Summary
Export of protein into the periplasm of Escherichia coli via the general secretory system requires that the transported polypeptides be devoid of stably folded tertiary structure. Capture of the precursor polypeptides before they fold is achieved by the promiscuous binding to the chaperone SecB. SecB delivers its ligand to export sites through its specific binding to SecA, a peripheral component of the membrane translocon. At the translocon the ligand is passed from SecB to SecA and subsequently through the SecYEG channel. We have previously used site-directed spin labeling and electron paramagnetic resonance spectroscopy to establish a docking model between SecB and SecA. Here we report use of the same strategy to map the pathway of a physiologic ligand, the unfolded form of precursor galactose-binding protein, on SecB. Our set of SecB variants each containing a single cysteine, which was used in the previous study, has been expanded to forty-eight residues which cover 49% of the surface of SecB. The residues on SecB involved in contacts were identified as those that, upon addition of the unfolded polypeptide ligand, showed changes in spectral line shape consistent with restricted motion of the nitroxide. We conclude that the bound precursor makes contact with a large portion of the surface of the small chaperone. The sites on SecB that interact with the ligand are compared with the previously identified sites that interact with SecA and a model for transfer of the ligand is discussed.
Keywords: precursor polypeptide, SecB, export, site-directed spin labeling, chaperone
Introduction
Export of polypeptides into the periplasmic space through the cytoplasmic membrane in the Gram negative bacterium Escherichia coli is mediated by two systems. One, the Tat system, handles proteins that fold into stable structures in the cytosol before they are translocated.1 The other, the general secretory, or Sec, system cannot handle folded proteins at all but must transfer polypeptides before they acquire their final folded structure.2 In addition to a pathway through the membrane, provided by the translocon comprising a SecYEG core and the accessory proteins SecDF, YajC, the general secretory system includes soluble chaperones that allow early capture of precursor polypeptides. SecB is such a chaperone. It binds to its ligands with high affinity selecting them by virtue of their nonnative state. The complex undergoes a rapid equilibrium between the bound and free states. The rate of folding of the polypeptide relative to the rate of binding SecB determines a kinetic partitioning between the irreversible nonproductive pathway of folding in the cytosol and the pathway of export.3 SecB and its bound ligand forms a complex with SecA which itself has affinity for SecY. Thus, the ternary complex of SecA:SecB:precursor engages the translocon. SecA, itself an ATPase, undergoes a cycle of ATP binding and hydrolysis to achieve transfer of the ligand from SecB to SecA and subsequently through the membrane. Elucidation of the mechanism of transfer requires a detailed description of the contacts between the members of the ternary complex.
We have previously determined the orientation of the docking of SecA onto SecB4 and now we investigate the binding of ligand. When physiologic ligands, precursors of polypeptides of 30 – 50 kDa in a non-native state, that are destined for export to the periplasmic space bind to SecB, the portion of the polypeptide that makes contact with the chaperone spans a minimal stretch of 150 amino acids.5–7 SecB is a small, homotetrameric chaperone of 69 kDa organized as a dimer of dimers,8,9 and thus to accommodate such a length of polypeptide the ligand must wrap around the surface. Examination of the X-ray crystal structure of SecB reveals a 70 Å long groove at the interface of the dimers on each side of the tetramer.8 It was proposed that the ligand binds in this groove. Our study described here indicates that in addition to residues in the putative binding groove there are many other regions of contact between SecB and its ligands. We have defined the binding interface by using site-directed spin labeling to examine variants of SecB each containing a nitroxide spin label at a unique position. Comparison of the spectral line shapes for SecB free in solution and in complex with precursor galactose-binding protein allowed identification of sites of contact between the ligand and the chaperone.
Results
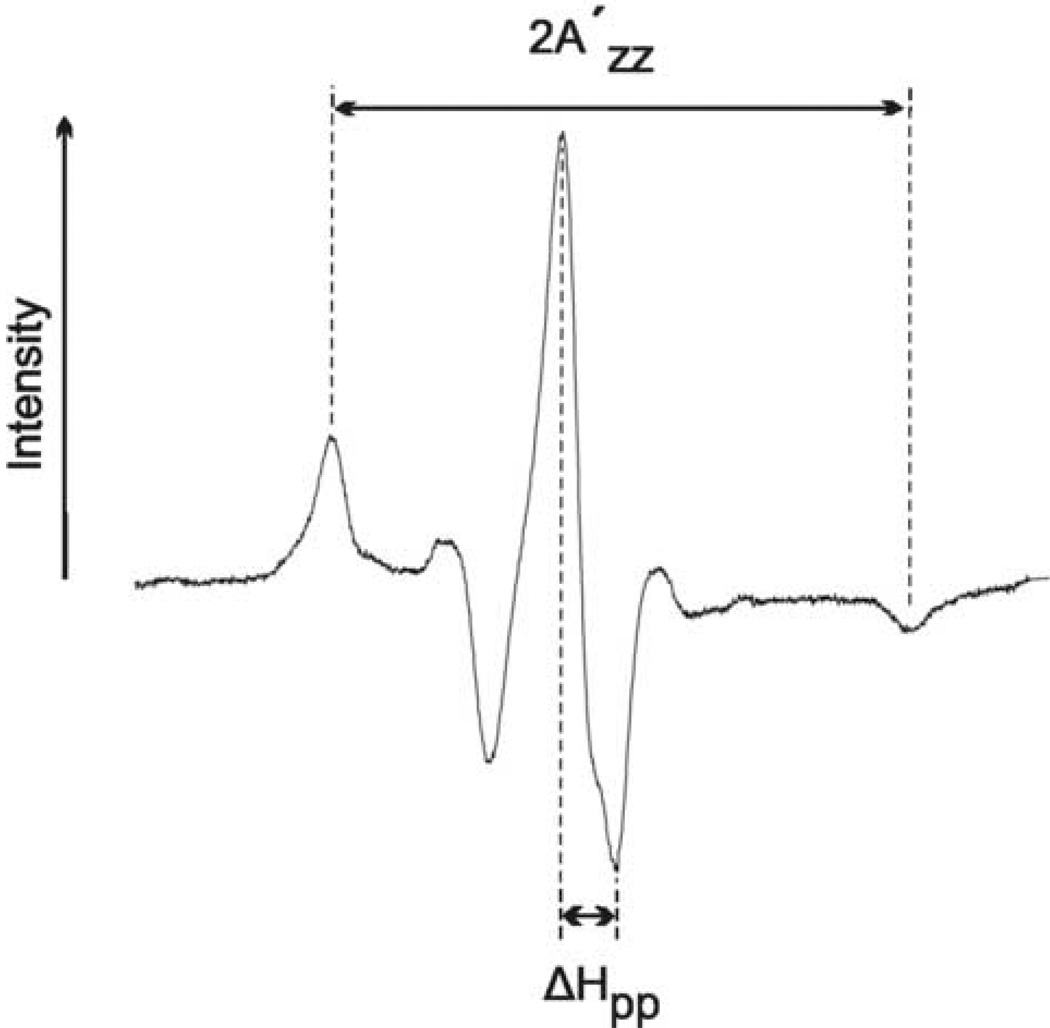
We have mapped the surface of contact between an unfolded polypeptide ligand that wraps around the chaperone by exploiting the fact that the line shape of an electron paramagnetic resonance (EPR) spectrum of spin-labeled SecB contains information about the mobility of the nitroxide side chain on the nanosecond timescale (for review see Schneider and Freed10 and Fajer11). If in a complex of ligand and chaperone the ligand either makes a direct contact with the nitroxide or is held in close vicinity one would expect the motion of the nitroxide to be constrained. As a nitroxide goes from highly mobile to constrained or immobile two features of the spectrum change. First, the central line width broadens (Figure 1, ΔHpp) which is also seen as a decrease in the intensity of the central line since all spectra are normalized. Second, the overall spectral breadth increases, that is, the total intensity is spread over a wider range of the magnetic field. For spectra reflecting slow motional states the hyperfine extrema are well resolved and the separation of the extrema can be used as a measure of the spectral breadth (Figure 1, 2A′zz). Here we use these simple indicators of mobility, which are apparent by visual inspection, to determine which areas on the surface of SecB interact with the ligand. It is a formal possibility that constraints on the nitroxide result not from a direct contact but from a conformational change. Nevertheless, determination of the sites that show constrained mobility should allow us to determine the surface of SecB that interacts with the precursor polypeptide ligand.
