Abstract
Understanding polymerase fidelity is an important objective towards ascertaining the overall stability of an organism's genome. Saccharomyces cerevisiae DNA polymerase η (yPolη), a Y-family DNA polymerase, is known to efficiently bypass DNA lesions (e.g., pyrimidine dimers) in vivo. Using pre-steady-state kinetic methods, we examined both full-length and a truncated version of yPolη which contains only the polymerase domain. In the absence of yPolη's C-terminal residues 514–632, the DNA binding affinity was weakened by 2-fold and the base substitution fidelity dropped by 3-fold. Thus, the C-terminus of yPolη may interact with DNA and slightly alter the conformation of the polymerase domain during catalysis. In general, yPolη discriminated between a correct and incorrect nucleotide more during the incorporation step (50-fold on average) than the ground-state binding step (18-fold on average). Blunt-end additions of dATP or pyrene nucleotide 5′-triphosphate revealed the importance of base stacking during the binding of incorrect incoming nucleotides.
1. Introduction
DNA polymerases are organized into seven families: A, B, C, D, X, Y, and reverse transcriptase [1, 2]. Among these families, DNA polymerases are involved in DNA replication, DNA repair, DNA lesion bypass, antibody generation, and sister chromatid cohesion [3]. Despite these diverse roles, DNA polymerases catalyze the nucleotidyl transfer reaction using a two divalent metal ion mechanism [4] with at least one positively charged residue [5] that functions as a general acid [6] at their active site, follow a similar minimal kinetic pathway [7], and share a similar structural architecture consisting of the fingers, palm, and thumb subdomains [8, 9]. Surprisingly, the polymerization fidelity of eukaryotic DNA polymerases spans a wide range: one error per one to one billion nucleotide incorporations (100 to10−9) [10].
The Y-family DNA polymerases are known for catalyzing nucleotide incorporation with low fidelity and poor processivity. These enzymes are specialized for translesion DNA synthesis which involves nucleotide incorporation opposite and downstream of a damaged DNA site. Lesion bypass can be either error-free or error-prone depending on the DNA polymerase and DNA lesion combination. To accommodate a distorted DNA substrate, Y-family DNA polymerases utilize several features: a solvent-accessible [11] and conformationally flexible active site [12], smaller fingers and thumb subdomains [11], an additional subdomain known as the little finger [11], the little finger and polymerase core domains move in opposite directions during a catalytic cycle [13], and a lack of 3′ → 5′ exonuclease activity [14]. Unfortunately, these features, which facilitate lesion bypass, may also contribute to the low fidelity of a Y-family DNA polymerase during replication of a damaged or undamaged DNA template. Thus, it is important to understand the mechanism and fidelity of the Y-family DNA polymerases.
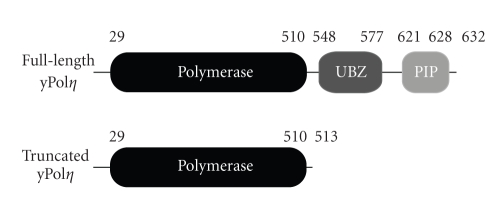
Saccharomyces cerevisiae DNA polymerase η (yPolη), a Y-family DNA polymerase, is critical for the error-free bypass of UV-induced DNA damage such as a cis-syn thymine-thymine dimer [15–19]. To date, Polη remains the only Y-family DNA polymerase with a confirmed biological function [20]. yPolη is organized into a polymerase domain, ubiquitin-binding zinc finger (UBZ) domain, and proliferating cell nuclear antigen- (PCNA) interacting peptide (PIP) motif (Figure 1). X-ray crystal structures of yPolη's catalytic core have been solved alone [21] as well as in complex with a cisplatin-DNA adduct and an incoming nucleotide [22]. Due to a lack of structures for full-length yPolη, it is unclear if the C-terminal residues 514–632 interact with DNA and contribute to the polymerase function of yPolη. Using pre-steady-state kinetic techniques, we have measured the base-substitution fidelity of full-length and truncated yPolη (Figure 1) catalyzing nucleotide incorporation into undamaged DNA. In addition, we have determined the DNA binding affinity of both full-length and truncated yPolη. Our results show that the C-terminus of yPolη has a minor effect on the DNA binding affinity and the base substitution fidelity of this lesion bypass DNA polymerase.
Figure 1.
Schematic illustration of yPolη. The polymerase domain of yPolη is at the N-terminus while a ubiquitin-binding zinc finger (UBZ) domain and PCNA-interacting peptide (PIP) motif is at the C-terminus. Residue numbers are denoted above each region. For this study, the truncated construct contains only the polymerase domain.
2. Materials and Methods
2.1. Materials
Materials were purchased from the following companies: [γ-32P] ATP, MP Biomedicals (Solon, OH); Biospin columns, Bio-Rad Laboratories (Herclues, CA); dNTPs, GE Healthcare (Piscataway, NJ); oligodeoxyribonucleotides, Integrated DNA Technologies, Inc. (Coralville, IA); and OptiKinase, USB (Cleveland, OH).
