9721–9732. doi: 10.1523/JNEUROSCI.22-22-09721.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pena V, Hothorn M, Eberth A, et al. The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. EMBO Rep. 2008;9:350–355. doi: 10.1038/embor.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Okano A, Tian QB, et al. Characterization of a novel synGAP isoform, synGAP-beta. J Biol Chem. 2001;276:21417–21424. doi: 10.1074/jbc.M010744200. [DOI] [PubMed] [Google Scholar]
- 19.Vazquez LE, Chen HJ, Sokolova I, Knuesel I, Kennedy MB. SynGAP regulates spine formation. J Neurosci. 2004;24:8862–8872. doi: 10.1523/JNEUROSCI.3213-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristiansen LV, Huerta I, Beneyto M, Meador-Woodruff JH. NMDA receptors and schizophrenia. Curr Opin Pharmacol. 2007;7:48–55. doi: 10.1016/j.coph.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Khajavi M, Inoue K, Lupski JR. Non-sense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 22.Angehagen M, Rönnbäck L, Hansson E, Ben-Menachem E. Topiramate reduces AMPA-induced Ca(2+) transients and inhibits GluR1 subunit phosphorylation in astrocytes from primary cultures. J Neurochem. 2005;94:1124–1130. doi: 10.1111/j.1471-4159.2005.03259.x. [DOI] [PubMed] [Google Scholar]
- 23.Du J, Gray NA, Falke CA, et al. Modulation of synaptic plasticity by antimanic agents: the role of AMPA glutamate receptor subunit 1 synaptic expression. J Neurosci. 2004;24:6578–6589. doi: 10.1523/JNEUROSCI.1258-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.