Figure 1.
Measures of mobility. The peak to peak width of the central resonance line (ΔHpp) is measured as indicated and is equivalent to the peak width at half-height of an absorbance spectrum. The spectral breadth (2A'zz) is the distance between the outermost hyperfine extrema. The spectrum used to illustrate these parameters is that of L67R1.
Forty-one of the forty-eight residues studied here were previously examined in our study of docking of SecA onto spin-labelled SecB.4 The reader is referred to that publication for a detailed description of whether the line shapes of those spectra reflect isotropic motion and whether resolved spectral components are due to multiple dynamic states of the nitroxide in slow exchange or are due to anisotropic motion of a nitroxide in one dynamic state.
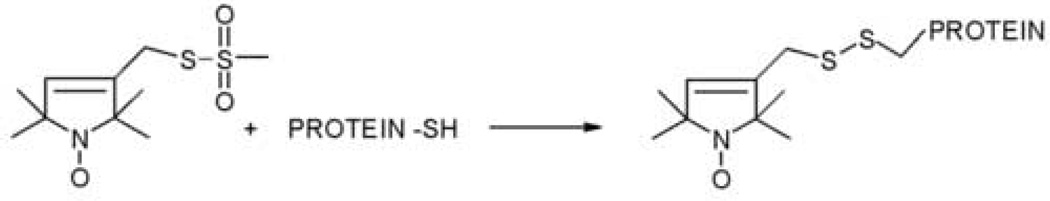
The crystal structure of SecB from E. coli9 was used as a guide in selection of side chains for labeling that are at solvent-exposed sites to avoid disruption of the protein structure and to maximize the probability of observing contacts. Cysteine was substituted for the native amino acid at sixty-five positions. Fifteen of the variant proteins aggregated during purification and were not pursued further. The remaining fifty variants were reacted with a sulfhydryl specific methanethiosulfonate reagent to generate a nitroxide side chain as shown in Figure 2. The spin-labeled proteins were shown to be properly folded and tested for activity in binding unfolded precursor galactose-binding protein, a natural ligand of SecB, by size-exclusion chromatography, (data not shown, see Activity Assays in Materials and Methods for details).
Figure 2.
The methanethiosulfonate spin label and the side chain (R1) it generates.
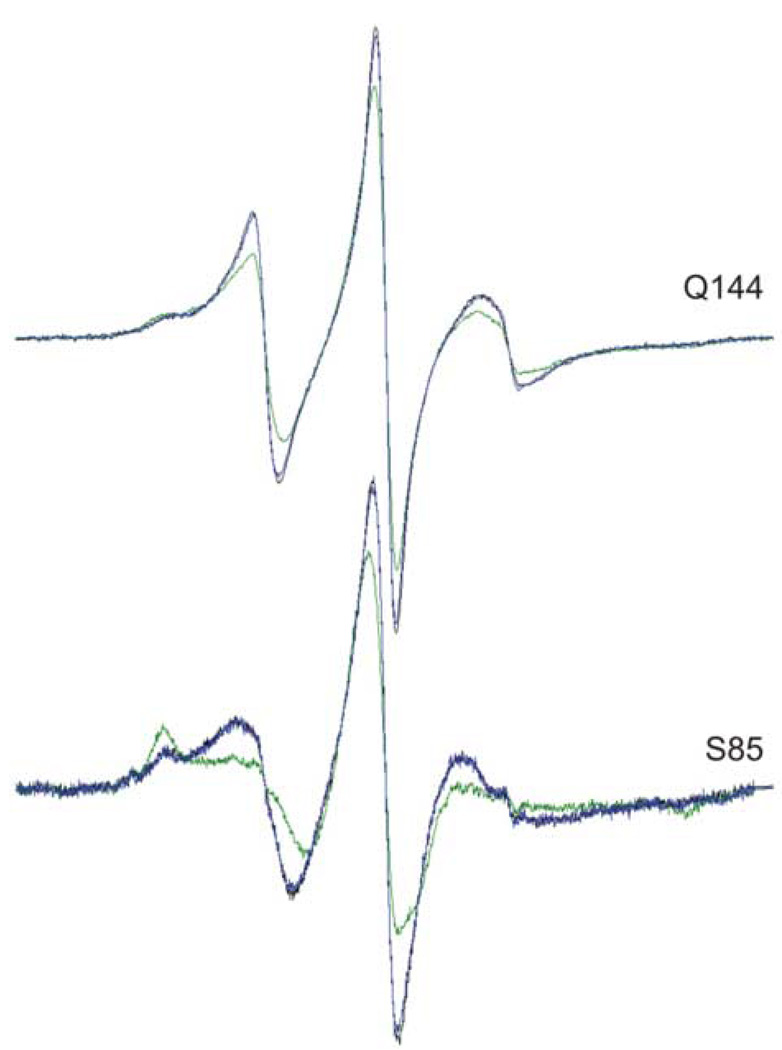
SecB forms a complex with its natural ligands only if the polypeptides are presented in a non-native state. The binding is of high affinity (Kd for unfolded precursor galactose-binding protein, the ligand used here is approximately 35 nM12 as determined by titration calorimetry in GnHCl at both 0.1 N and 0.2 N), but the system is in a rapid dynamic equilibrium. Binding to SecB is readily reversible and each time the ligand is released it undergoes a kinetic partitioning between folding to its native state and rebinding to SecB.13,14 Precursor galactose-binding protein is an ideal ligand for these studies because the rate of folding can be modulated by temperature, as is true for the folding of all proteins, and also by calcium. The native structure of galactose-binding protein is stabilized by a calcium ion and the rate of folding of the polypeptide is drastically decreased in the absence of calcium.14 In the experimental design used here the precursor, unfolded in 1 N GnHCl, was added to the SecB by rapid dilution into a solution held on ice so that the final concentration of the GnHCl was 0.17 N and calcium was chelated by 1 mM EGTA. The spectra were collected at 6°C. Under these conditions the rate of folding of the ligand is sufficiently slow so that the partitioning is poised to favor complex formation. Addition of CaCl2 to 3 mM to the mixture results in a shift of the partitioning to favor folding. Thus, the changes in spectral line shape observed upon addition of precursor galactose-binding protein to spin-labeled SecB can be shown to be specific for binding of an unfolded ligand by adding CaCl2 to reverse the change. Figure 3 shows two examples of such a reversal of the line shape (compare black traces, free SecB; green traces, SecB and precursor galactose-binding protein; blue traces, SecB and precursor galactose-binding protein with CaCl2, note that the black and blue traces overlay).
Figure 3.
Reversal of complex formation between precursor galactose-binding protein and SecB. Black traces are free SecB, green traces are SecB and galactose-binding protein, blue traces are SecB and galactose-binding protein after addition of 3 mM CaCl2 and incubation at room temperature for 30 minutes before acquiring the spectrum at 6°C.
The spectra of the spin-labeled variants of SecB free in solution and those of SecB in complex with precursor were compared to identify areas at which motion of the nitroxide side chain was constrained indicating contact with the ligand. We have grouped the sites examined into three classes: those with a high degree of visible change in the spectra, those with significant changes that are less readily seen and those showing very small change which we consider insignificant. It should be noted that the groupings are qualitative and although constraint of motion of a nitroxide does indicate that the residue lies on or close to the pathway taken by the ligand, we cannot assign contributions to the energy of stabilization of the complex to any particular contacts. The goal of this study and of our previous study of spin-labeled SecB is to map the binding interface between SecB and each of its binding partners, the unfolded ligand and SecA, so that we may gain insight into the role of the ternary complex in export. To facilitate comparison of the surface of contact with the precursor to that with SecA we have included some data from our previous work. The combined results from both studies indicate that the binding sites for precursor and SecA show considerable overlap.