2.2. Preparation of Substrates and Enzymes
The synthetic oligodeoxyribonucleotides listed in Table 1 were purified as described previously [23]. The primer strand 21-mer or blunt-end 16-mer was 5′-radiolabeled with [γ-32P]ATP and OptiKinase. Then, the 21-mer was annealed to the appropriate 41 mer template (Table 1) and the palindromic blunt-end substrates were annealed as described previously [23]. The catalytic core of yPolη (1–513) containing an N-terminal MGSSH6SSGLVPRGSH tag was purified as described previously [24]. The full-length yPolη (1–632) was expressed and purified from yeast [25]. Pyrene 5′-triphosphate (dPTP) was synthesized as described previously [26].
Table 1.
Sequences of DNA substratesa.
| D-1 | 5′-CGCAGCCGTCCAACCAACTCA-3′ 3′-GCGTCGGCAGGTTGGTTGAGT A GCAGCTAGGTTACGGCAGG-5′ |
| D-6 | 5′-CGCAGCCGTCCAACCAACTCA-3′ 3′-GCGTCGGCAGGTTGGTTGAGT G GCAGCTAGGTTACGGCAGG-5′ |
| D-7 | 5′-CGCAGCCGTCCAACCAACTCA-3′ 3′-GCGTCGGCAGGTTGGTTGAGT T GCAGCTAGGTTACGGCAGG-5′ |
| D-8 | 5′-CGCAGCCGTCCAACCAACTCA-3′ 3′-GCGTCGGCAGGTTGGTTGAGT C GCAGCTAGGTTACGGCAGG-5′ |
| F-8 | 5′-CGCAGCCGTCCAACCAACTCA-3′ 3′-GCGTCGGCAGGTTGGTTGAGTC X CAGCTAGGTTACGGCAGG-5′ |
| BE1 | 5′-ATGAGTTGCAACTCAT-3′ 3′-TACTCAACGTTGAGTA-5′ |
| BE2 | 5′-TTGAGTTGCAACTCAA-3′ 3′-AACTCAACGTTGAGTT-5′ |
| BE3 | 5′-CTGAGTTGCAACTCAG-3′ 3′-GACTCAACGTTGAGTC-5′ |
| BE4 | 5′-GTGAGTTGCAACTCAC-3′ 3′-CACTCAACGTTGAGTG-5′ |
aThe template base highlighted in bold is unique to each strand and X denotes 2-aminopurine.
2.3. Pre-Steady-State Kinetic Assays
All experiments were performed in reaction buffer A which contained 40 mM Tris-HCl pH 7.5 at 23°C, 5 mM MgCl2, 1 mM DTT, 10 μg/mL BSA, and 10% glycerol. A rapid chemical-quench flow apparatus (KinTek, PA, USA) was used for fast reactions. For burst assays, a preincubated solution of yPolη (320 nM) and 5′-[32P]-labeled D-1 DNA (480 nM) was mixed with dTTP·Mg2+ (100 μM). To measure the dissociation rate of the yPolη·DNA binary complex, a preincubated solution of yPolη (50 nM) and 5′-[32P]-labeled D-1 DNA (100 nM) was mixed with a molar excess of unlabeled D-1 DNA (2.5 μM) for various time intervals prior to initiating the polymerization reaction with dTTP·Mg2+ (150 and 400 μM for truncated and full-length yPolη, resp.) for 15 s. For single-turnover kinetic assays, a preincubated solution of yPolη (150 nM) and 5′-[32P]-labeled DNA (30 nM) was mixed with an incoming dNTP·Mg2+ (0.4–800 μM). Reactions were quenched at the designated time by adding 0.37 M EDTA. Reaction products were analyzed by sequencing gel electrophoresis (17% acrylamide, 8 M urea, 1 × TBE running buffer), visualized using a Typhoon TRIO (GE Healthcare), and quantitated with ImageQuant software (Molecular Dynamics).
2.4. DNA Binding Assays
The equilibrium dissociation constant (K d DNA) of the yPolη·DNA binary complex was determined using two techniques. First, an electrophoretic mobility shift assay (EMSA) was employed by adding increasing concentrations of yPolη (10–450 nM) into a fixed concentration of 5′-[32P]-labeled D-1 DNA (10 nM) in buffer A. The solution established equilibrium during a 20-minute incubation period. Then, the binary complex was separated from unbound DNA using a 4.5% native polyacrylamide gel and running buffer as previously described except the final concentration of Tris was adjusted to 40 mM [27]. Second, a fluorescence titration assay was used. Increasing concentrations of yPolη (2–300 nM) were titrated into a fixed concentration of F-8 DNA (25 nM) in buffer A (devoid of BSA). The F-8 DNA substrate (Table 1) was excited at a wavelength of 312 nm with emission and excitation slit widths of 5 nm. The emission spectra were collected at 1 nm intervals from 320 to 500 nm using a Fluoromax-4 (Jobin Jvon Horiba). Emission background from the buffer and intrinsic protein fluorescence were subtracted from each spectrum.
2.5. Data Analysis
For the pre-steady-state burst assay, the product concentration was graphed as a function of time (t) and the data were fit to the burst equation (1) using the nonlinear regression program, KaleidaGraph (Synergy Software):
| (1) |
A represents the fraction of active enzyme, k 1 represents the observed burst rate constant, and k 2 represents the observed steady-state rate constant.
Data for the EMSA were graphed by plotting the concentration of the binary complex as a function of enzyme concentration (E 0) and fitting it to a quadratic equation (2):
| (2) |
D 0 is the DNA concentration.