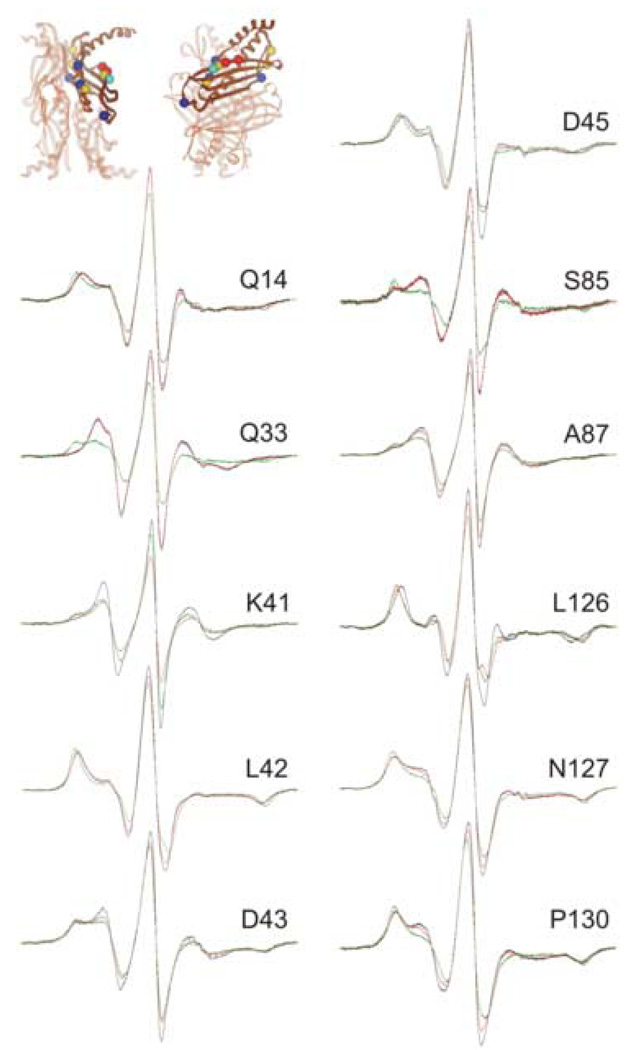
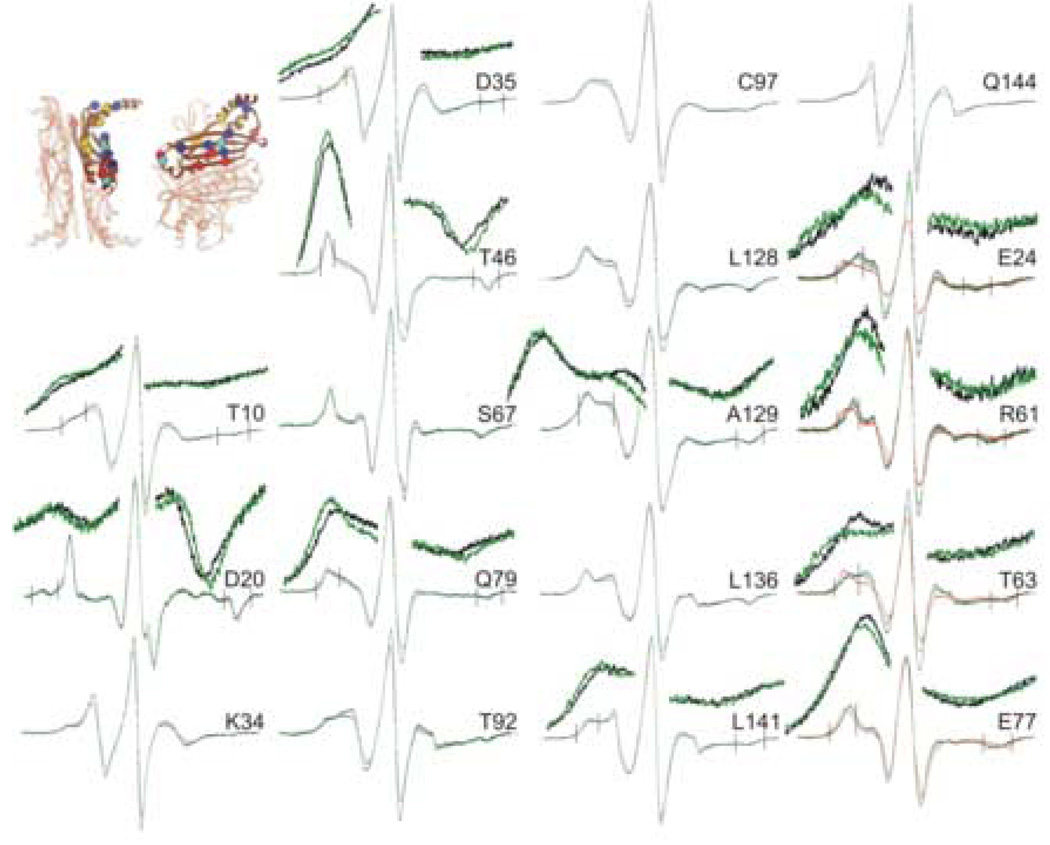
The group of residues that show the highest degree of constraint when precursor makes a complex with SecB are shown in Figure 4 (compare the spectral lines shown in black, free SecB with those shown in green, SecB in complex with precursor). The constraints on the motion of the nitroxides when the unfolded ligand binds can be readily seen as an increase in spectral breadth that is obvious both from the outward movement of the hyperfine extrema and the decreases in amplitude of the center lines. The nitroxides at four of these positions, K41R1, D43R1, D45R1 and L126R1 show the same degree of constraint when SecB is in complex with either the ligand or with SecA. The other seven positions show either very little contact (A87R1 and P130R1) or no contact (Q14R1, Q33R1, L42R1, S85R1 and N127R1) when SecA binds (Figure 4, compare the black traces, free SecB with the red traces, SecB in complex with SecA).
Figure 4.
Positions on SecB that show a high degree of constraint with precursor galactose-binding protein bound. The black traces are spectra of spin-labeled SecB alone, the red are spectra of an equimolar mixture of SecA and spin-labeled SecB and the green are an equimolar mixture of spin-labeled SecB and unfolded precursor galactose-binding protein. The positions of the residues examined are shown on the structure of SecB as spheres at the site of the α-carbon atom. The color indicates the nature of the original residue: gold, hydrophobic; blue, polar; bright blue, negative charge; red, positive charge. The structure in this and all subsequent structures is that generated by threading the E. coli sequence through the H. influenzae structure because the C-terminal residues are resolved. The PDB code for SecB is 1FX3.
Figure 5 shows the spectra of an additional eighteen positions that showed line shape changes upon binding precursor galactose-binding protein. Although the spectral line changes of the nitroxides are not as obvious to the eye as those shown in Figure 4 they represent significant decreases in mobility. In order to see the movement outward of the hyperfine extrema for several positions that region is displayed at a four-fold magnification. Four of these positions (T46R1, C97R1, L128R1 and A129R1) do not show any change in spectral line shape when SecA binds whereas the remaining fourteen show constraints when either SecA or precursor binds. For those positions where the degree of spectral change is the same with the two binding partners we show only the spectra of free SecB and those complexed with precursor galactose-binding protein. For the four positions E24R1, R61R1, T63R1 and E77R1 where a larger change occurs upon SecA binding relative to the unfolded ligand we show all three spectra, SecB free (black traces), SecB with precursor (green traces) and SecB with SecA (red traces).
Figure 5.
Positions on SecB that show significant constraint. The black traces are spectra of spin-labeled SecB alone, the red are spectra of an equimolar mixture of SecA and spin-labeled SecB and the green are an equimolar mixture of spin-labeled SecB and unfolded precursor galactose-binding protein. The positions of the residues examined are shown on the structure of SecB as spheres at the site of the α-carbon atom. The key to the colors is described in Figure 4. The insets show the region of the extrema, indicated by the vertical lines, magnified by a factor of four for both the intensity and the field.
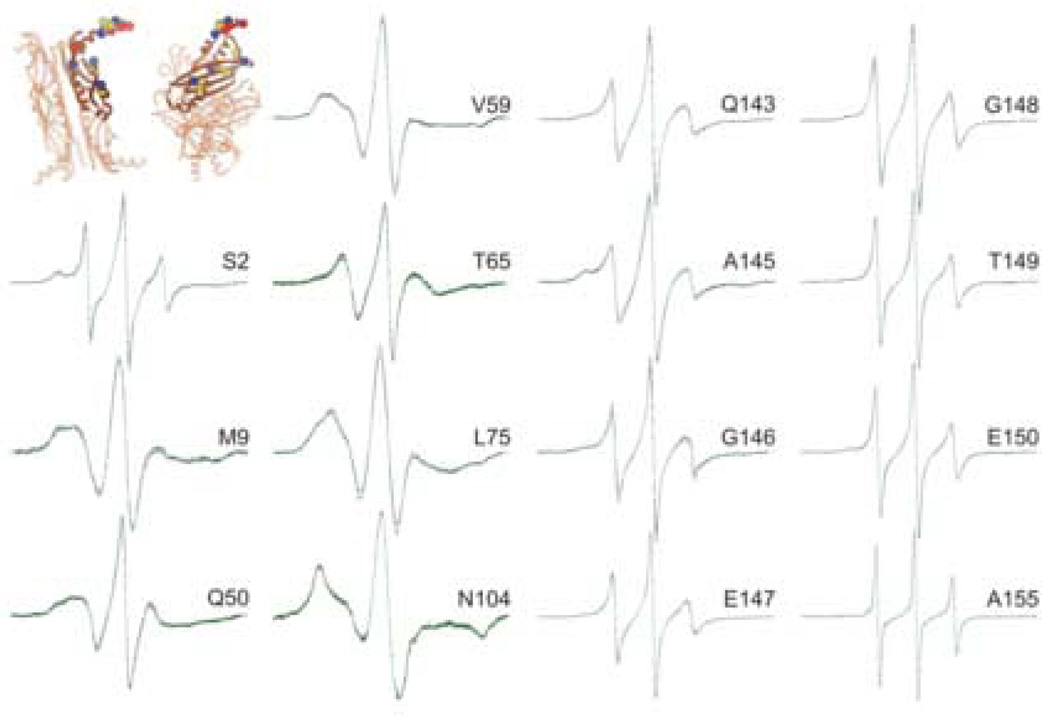
The residues tested that showed either no change or very small changes in line shape when precursor galactose-binding protein was added to SecB are grouped in Figure 6. Three of these sites S2R1, M9R1 and Q50R1, also showed no change when SecA was bound. Each of the spin-labeled variants of SecB shown in Figure 6 as well as variants displayed in Figures 4 and 5 were demonstrated to form a complex with precursor galactose-binding protein and with SecA by size-exclusion chromatography (data not shown). It is important to note that the narrow line widths of residues G146 through the C-terminal A155 are not consistent with well-ordered α-helical structure.15,16 The helices in the crystal structure are likely to be the result of crystal contacts whereas in solution the region is very flexible on the nanosecond timescale. A solution study of SecB by NMR also indicates a high degree of mobility for the C-terminal 13 aminoacyl residues.17
Figure 6.
Positions on SecB that show no significant change when precursor galactose-binding protein binds. The black traces are spectra of spin-labeled SecB alone, the red are spectra of an equimolar mixture of SecA and spin-labeled SecB and the green are an equimolar mixture of spin-labeled SecB and unfolded precursor galactose-binding protein. The positions of the residues examined are shown on the structure of SecB as spheres at the site of the α-carbon atom. The key to the colors is described in Figure 4.
At four positions L44R1, E90R1, Q125R1 and F137R1 substitution of a cysteine followed by derivatization with the nitroxide eliminated the ability of SecB to bind the precursor galactose-binding protein. The modifications did not have a global effect on the structure of SecB since each of the four species eluted as a well-folded tetramer that formed a complex with SecA. Even though we can draw no conclusions from the EPR spectra of these SecB variants we can conclude that the four residues are important in binding precursor and, in fact, are likely to be residues that provide binding energy to stabilize the complex.
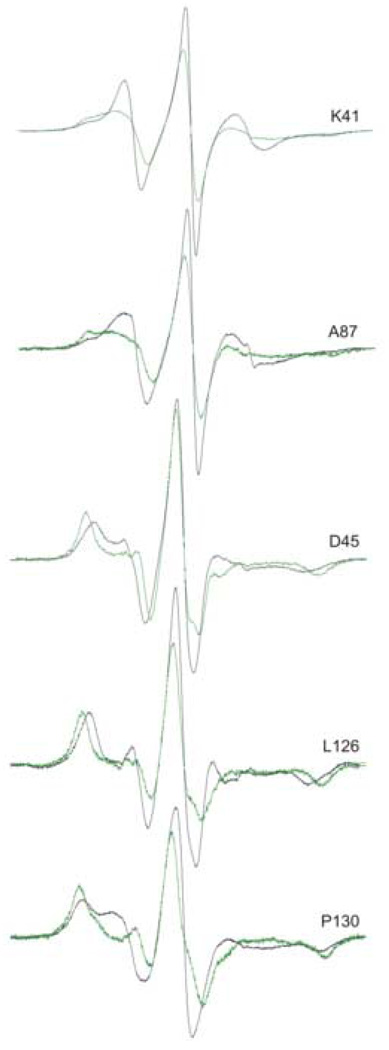
Each of the spin-labeled variants of SecB was stoichiometrically labeled, as was verified by MALDI mass spectrometry, and thus would carry four nitroxide side chains. Unfolded polypeptides bind to SecB with a stoichiometry of one polypeptide per tetramer.18 The portion of the ligand in contact with SecB has been estimated to be approximately 150 aminoacyl residues.5–7 It seems unlikely that this stretch of extended polypeptide could simultaneously make contact with all four symmetrically related residues. If all four side chains are not constrained then the observed spectrum would be a composite of the corresponding spectrum of the nitroxide on free SecB and the spectrum of the same position constrained by the bound ligand. We can eliminate the contribution to the spectrum of the free residue to reveal the spectrum of the constrained site by examining a difference between the normalized composite spectrum of the complex and the normalized spectrum of free SecB reduced incrementally until an obvious endpoint is reached. The endpoint of subtraction is recognized because oversubtraction results in a difference spectrum that contains features that are not reasonable for a nitroxide side chain on a protein. All of the spectra shown in Figure 4 have changes that are large enough to allow titration that reveals the new states resulting from contact within the complex. Figure 7 shows five examples. For some sites such as K41R1 and A87R1 the constrained states (green traces) show a line shape with two spectral components whereas at the other sites (D45R1, L126R1 and P130R1) the constrained spectrum has one component. The similarity of the spectral line shape in the latter group reflects similar constraint of motion. Comparison of the titrated spectra with the corresponding spectra of SecB only (Figure 7, black traces) shows an increase in spectral breadth. These residues are highly constrained showing a splitting of outer hyperfine extrema between 66 and 70G, near the maximal values of 66G in hydrophobic solvent and 74G in aqueous solution in the absence of motion.19 Titration of most residues reached an endpoint after subtraction of approximately 40% of the uncomplexed spectrum indicating that 60% of the spin-labeled sites available were occupied.
Figure 7.
Titration of spectra to reveal the state of nitroxide in contact sites. The spectra shown were generated by taking the difference between the normalized spectrum of the complex and the normalized spectrum of SecB reduced as described in the text. The amount of the reduction of the SecB spectrum was 41% for K41R1, 39% for A87R1, 31% for D45R1, 43% for L126R1 and 52% for P130R1. The titrated spectra were normalized to have the same total spin as the spectra of the complexes.