For the fluorescence titration experiments, a modified quadratic equation (3) was applied to a plot of the fluorescence intensity (F) measured at 370 nm versus enzyme concentration:
| (3) |
Fmax and Fmin represent the maximum and minimum fluorescence intensity, respectively.
For the rate of DNA dissociation from the binary complex, a single-exponential equation (4) was applied to a plot of product concentration versus time:
| (4) |
A represents the reaction amplitude, k off is the observed rate constant of DNA dissociation, and C is the concentration of the radiolabeled DNA product in the presence of a DNA trap for unlimited time.
For the single-turnover kinetic assays, a plot of product concentration versus time was fit to a single-exponential equation (5) to extract the observed rate constant of nucleotide incorporation (k obs):
| (5) |
To measure the maximum rate constant of incorporation (k p) and the apparent equilibrium dissociation constant (K d) of an incoming nucleotide, the extracted k obs values were plotted as a function of nucleotide concentration and fit to a hyperbolic equation (6):
| (6) |
The free energy change (∆∆G) for a correct and incorrect nucleotide substrate dissociating from the E·DNA·dNTP complex was calculated according to (7).
| (7) |
Here, R is the universal gas constant and T is the reaction temperature in Kelvin.
3. Results and Discussion
3.1. Truncated and Full-Length yPolη Display Biphasic Kinetics
Previously, transient state kinetic techniques have been used to characterize full-length yPolη at 30°C [28]. Therefore, we first performed a burst assay (see Section 2) to ensure that our purified proteins, truncated and full-length yPolη (Figure 1), behaved in a similar manner at 23°C. Compared to wild-type yPolη, the truncated construct contains only the polymerase domain (Figure 1). A preincubated solution of yPolη (320 nM) and 5′-[32P]-labeled 21/41 mer D-1 DNA (480 nM) was mixed with dTTP·Mg2+ (100 μM) and quenched with EDTA at various times. Product concentration was plotted as a function of time and was fit to (1), since there were two distinct kinetic phases: a rapid, exponential phase and a slow, linear phase (data not shown). These burst results were similar to those previously published [28]. Biphasic kinetics of nucleotide incorporation indicated that the first turnover rate was the rate of nucleotide incorporation occurring at the enzyme's active site while subsequent turnovers (i.e., linear phase) were likely limited by the DNA product release step as demonstrated by full-length yPolη at 30°C [28] and other DNA polymerases [23, 29, 30].
3.2. The C-Terminal 119 Residues Slightly Enhance DNA Binding Affinity of yPolη
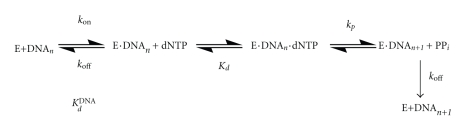
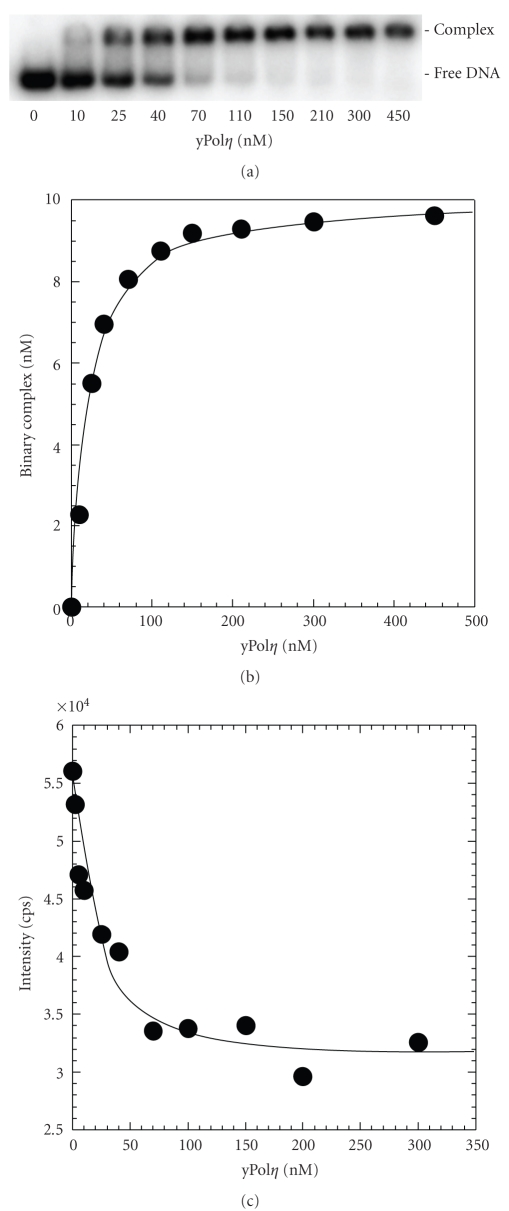
The equilibrium dissociation constant for the binary complex of yPolη·DNA (K d DNA) was measured to determine if the C-terminus of yPolη affects DNA binding affinity (Scheme 1). First, the K d DNA was estimated using the EMSA (see Section 2). For example, varying concentrations of full-length yPolη (10–450 nM) were incubated with a fixed concentration of 5′-[32P]-labeled D-1 DNA (10 nM) before separating the binary complex from the unbound DNA on a native gel (Figure 2(a)). Then, a quadratic equation (2) was applied to a plot of the binary complex concentration versus yPolη concentration which resolved a K d DNA of 16 ± 1 nM (Figure 2(b) and Table 2). Under similar reaction conditions, the K d DNA of truncated yPolη was estimated to be 34 ± 3 nM, a binding affinity (1/K d DNA) value that is 2-fold weaker than that of full-length yPolη (Table 2).
Scheme 1.
Figure 2.
Equilibrium dissociation constant for full-length yPolη. (a) Gel image showing binary complex formation at various concentrations of full-length yPolη (10–450 nM) in the presence of 5′-[32P]-labeled D-1 DNA (10 nM). (b) The concentration of the binary complex was plotted as a function of full-length yPolη concentration and fit to (2) to yield a K d DNA = 16 ± 1 nM. (c) For the fluorescence titration assay, a plot of fluorescence intensity versus full-length yPolη concentration was fit to (3) which resolved a K d DNA = 7 ± 4 nM.
Table 2.
Rate and equilibrium dissociation constants for the binary complex yPolη·DNA at 23°C.
| Kinetic Parameter | Truncated yPolη | Full-length yPolη |
|---|---|---|
| k on (μM−1 s−1)a | 0.62 | 0.59 |
| k off (s−1) | 0.008 ± 0.001 | 0.0041 ± 0.0008 |
| K d DNA (nM)b | 34 ± 3 | 16 ± 1 |
| K d DNA (nM)c | 13 ± 5 | 7 ± 4 |
aCalculated as k off/K d DNA. The K d DNA value was measured from a fluorescence titration assay.
bEstimated using EMSA.
cMeasured using a fluorescence titration assay.
To corroborate these estimated K d DNA values, we measured the true K d DNA for the yPolη·DNA complex using a fluorescence titration assay. An analog of dA, 2-aminopurine, was embedded into the 41 mer template of F-8 DNA which is identical to 21/41 mer D-8 DNA except that 2-aminopurine flanks the 5′ end of the templating dC base (Table 1). The F-8 DNA substrate (25 nM) was excited at 312 nm, and the emission spectrum was collected from 320 to 500 nm. After serial additions of full-length or truncated yPolη in independent titrations, a decrease in the fluorescence intensity of F-8 was observed. These changes in fluorescence intensity at 370 nm were plotted as a function of the yPolη concentration and were fit to (3) to extract a K d DNA equal to 7 ± 4 nM for full-length yPolη (Figure 2(c)) and 13 ± 5 nM for truncated yPolη (Table 2). These K d DNA measurements were tighter than those determined using EMSA, since the fluorescence titration assay allows yPolη to associate and dissociate during data collection. In contrast, EMSA does not maintain a constant equilibrium because dissociated yPolη cannot reassociate with DNA during electrophoresis separation. Nonetheless, there was a confirmed ~2-fold difference in the DNA binding affinity between full-length and the catalytic core of yPolη which indicates that the C-terminal 119 amino acid residues of yPolη slightly enhance the binding of the enzyme to DNA.
Next, we directly measured the rate of DNA dissociation from the yPolη·DNA complex (see Section 2). A preincubated solution of yPolη (50 nM) and 5′-radiolabeled D-1 DNA (100 nM) was combined with a 50-fold molar excess of unlabeled D-1 DNA for various time intervals before dTTP was added for 15 s to allow ample extension of the labeled D-1 DNA that remained in complex with yPolη. A plot of product concentration versus the incubation time with the unlabeled DNA trap (data not shown) was fit to (4) which yielded DNA dissociation rates (k off) of 0.008 ± 0.001 s−1 and 0.0041 ± 0.0008 s−1 for truncated and full-length yPolη, respectively (Table 2 and Scheme 1). Interestingly, the rate of DNA dissociation from full-length yPolη is 2-fold slower than that from truncated yPolη, which indicated that the C-terminus of yPolη may slightly contribute to this polymerase's DNA binding affinity.
Based on the measured K d DNA from Figure 2(c) and k off values, the apparent second-order association rate constant (k on = k off/K d DNA) of the binary complex yPolη·DNA was calculated to be 0.62 and 0.59 μM−1 s−1 for truncated and full-length yPolη, respectively (Table 2). These similar k on values indicate that the slightly stronger DNA binding affinity of full-length yPolη is mainly due to a slightly slower rate of DNA dissociation (k off). Taken together, the data in Table 2 suggest that the C-terminal 119 amino acid residues of yPolη slightly hinder the dissociation of DNA from the binary complex yPolη·DNA. This hindrance is through either direct physical interactions between the C-terminus of yPolη and DNA, modulation of the conformation of the polymerase domain by the C-terminus of yPolη, or both.