Discussion
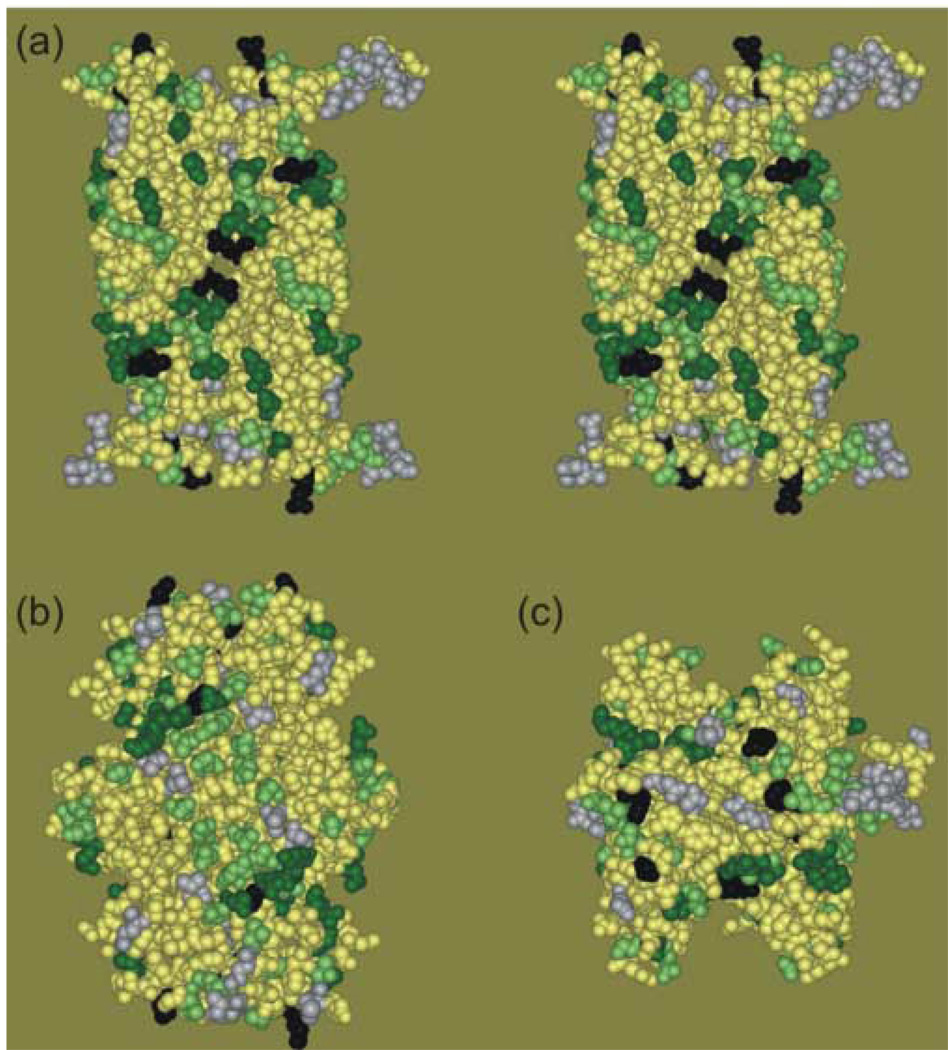
The sites of interaction between precursor galactose-binding protein and the chaperone as determined by EPR spectroscopy of spin-labeled residues are displayed on three views of SecB in Figure 8. Panel (a) of the figure is in stereo to illustrate the depth of the channel at the interface of the dimers. Panel (b) shows the contact sites on the flat 8-stranded β-sheet that forms the sides of the tetramer and panel (c) shows residues on the top. All sites that have been examined are shown as CPK models of the original side chains that were substituted by cysteine and modified by the spin label. If there was no change or very small change in line shape when the unfolded precursor formed a complex with SecB (data in Figure 6) the residue is displayed in gray. The positions that showed changes, indicative of contact, are shown in dark green for those with the highest degree of change in line shape (data in Figure 4) and blue-green for the remaining eighteen positions that showed significant changes in mobility (data in Figure 5). The four residues that were implicated as part of the binding site because conversion to the nitroxide side chain resulted in an inability to bind ligand are shown in black.
Figure 8.
Contact sites for precursor galactose-binding protein on SecB. The residues analyzed by site-directed spin labeling are shown on the structure as CPK models of the residue that was substituted by cysteine and derivatized with the spin label. The positions that showed a high degree of change in spectral line shape when precursor galactose-binding protein was added are shown in dark green, those that showed a significant change in blue green and those that showed no change in gray. The residues in black are assigned to the binding site because derivatization inactivated SecB for binding precursor. Panel (a) is a stereo view with the dimer interface facing forward. Panel (b) is related to panel (a) by a 90° rotation about the vertical axis to the right. This view shows the flat β-sheet on the side of the tetramer. Panel (c) is related to panel (a) by a 90° rotation about the horizontal axis toward the viewer. This view shows the top of the tetramer.
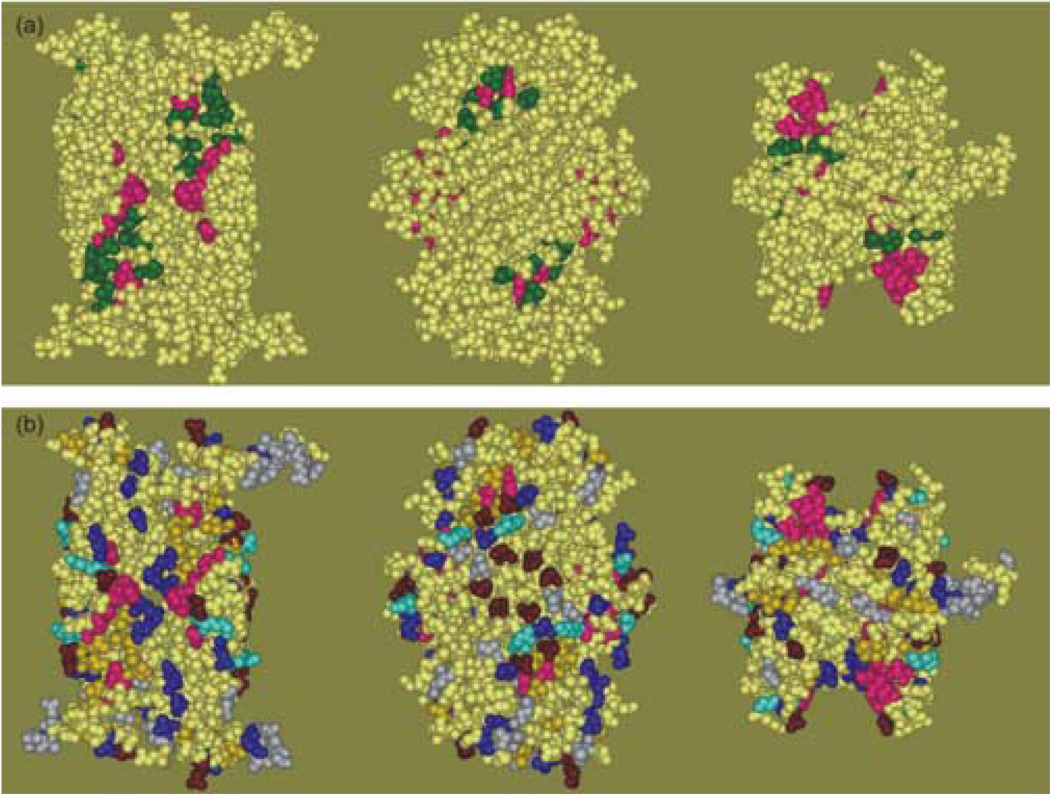
It is of interest to compare the sites of interaction identified in this study with the putative binding site proposed by Xu and his colleagues8 based on the examination of the crystal structure of SecB. They proposed that the unstructured polypeptide ligand binds into the channel at the interface of the dimers. It was assumed that binding of the unfolded proteins would be through hydrophobic interactions and therefore only hydrophobic residues were assigned to the putative binding site. The binding channel was described as comprising two subsites: subsite 1 is situated in the deep central region of the channel whereas subsite 2 is located at the shallow ends of the cleft. We were able to test seven residues in this putative site, all located in subsite 2 (Figure 9(a), green) and all showed interaction with the ligand. The residues that we were unable to assess because substitution by cysteine caused aggregation included all residues assigned to subsite 1 (F32, W36, P38, F123 and P124) as well as V40, V131 and F133, which are located in subsite 2. These residues are displayed in magenta on this structure and in all subsequent figures to facilitate comparison of the structures. The residues identified in this study as making contact with ligand are colored in Figure 9(b) to illustrate the nature of the side chain. It is clear from a comparison of Figure 9(a) with Figures 8 and 9(b) that although the channel does serve to bind the ligand, the contacts are not restricted to the channel nor are they all hydrophobic as proposed by Xu and his colleagues.8 In addition to hydrophobic residues (gold), the ligand makes contact with polar residues (blue), positively charged residues (bright blue) and negatively charged residues (red brown). If the ligand is to simultaneously occupy both channels on the tetramer, it must wrap around the chaperone to reach from one side to the other; thus, it is reasonable to expect contacts along the pathway that is taken across the sides or the top and bottom.
Figure 9.
Panel (a) Binding site previously proposed based on the crystal structure. The structure on the left is a view of the dimer interface. The view in the center was achieved by a 90° rotation about the vertical axis to show the flat -sheet on the side of the tetramer. The view on the right is related to that on the left by a 90° rotation about the horizontal axis toward the viewer. Panel (b) Contact sites identified in this study colored to indicate the nature of the residue. The color scheme is described in the text. The orientation of the structure is as described in panel (a).