3.3. Base Substitution Fidelity of Truncated yPolη
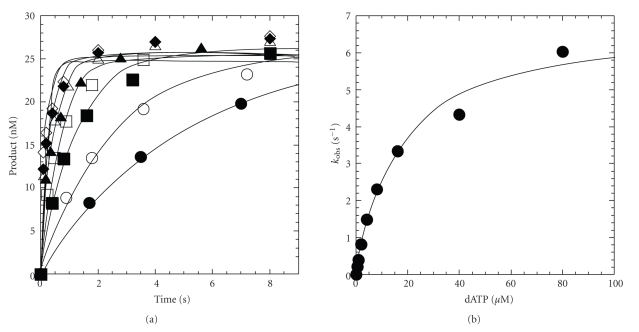
Since a pre-steady-state burst was observed for truncated yPolη, we continued to investigate the nucleotide incorporation efficiency (k p/K d) by measuring the maximum rate of nucleotide incorporation (k p) and the apparent equilibrium dissociation constant (K d) of an incoming nucleotide under single-turnover conditions [31]. By performing these experiments with yPolη in molar excess over DNA, the conversion of D-DNAn to D-DNAn+1 (Scheme 1) was directly observed in a single pass through the enzymatic pathway [32]. A preincubated solution of truncated yPolη (150 nM) and 5′-[32P]-labeled D-7 DNA (30 nM) was mixed with varying concentrations of dATP·Mg2+ (0.4–80 μM) and quenched with EDTA at various times (see Section 2). A plot of product concentration versus time was fit to (5) to extract the observed rate constant (k obs) for dATP incorporation (Figure 3(a)). Then, the k obs values were plotted as a function of dATP concentration and fit to a hyperbolic equation (6) which resolved a k p of 6.9 ± 0.4 s−1 and an apparent K d of 17 ± 3 μM (Figure 3(b)). The pre-steady-state kinetic parameters for the remaining 15 possible dNTP:dN base pair combinations were determined under single-turnover conditions and were used to calculate the substrate specificity constant (k p/K d), discrimination factor ((k p/K d)correct/(k p/K d)incorrect), and fidelity ((k p/K d)incorrect/[(k p/K d)correct + (k p/K d)incorrect]) of truncated yPolη (Table 3).
Figure 3.
Concentration dependence on the pre-steady-state rate constant of nucleotide incorporation catalyzed by truncated yPolη. (a) A preincubated solution of truncated yPolη (150 nM) and 5′-[32P]-labeled D-7 DNA (30 nM) was mixed with dATP·Mg2+ (0.4 μM, ⬤; 0.8 μM, ⚪; 2 μM, ■; 4 μM, □; 8 μM, ▲; 16 μM, △; 40 μM, ◆; 80 μM, ◊) and quenched with EDTA at various time intervals. The solid lines are the best fits to a single-exponential equation which determined the observed rate constant, k obs. (b) The k obs values were plotted as a function of dATP concentration. The data (⬤) were then fit to a hyperbolic equation, yielding a k p of 6.9 ± 0.4 s−1 and a Kd of 17 ± 3 μM.
Table 3.
Kinetic parameters of nucleotide incorporation into D-DNA catalyzed by truncated yPolη at 23°C.
| dNTP | kp (s−1) | Kd (μM) | kp/Kd(μM−1s−1) | Discrimination Factora | Fidelityb |
|---|---|---|---|---|---|
| Template dA (D-1) | |||||
| dTTP | 3.9 ± 0.2 | 15 ± 2 | 2.6 × 10−1 | ||
| dATP | 0.089 ± 0.005 | 80 ± 20 | 1.1 × 10−3 | 230 | 4.3 × 10−3 |
| dCTP | 0.43 ± 0.06 | 210 ± 60 | 2.0 × 10−3 | 130 | 7.8 × 10−3 |
| dGTP | 0.15 ± 0.01 | 80 ± 20 | 1.9 × 10−3 | 140 | 7.2 × 10−3 |
|
| |||||
| Template dG (D-6) | |||||
| dCTP | 15.6 ± 0.3 | 11.2 ± 0.8 | 1.4 | ||
| dATP | 0.071 ± 0.002 | 138 ± 9 | 5.1 × 10−4 | 2700 | 3.7 × 10−4 |
| dGTP | 0.116 ± 0.006 | 80 ± 10 | 1.5 × 10−3 | 960 | 1.0 × 10−3 |
| dTTP | 0.92 ± 0.07 | 330 ± 40 | 2.8 × 10−3 | 500 | 2.0 × 10−3 |
|
| |||||
| Template dT (D-7) | |||||
| dATP | 6.9 ± 0.4 | 17 ± 3 | 4.1 × 10−1 | ||
| dCTP | 1.00 ± 0.04 | 210 ± 20 | 4.8 × 10−3 | 85 | 1.2 × 10−2 |
| dGTP | 0.55 ± 0.01 | 46 ± 3 | 1.2 × 10−2 | 30 | 2.9 × 10−2 |
| dTTP | 0.62 ± 0.02 | 280 ± 20 | 2.2 × 10−3 | 180 | 5.4 × 10−3 |
|
| |||||
| Template dC (D-8) | |||||
| dGTP | 6.3 ± 0.1 | 6.8 ± 0.4 | 9.3 × 10−1 | ||
| dATP | 0.087 ± 0.003 | 90 ± 10 | 9.7 × 10−4 | 960 | 1.0 × 10−3 |
| dCTP | 0.127 ± 0.007 | 200 ± 30 | 6.4 × 10−4 | 1500 | 6.9 × 10−4 |
| dTTP | 1.39 ± 0.06 | 460 ± 40 | 3.0 × 10−3 | 310 | 3.3 × 10−3 |
aCalculated as (k p/K d)correct/(k p/K d)incorrect.
bCalculated as (k p/K d)incorrect/[(k p/K d)correct + (k p/K d)incorrect].