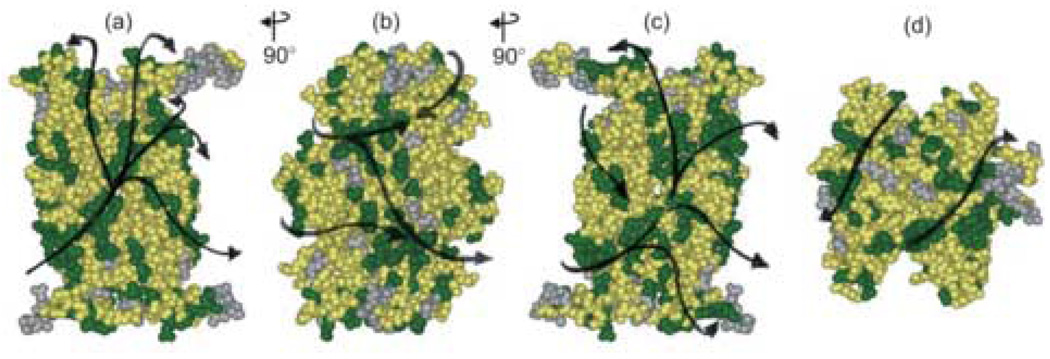
SecB binds promiscuously to many unfolded polypeptides and therefore it is unlikely that there is a unique pathway around the chaperone taken by all ligands. Figure 10 and a movie provided in the supplementary materials illustrate several of the numerous possible routes. All sites of contact are shown in green to simplify tracing the putative pathways. A ligand following one of the arrows in Figure 10 through the central channel shown in panel (a) would emerge on the side as shown in panel (b), which is related to panel (a) by a rotation of 90° about the vertical axis. Several routes are available across the flat β-sheet that forms the side and these paths emerge onto the face of SecB that is related to panel A by 180° and is shown in panel (c). The extended polypeptide might wrap around the chaperone using any of the branches of the pathway and in fact there are likely to be routes we have not indicated that traverse the surfaces that we have not tested (shown in yellow). The view of the top of SecB (panel (d)) shows two obvious paths that angle over the top of the C-terminal regions. Symmetrically related routes would exist across the bottom (not shown). The titrations of spectra indicated that the four symmetrically related residues that carry spin labels are not all constrained by bound ligand. This could be explained if the complexes were heterogeneous with respect to the routes taken.
Figure 10.
Possible pathways around SecB. All contact sites identified in this study are shown in green. The arrows indicate possible routes around the chaperone. (a), (b) and (c) are each related to the previous structure by a 90° rotation about the vertical axis to the left. (d) is related to (a) by a 90° rotation about the horizontal axis toward the viewer. (a) front view of the deep channel; the interface of the dimers that form the tetramer is aligned with the vertical axis. (b) flat 8-stranded β-sheet that is the side of the tetramer. (c) channel of the dimer interface at the face opposite that shown in (a). (d) end view of the tetramer.
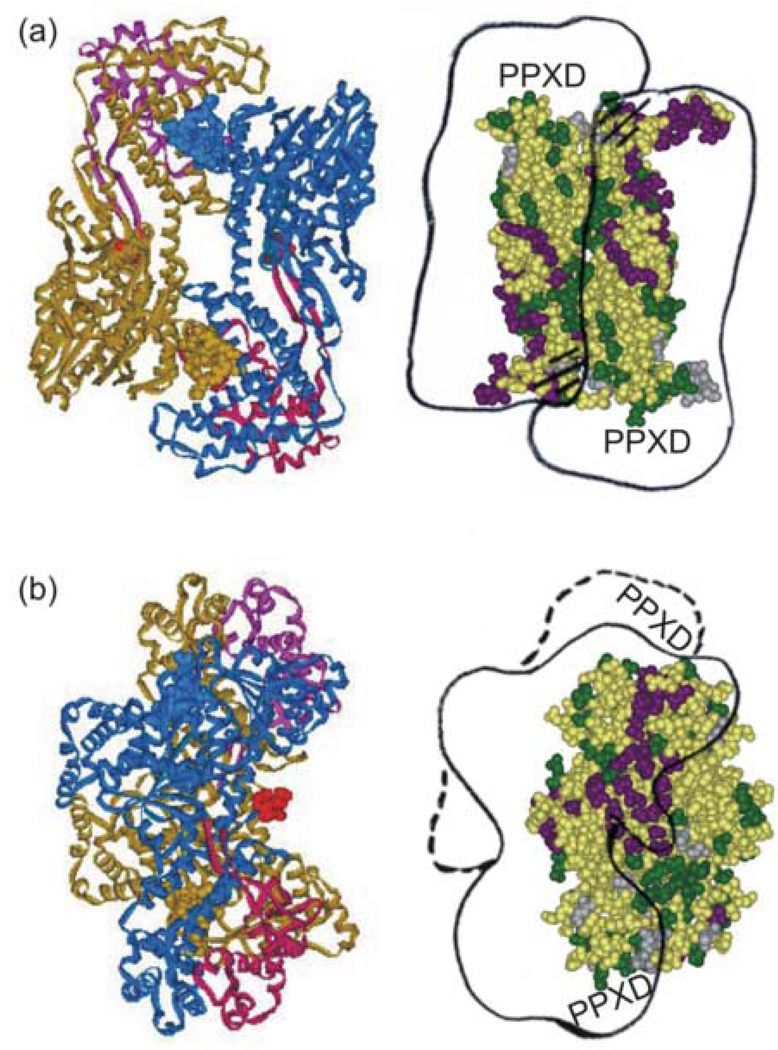
Flexibility in choice of pathway may be important for function since the complex of SecB and ligand must bind SecA to be directed to the translocon. SecA docks onto SecB across the channel and interacts with many residues that are also capable of interaction with the precursor (contacts with SecA4 are shown in purple in Figure 11). Presumably precursor would be excluded from these sites when SecA is bound. However, since only one dimer of SecA binds a tetramer of SecB20 the corresponding residues on the opposite face would be available for binding of precursor. It is of particular interest to examine those sites on SecB in the ternary complex that make contact with precursor but not with SecA (shown in green, Figure 11(a)) even though when docked onto SecB a large domain of SecA, the precursor binding domain (PPXD)21 would be positioned directly over them. This domain has been shown to bind precursors by several laboratories using a variety of techniques including cross-linking studies,22 mutational analysis23 and direct binding.24,25 In the dimeric form of SecA the PPXD is packed onto the core of the protein. The crystal structure of a monomeric form reported by Osborne et al.26 reveals a dramatic 60° rigid body movement of that domain which breaks the interface with the core of the protein and results in formation of a large groove. The authors proposed that this groove could provide a binding site for the unfolded polypeptide ligand. We have previously shown that when SecB binds SecA, the flexible C-terminal regions of SecB make contact with the interfacial region of SecA4 that includes the aminoacyl residues 1–1120 indicated on the line drawing by the hatched lines. In the complex the PPXD of one protomer of SecA is situated directly opposite the site of interaction between the C terminus of SecB and the other subunit of SecA. If the interfacial contacts were broken by the binding of SecB, the PPXD would be free to take on the open conformation observed by Rapoport and his colleagues26 and reveal the deep groove. Since the residues on SecB that would underlie this open domain of SecA (T10, Q14, Q33, S85 and A87) are involved in contacts with precursor, the opening of the interface and subsequent creation of the groove would provide a mechanism for the transfer of ligand from SecB to SecA.
Figure 11.
Simultaneous binding of precursor galactose-binding protein and SecA. Panel (a) left side, structure of the SecA dimer oriented so that the extreme C-termini, which are not resolved, and the PPXD (magenta and pink) are on the lower surface. Right side, an outline of the SecA dimer is docked across SecB as described in the text. The hatched lines indicate the binding sites on SecA that interact with the flexible C-terminal regions of SecB (shown in structure as helical). Panel (b) structures shown are related to those in panel (a) by a 90° rotation about the vertical axis to the left. The PDB code for SecA is 1M74.
Our application of site-directed spin labeling and EPR to the study of SecB has previously allowed us to map the docking interface between SecB and its specific binding partner SecA,4 and now we have examined the promiscuous interactions of SecB as a chaperone. The large area of the chaperone that is involved in contacts with ligand indicates that numerous routes are available on the surface. Interaction sites on SecB thus have the inherent flexibility to allow simultaneous binding of a ligand and SecA. The transfer of unfolded polypeptides from SecB to SecA may rely on this flexibility as well as on the unusual properties of the binding between SecA and SecB. Biochemical and biophysical studies have shown that the contacts between these two symmetric molecules are asymmetric20 and that the loss of either one of two contact areas out of three27 results in no decrease in affinity. Perhaps transfer of the ligand involves making and breaking contacts so that the polypeptide, trapped between SecA and SecB, can be transferred without complete dissociation of the complex.
To date we have used the capacity of EPR to detect constraint of motion of individual residues when complexes are formed and thereby map the interfaces of interaction. In future work, based on the information acquired from these studies, we plan to exploit the power of EPR to measure distance using spin coupling between two nitroxide side chains. Our hope is to elucidate the movement of regions of the proteins during the dynamic and complex transfer of the ligand.
Materials and Methods
Mutagenesis and Protein Purification
The strategy of site-directed spin labeling requires that only the cysteine at the site of interest be modified by the nitroxide reagent via sulfhydryl chemistry. Ideally all native cysteines in the protein of interest are replaced and a unique cysteine is introduced at the desired site. SecB contains four native cysteines (C76, C97, C102 and C113). We were unable to replace C76 or C113 without disrupting the structure of SecB. Fortunately only C97 showed reactivity in the native protein. Therefore to create our base protein for this study we substituted C97 with serine and C102 with leucine. This base protein forms a wild-type complex with precursor galactose-binding protein as assessed by size exclusion chromatography (see Size exclusion chromatography in Materials and Methods), but does not react with the spin label as shown by MALDI mass spectrometry. The plasmid pAL204 carrying the gene for the base protein under the T7 promoter was modified by standard recombinant DNA techniques (Quickchange, Strategene) to generate the cysteine mutants used here. Each mutant subjected to DNA sequencing to confirm the change. The plasmids were transformed into BL21(DE3) (Novagen) or BL21(DE3) secB::Tn10 srl::Tn10 recA (a secB null strain obtained from Carol Kumamoto). The proteins were purified from the appropriate strains as described for wild-type SecB.20 Precursor galactose-binding protein was purified as described.14 The protein concentrations were determined spectrophotometrically at 280 nm using extinction coefficients of 47,600 M−1 cm−1 for SecB tetramer and 37410 M−1 cm−1 for precursor galactose-binding protein.
Spin-labeling of SecB Variants
Immediately before labeling with the nitroxide reagent (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (Toronto Research Chemicals Inc.), the reducing agent, tris-(2-carboxyethyl) phosphine hydrochloride (TCEP, Molecular Probes) was removed by exchange from the usual storage buffer (10 mM HEPES-KOH, 300 mM KOAc, 2 mM TCEP pH 7.6) into 10 mM HEPES-KOH, 300 mM KOAc pH 6.7 using a Nap10 column (Amersham). The proteins were concentrated using a Centricon 30 (Millipore). Typically 0.2 mL of each SecB variant, at a concentration of approximately 1.3 mM monomer, was incubated with an equimolar amount of the spin labeling reagent. To generate C97R1 we used the native, wild-type protein. In the case of residues that were close enough to show spin-spin interaction (D20, E24, E77and Q79) the labeling was carried out with a mixture of the spin label and an analogue (the generous gift of Kálmán Hideg), which has no free electron, at a ratio of 1:1.5. D35 shows spin-spin interaction across the interface of the dimer of dimers. This interaction was reduced by mixing labeled SecBD35R1 with an equal amount of unlabeled SecB and incubating to allow the spontaneous exchange of dimers. The spin label reagent and the analogue were each prepared as a stock of 100 mM in acetonitrile and stored in the dark at −80°C. The reaction was allowed to proceed on ice in the dark for 2 – 3 hours. To remove excess spin label and to separate the spin-labeled species that were tetrameric from aggregated species the entire reaction mixture was applied to a BioSep-SEC-S 3000 column (600 × 7.8 mm, Phenomenex), equilibrated in 10 mM HEPES-KOH, 300 mM KOAc, 5 mM Mg(OAc)2 pH 6.7. The proteins that eluted in the position of tetramer were concentrated using a Nanosep filter concentrator, 10,000 MW cut off (Pall Life Sciences). The concentration of each spin-labeled protein was determined by absorbance at 280 nm. Each spin-labeled variant was analyzed by MALDI mass spectrometry and in all cases the labeling was near stoichiometric. The proteins were stored at −80°C. The spin-labeled variants are designated by the amino acid and residue number that has been substituted by cysteine and modified by the nitroxide side chain, R1. Thus, K41R1 indicates that lysine at position 41 was converted to cysteine and reacted with the spin label reagent.
Activity Assays
To ascertain that introduction of the nitroxide did not disrupt structure all spin labeled variants were subjected to analysis by HPLC using a size-exclusion column (TSK G3000SW, TosoHaas). At six positions (N104, R111, L128, A129, D134 and F137) derivatization of the cysteine by the spin label resulted in considerable aggregation (~50% in some cases). However, the protein that eluted as a tetramer could be isolated by size-exclusion chromatography and if used immediately did not repopulate the aggregate. All spin-labeled variants included in this EPR study were shown to be tetrameric and to form complexes with precursor galactose-binding protein as well as with SecA by size-exclusion chromatography as described (data not shown).20
EPR Measurements
EPR spectroscopy was performed on an X-band spectrometer, a Bruker Elexsys E500 or Bruker EMX, with a high sensitivity resonator. Protein samples of 5 µL (spin label was either 80 µM or 160 µM) were loaded into synthetic silica capillaries (0.6 mm i.d.× 0.84 mm o.d.) sealed at one end. All spectra were acquired using incident microwave power at 20 mW, and a 100 kHz field modulation of 1 to 4 gauss as appropriate. All spectra were recorded at 6°C with a scan width of 100 gauss centered at 3356 gauss. Between 15 and 25 scans were averaged for each spectral line. All spectra were normalized and further analyzed using the Labview programs written by Christian Altenbach (UCLA).
Precursor galactose-binding protein was chosen as the natural ligand used in this study because its rate of folding can be modulated by temperature and the absence of calcium. For each experiment equimolar precursor galactose-binding protein, unfolded in 10 mM HEPES-KOH, 300 mM KOAc, 5 mM Mg(OAc)2, 1 mM EGTA, 1 N GnHCl pH 7.6, was added to the spin-labeled SecB variant (20 or 40 µM) in 10 mM HEPES-KOH, 300 mM KOAc, 5 mM Mg(OAc)2, 1 mM EGTA, pH 6.7 by rapid dilution into a solution held on ice so that the final concentration of the GnHCl was 0.17 N. The spectra were collected at 6°C in the presence of 1 mM EGTA to chelate the calcium in order to slow the rate of folding of precursor. As a control for the effect of GnHCl buffer alone was added to spin-labeled SecB in place of precursor galactose-binding protein so that the final concentration of GnHCl was 0.17 N GnHCl. The Kd for the complex formed between SecB and precursor galactose-binding protein is approximately 35 nM.12
Calculation of Timescale of Tumbling
EPR is sensitive to motion of the nitroxide side chain in the timescale of 0.1 nsec to 20 nsec. The molecular tumbling of SecB (τc = 60 nsec) is too slow to be averaged into the spectra reported here and thus the spectra are interpreted in terms of local fluctuations of the backbone and internal motion of the nitroxide side chain. The timescale of tumbling (τc) for SecB was calculated from the rotational diffusion constant (Dr) of the protein as follows:
where k is Boltzmann’s constant, T is temperature, η is the viscosity of the solvent, and a is the radius of hydration. We have determined the radius of hydration of SecB to be 3.3 nm by quasi-elastic light scatter using an in-line detector (Wyatt QELS, Wyatt Technology Corporation) following chromatography on a BioSep-SEC-S4000 size exclusion column (7.8 mm internal diameter × 30 cm, Phenomenex) in 10 mM HEPES-KOH, 300 mM KOAc, 5 mM Mg(OAc)2 pH 7.6 at 7° C. The ASTRA software provided with the instrument was used to calculate the hydrodynamic radius from the diffusion coefficient measured for material at the apex of the chromatographic peak. Using 3.3 nm as the radius, τc for SecB is calculated to be 60 nsec.