Overall, the base substitution fidelity of truncated yPolη was in the range of 10−2 to 10−4 which translates into 1 misincorporation per 100 to 10,000 nucleotide incorporations (Table 3). Depending on the mispair, truncated yPolη catalyzed a misincorporation with 30- to 2,700-fold (640-fold on average) lower efficiency than the corresponding correct base pair. To better understand the mechanistic basis of truncated yPolη's fidelity, the equation for polymerase fidelity can be simplified as follows:
| (8) |
Thus, fidelity is inversely proportional to the rate difference and apparent binding affinity difference between correct and incorrect nucleotide incorporation. In general, the mechanistic basis of yPolη's discrimination was due to a 3- to 68-fold (18-fold on average) weaker apparent binding affinity (1/Kd) and 5- to 220-fold (50-fold on average) slower rate constant of incorporation for a mismatched dNTP.
3.4. Kinetic Significance of Base Stacking Contributing to the Binding Affinity of an Incoming Nucleotide
Although all four correct dNTPs were bound with similarly high affinity (Table 3), mismatched purine deoxyribonucleotides have 2- to 6-fold lower apparent Kd values than mismatched pyrimidine deoxyribonucleotides. Because 5′-protruding purines have been found to have stronger stacking interactions with a terminal DNA base pair than 5′-protruding pyrimidines [33], the difference in apparent Kd values suggests that base-stacking interactions between an incorrect dNTP and the terminal primer/template base pair dA:dT (Table 1) play a role on the binding of dNTP by truncated yPolη. Interestingly, we have previously demonstrated that the preferred nucleotide for template-independent nucleotide incorporation catalyzed by Dpo4, another Y-family DNA polymerase, is dATP mainly due to its strong intrahelical base-stacking ability [26]. To further evaluate the role of base stacking, we first examined if truncated yPolη can catalyze template-independent nucleotide incorporation of dATP or dPTP (Figure 4) onto four palindromic, blunt-end DNA substrates (BE1, BE2, BE3, and BE4 in Table 1). The base of dPTP, a dNTP analog, has four conjugated benzene rings but possesses no hydrogen-bonding abilities. The DNA substrates possess all four possible terminal base pairs and each molecule of them can be bound by a single polymerase molecule. Our radioactive experiments showed that truncated yPolη was able to incorporate dATP and dPTP (data not shown). Then, we individually measured the kinetic parameters for dATP and dPTP incorporation under single-turnover reaction conditions (Table 4). Interestingly, the apparent Kd values of dATP were 3- to 5-fold smaller with a purine than those with a pyrimidine on the primer's 3′-base, indicating that base stacking is also important for the binding of dATP to the binary complex of yPolη·blunt-end DNA. This base-stacking effect is more dramatic for dPTP incorporation onto blunt-end DNA because the apparent Kd values of dPTP are 10- to 80-fold tighter than dATP incorporation onto the same blunt-end DNA substrate (Table 4). Thus, the binding free energy difference between dATP and dPTP is 1.4 to 2.6 kcal/mol. Previously, we have obtained a comparable binding free energy difference of 2.3 kcal/mol for similar blunt-end dATP and dPTP incorporation at 37°C catalyzed by Dpo4 [26]. Although neither dATP nor dPTP forms any hydrogen bonds with a template base when bound by yPolη·blunt-end DNA, the bases of these two nucleotides should have different base-stacking interactions with a terminal base pair of a blunt-end DNA substrate considering that a dangling pyrene base (1.7 kcal/mol) has previously been found to possess a higher base-stacking free energy than a dangling adenosine (1.0 kcal/mol) [33]. However, the base-stacking free energy difference (0.7 kcal/mol) between pyrene and adenosine is smaller than the aforementioned binding free energy difference (1.4–2.6 kcal/mol) between dPTP and dATP. Thus, other sources likely contribute to the tighter binding of dPTP over dATP. One possible source is favorable van der Waals interactions between pyrene and active site residues of truncated yPolη. In addition, the base-stacking effect and van der Waals interactions may stabilize the ternary complex of yPolη·blunt-end DNA·nucleotide and facilitate catalysis, leading to much higher kp values with dPTP than those with dATP (Table 4). Due to the differences in kp and apparent Kd, the substrate specificity values of dPTP are 100- to 1,000-fold higher than those of dATP with blunt-end DNA (Table 4) and 10- to 100-fold higher than mismatched dATP with regular DNA (Table 3).
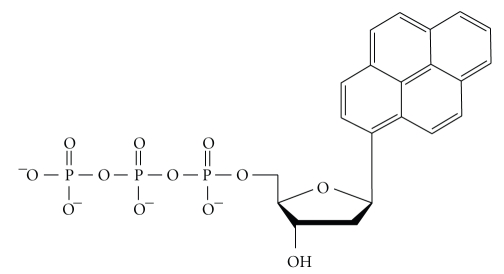
Figure 4.
Chemical structure of a nonnatural nucleotide analog, dPTP.
Table 4.