Calculation of surface area
The Hemophilus influenzae structure has more residues resolved then does the E. coli structure. Therefore, for surface area calculations a homology model was created by threading the sequence of E. coli SecB through the structure of the H. influenzae SecB. The accessible surface area of individual residues of SecB was calculated using Calc-surface Program28 (available at the NIH Computational Molecular Biology web site, http://molbio.info.nih.gov/molbio/) with a probe size of 1.4 Å. Because of limitations in the electron density of the crystal structure, it was possible to obtain surface areas for only 176 of the 192 mutated residues in the tetramer (48 mutations per protomer). The surface area of missing residues was reported as the average of the surface area for that residue on the other protomers.
Generation of movie provided in supplementary materials
Still images were generated using the PyMol molecular graphics system v0.99. The Connolly solvent surface was generated using a probe with radius 1.4 Å.
Supplementary Material
Description of movie
Clockwise rotation of the SecB tetramer about the vertical axis. All sites of contact with precursor galactose-binding protein are shown in green.
Acknowledgments
We thank Gseping Liu for purifying the SecA used in this study, Beverly B. DaGue of The Proteomics Center at the University of Missouri for MALDI mass spectrometry, Christian Altenbach of UCLA for providing the software used for data analysis, Hilary C. Vasel for technical assistance and Kálmán Hideg, University of Pécs, Hungary, for the generous gift of the analogue of the spin label. We are grateful to Dylan B. Cooper for generation of the movie in supplementary materials. This work was supported in part by NIH research grants GM29798 (L.L.R.) and EY05216 (W.L.H.), an endowment from the Hugo Wurdack Trust at the University of Missouri (L.L.R.), the Jules Stein Professor Endowment (W.L.H.), and the Bruce Ford Bundy and Anne Smith Bundy Foundation (W.L.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berks BC, Sargent F, Palmer T. The Tat protein export pathway. Mol. Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 2.Economou A. Bacterial secretome: the assembly manual and operating instructions. Molecular Membrane Biology. 2002;19:159–169. doi: 10.1080/09687680210152609. [DOI] [PubMed] [Google Scholar]
- 3.Randall LL, Hardy SJS. High selectivity with low specificity: how SecB has solved the paradox of chaperone binding. Trends Biochem. Sci. 1995;20:65–69. doi: 10.1016/s0968-0004(00)88959-8. [DOI] [PubMed] [Google Scholar]
- 4.Crane JM, Mao C, Lilly AA, Smith VF, Suo Y, Hubbell WL, Randall LL. Mapping of the docking of SecA onto the chaperone SecB by site-directed spin labeling: insight into the mechanism of ligand transfer during protein export. J. Mol. Biol. 2005;353:295–307. doi: 10.1016/j.jmb.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 5.Topping TB, Randall LL. Determination of the binding frame within a physiological ligand for the chaperone SecB. Protein Sci. 1994;3:730–736. doi: 10.1002/pro.5560030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khisty VJ, Munske GR, Randall LL. Mapping of the binding frame for the chaperone SecB within a natural ligand, galactose-binding protein. J. Biol. Chem. 1995;270:25920–25927. doi: 10.1074/jbc.270.43.25920. [DOI] [PubMed] [Google Scholar]
- 7.Smith VF, Hardy SJS, Randall LL. Determination of the binding frame of the chaperone SecB within the physiological ligand oligopeptide-binding protein. Protein Sci. 1997;6:1746–1755. doi: 10.1002/pro.5560060815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Z, Knafels JD, Yoshino K. Crystal structure of the bacterial protein export chaperone SecB. Nat. Struct. Biol. 2000;7:1172–1177. doi: 10.1038/82040. [DOI] [PubMed] [Google Scholar]
- 9.Dekker C, de Kruijff B, Gros P. Crystal structure of SecB from Escherichia coli. J. Struct. Biol. 2003;144:313–319. doi: 10.1016/j.jsb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Schneider D, Freed J. Calculating slow motional resonance spectra: A user's guide. In: Berliner L, Reuben J, editors. Spin Labeling: Theory and Application, Biological Magnetic Resonance. Vol. 8. New York: Plenum; 1989. [Google Scholar]
- 11.Fajer PG. Electron spin resonance spectroscopy labeling in peptide and protein analysis. In: Meyers RA, editor. Encyclopedia of Analytical Chemistry. London: John Wiley & Sons Ltd.; 2000. pp. 5725–5761. [Google Scholar]
- 12.Randall LL, Topping TB, Suciu D, Hardy SJS. Calorimetric analyses of the interaction between SecB and its ligands. Protein Sci. 1998;7:1195–1200. doi: 10.1002/pro.5560070514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy SJS, Randall LL. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science. 1991;251:439–443. doi: 10.1126/science.1989077. [DOI] [PubMed] [Google Scholar]
- 14.Topping TB, Randall LL. Chaperone SecB from Escherichia coli mediates kinetic partitioning via a dynamic equilibrium with its ligands. J. Biol. Chem. 1997;272:19314–19318. doi: 10.1074/jbc.272.31.19314. [DOI] [PubMed] [Google Scholar]
- 15.Isas JM, Langen R, Haigler HT, Hubbell WL. Structure and dynamics of a helical hairpin and loop region in annexin 12: a site-directed spin labeling study. Biochemistry. 2002;41:1464–1473. doi: 10.1021/bi011856z. [DOI] [PubMed] [Google Scholar]
- 16.Columbus L, Kalai T, Jeko J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in alpha-helices: analysis by variation of side chain structure. Biochemistry. 2001;40:3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 17.Volkert TL, Baleja JD, Kumamoto CA. A highly mobile C-terminal tail of the Escherichia coli protein export chaperone SecB. Biochem. Biophys. Res. Commun. 1999;264:949–954. doi: 10.1006/bbrc.1999.1590. [DOI] [PubMed] [Google Scholar]
- 18.Bruce JE, Smith VF, Liu C, Randall LL, Smith RD. The observation of chaperone-ligand noncovalent complexes with electrospray ionization mass spectrometry. Protein Sci. 1998;7:1180–1185. doi: 10.1002/pro.5560070512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross A, Columbus L, Hideg K, Altenbach C, Hubbell WL. Structure of the KcsA potassium channel from Streptomyces lividans: a site-directed spin labeling study of the second transmembrane segment. Biochemistry. 1999;38:10324–10335. doi: 10.1021/bi990856k. [DOI] [PubMed] [Google Scholar]
- 20.Randall LL, Crane JM, Lilly AA, Liu G, Mao C, Patel CN, Hardy SJS. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J. Mol. Biol. 2005;348:479–489. doi: 10.1016/j.jmb.2005.02.036. [DOI] [PubMed] [Google Scholar]
- 21.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 22.Kimura E, Akita M, Matsuyama S, Mizushima S. Determination of a region in SecA that interacts with presecretory proteins in Escherichia coli. J. Biol. Chem. 1991;266:6600–6606. [PubMed] [Google Scholar]
- 23.Kourtz L, Oliver D. Tyr-326 plays a critical role in controlling SecA-preprotein interaction. Mol. Microbiol. 2000;37:1342–1356. doi: 10.1046/j.1365-2958.2000.02078.x. [DOI] [PubMed] [Google Scholar]
- 24.Baud C, Karamanou S, Sianidis G, Vrontou E, Politou AS, Economou A. Allosteric communication between signal peptides and the SecA 25 protein DEAD motor ATPase domain. Journal of Biological Chemistry. 2002;277:13724–13731. doi: 10.1074/jbc.M200047200. [DOI] [PubMed] [Google Scholar]
- 25.Papanikou E, Karamanou S, Baud C, Frank M, Sianidis G, Keramisanou D, Kalodimos CG, Kuhn A, Economou A. Identification of the preprotein binding domain of SecA. J. Biol. Chem. 2005;280:43209–43217. doi: 10.1074/jbc.M509990200. [DOI] [PubMed] [Google Scholar]
- 26.Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA. PNAS, USA. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel CN, Smith VF, Randall LL. Characterization of three areas of interactions stabilizing complexes between SecA and SecB, two proteins involved in protein export. Protein Sci. 2006;15 doi: 10.1110/ps.062141006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerstein M. A resolution-sensitive procedure for comparing protein surfaces and its application to the comparison of antigen-combining sites. Acta Crystallographica. 1992;A48:271–276. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of movie
Clockwise rotation of the SecB tetramer about the vertical axis. All sites of contact with precursor galactose-binding protein are shown in green.