Kinetic parameters for nucleotide incorporation onto blunt-end DNA catalyzed by truncated yeast Polη at 23°C.
| DNA (Terminal base pair) | dNTP | kp (s−1) | Kd (μM) | kp/Kd (μM−1 s−1) | Efficiency Ratioa |
|---|---|---|---|---|---|
| BE1 (dT : dA) | dATP | 0.026 ± 0.002 | 1200 ± 200 | 2.2 × 10−5 | — |
| dPTP | 1.27 ± 0.08 | 60 ± 10 | 2.1 × 10−2 | 980 | |
| BE2 (dA : dT) | dATP | 0.036 ± 0.002 | 220 ± 30 | 1.6 × 10−4 | — |
| dPTP | 0.68 ± 0.03 | 23 ± 3 | 3.0 × 10−2 | 180 | |
| BE3 (dG : dC) | dATP | 0.0087 ± 0.0003 | 360 ± 30 | 2.4 × 10−5 | — |
| dPTP | 0.22 ± 0.01 | 9 ± 2 | 2.4 × 10−2 | 1000 | |
| BE4 (dC : dG) | dATP | 0.032 ± 0.001 | 930 ± 70 | 3.4 × 10−5 | — |
| dPTP | 0.74 ± 0.03 | 12 ± 2 | 6.2 × 10−2 | 1800 |
aCalculated as (k p/K d)dPTP/(k p/K d)dATP.
3.5. Base Substitution Fidelity of Full-Length yPolη
The base substitution fidelities of full-length and truncated yPolη may differ because the C-terminal, nonenzymatic regions may alter the polymerization fidelity. For example, the proline-rich domain of human DNA polymerase λ has been shown to upregulate the polymerase fidelity up to 100-fold [34]. To determine if the C-terminus of yPolη influences polymerization fidelity, we measured the pre-steady-state kinetic parameters for dNTP incorporation into D-1 DNA (template dA) catalyzed by full-length yPolη (Table 5). The fidelity was calculated to be in the range of (1.4 to 2.6) × 10−3 for full-length yPolη (Table 5). Relative to the fidelity of truncated yPolη with D-1 (Table 3), full-length yPolη has a 3-fold higher fidelity. Therefore, the C-terminus of yPolη slightly affects the base substitution fidelity. Moreover, truncated yPolη discriminated between a correct and incorrect dNTP by ~30-fold on average based on the kp difference while the discrimination for full-length yPolη was ~170-fold on average for incorporation into D-1 DNA (Tables 3 and 5). The incorporation rate constant for correct dTTP was ~4 s−1 for both yPolη enzymes, but the misincorporation rate was 3- to 23-fold faster for truncated yPolη. This rate enhancement for truncated yPolη is partially offset by a greater discrimination at the apparent ground-state binding level so that the fidelity of truncated yPolη was only 3-folder lower than that of full-length yPolη.
Table 5.
Kinetic parameters of nucleotide incorporation into D-1 DNA catalyzed by full-length yPolη at 23°C.
| dNTP | kp (s−1) | Kd (μM) | kp/Kd (μM−1 s−1) | Discrimination Factora | Fidelityb |
|---|---|---|---|---|---|
| Template dA (D-1) | |||||
| dTTP | 4.2 ± 0.5 | 40 ± 10 | 1.1 × 10−1 | ||
| dATP | 0.0235 ± 0.0003 | 156 ± 7 | 1.5 × 10−4 | 700 | 1.4 × 10−3 |
| dCTP | 0.019 ± 0.001 | 70 ± 10 | 2.7 × 10−4 | 390 | 2.6 × 10−3 |
| dGTP | 0.043 ± 0.003 | 170 ± 40 | 2.5 × 10−4 | 420 | 2.4 × 10−3 |
aCalculated as (k p/K d)correct/(k p/K d)incorrect.
bCalculated as (k p/K d)incorrect/[(k p/K d)correct + (k p/K d)incorrect].
3.6. Effect of the Nonenzymatic C-Terminus of yPolη on Its Polymerase Activity
Our above studies demonstrated that the C-terminus of yPolη enhances this enzyme's DNA binding affinity and base substitution fidelity by 2- and 3-fold, respectively. These results suggest that the nonenzymatic, C-terminal region of yPolη (Figure 1) has a mild impact on the N-terminal polymerase domain and its activity. This conclusion is inconsistent with previous studies which have qualitatively demonstrated that mutations or deletions in the UBZ domain or PIP motif do not affect polymerase activity [35–37]. However, these reported qualitative assays are not sufficiently sensitive to detect the small perturbation on polymerase activity as described in this paper. The presence of the C-terminal 119 residues of yPolη may either interact with DNA, slightly alter the conformation of the polymerase domain, or both (see above discussion), thereby enhancing its DNA binding affinity and polymerase fidelity.
3.7. Kinetic Comparison among Y-Family DNA Polymerases
The fidelity of several Y-family DNA polymerases synthesizing undamaged DNA has been determined by employing steady-state [38–48], pre-steady-state [28, 30, 49–53], or M13-based mutation assays [39, 41, 42, 45, 54, 55]. From these studies, the fidelity ranges from 100 to 10−4. Under steady-state reaction conditions, the base substitution fidelity of yPolη and human Polη has been measured to be in the range from 10−2 to 10−4and 10−2 to 10−3, respectively [38, 40], which is similar to our pre-steady-state kinetic results. Consistently, Polη displays the highest substrate specificity for the dCTP : dG base pair under both steady-state and pre-steady-state reaction conditions (Table 3 and unpublished data, Brown and Suo) [38, 40]. This may seem surprising, since Polη participates in the efficient bypass of UV-induced DNA damage such as a cis-syn thymine-thymine dimer (i.e., a dATP:dT base pair) [15–20, 56, 57]. However, Polη has also been shown to be efficient at bypassing guanine-specific damage such as 8-oxo-7,8-dihydro-dG [58, 59], 1,2-cis-diammineplatinum(II)-d(GpG) intrastrand cross-links [60–63], and various N2-dG lesions [64, 65].
Among the four eukaryotic Y-family DNA polymerases (i.e., Polη, DNA polymerase κ, DNA polymerase ι (Polι), and Rev1), Rev1 exhibits low fidelity on undamaged DNA due to its strong preference for inserting dCTP [46, 52] while Polι has an unusual preference for dGTP:dT mispairs over dATP:dT due to Hoogsteen base pair formation [51, 69]. Interestingly, the lowest fidelity base pair for truncated yPolη was dGTP:dT (Table 3). This observation likely results from the formation of a wobble base pair. The two hydrogen bonds established in the wobble base pair may enhance the catalytic efficiency of yPolη since hydrogen bonding is important for the efficiency and accuracy of yPolη [70]. Also noteworthy, the truncated versions of eukaryotic Y-family DNA polymerases have been used for many biochemical studies in literature. Based on our quantitative kinetic analysis of yPolη, these results suggested the nonenzymatic regions of Y-family DNA polymerases do not alter the polymerase activity significantly.
3.8. Fidelity Comparison among Various DNA Polymerase Families
As a Y-family DNA polymerase, yPolη displays low fidelity on undamaged DNA (Tables 3 and 5) [38]. In contrast, replicative DNA polymerases in the A- and B-families have a polymerization fidelity that is 1–3 orders of magnitude greater than the Y-family DNA polymerases (Table 6). DNA polymerases with higher fidelity are more proficient at using the ground-state binding affinity to discriminate between a correct and incorrect dNTP. The Y-family DNA polymerases provide little to no discrimination based on the Kd difference while replicative DNA polymerases discriminate up to almost three orders of magnitude. This lack of selection in the ground state by the Y-family DNA polymerases may be due to the relatively loose and solvent-accessible active site which has minimal contacts with the nascent base pair [11, 21, 71]. Moreover, nucleotide selection by the Y-family DNA polymerases in the ground state may be mainly governed by Watson-Crick base pairing, since the calculated ∆∆G values (0.95–1.7 kcal/mol) are similar to the free energy differences between correct and incorrect base pairs (0.3–1.0 kcal/mol at 37°C) at the primer terminus based on DNA melting studies (Table 6) [72]. However, with ∆∆G values ≥3.0 kcal/mol, the replicative DNA polymerases harness the additional 2.0 kcal/mol of energy from other sources such as a tight active site or close contacts with the nascent base pair. One common fidelity checkpoint among DNA polymerases is the varying rate differences between a matched and mismatched base pair. These large differences may correspond to different rate-limiting steps (e.g., protein conformational change, or phosphodiester bond formation) during nucleotide incorporation [9, 30, 71]. For yPolη, kinetic data suggest that correct and incorrect dNTPs are limited by a conformational step preceding chemistry, although, additional studies are needed to confirm these results [28].
Table 6.
Comparison of base substitution fidelity for various DNA polymerases.
| Polymerase | Polymerase Family | Fidelitya | K d Differenceb | k p Differenceb | ∆∆G (kcal/mol)c |
|---|---|---|---|---|---|
| Truncated yPolη d | Y | 3.7 × 10−4 to 2.9 × 10−2 | 3 to 68 | 5 to 220 | 1.6 |
| Dpo4e | Y | 1.5 × 10−4 to 3.2 × 10−3 | 1 to 18 | 240 to 1700 | 0.95 |
| rPolβ f | X | 1.1 × 10−5 to 5.9 × 10−4 | 35 to 342 | 28 to 708 | 3.0 |
| PolB1 exo-g | B | 3.5 × 10−6 to 1.2 × 10−4 | 109 to 918 | 4 to 589 | 3.7 |
| hPolγ h | A | 4.6 × 10−7 to 2.9 × 10−4 | 42 to 900 | 39 to 12000 | 3.4 |
aCalculated as (k p/K d)incorrect/[(k p/K d)correct + (k p/K d)incorrect].bCalculated as defined in equation (8). cCalculated using equation (7). dAt 23°C (this work). eAt 37°C [50]. fAt 37°C [66]. gAt 37°C, excluding the fidelity contribution from the 3′ → 5′ exonuclease activity [67]. hAt 37°C, excluding the fidelity contribution from the 3′ → 5′ exonuclease activity [68].
4. Conclusions
This work presents the mechanistic basis of the base substitution fidelity of yPolη on undamaged DNA, which examined all possible dNTP:dN base pair combinations for the first time. yPolη discriminates against incorrect nucleotides at both the ground-state nucleotide binding and incorporation steps. Furthermore, base stacking contributes to tighter binding for a misincorporation. Finally, the 119 residues at the C-terminus have a mild impact on the kinetic mechanism of yPolη.
Acknowledgments
This work was supported by the National Institutes of Health Grants CA040463 (to Zucai Suo and John-Stephen Taylor) and GM032431 (to Peter M.J. Burgers). Jessica A. Brown was supported by an American Heart Association Pre-doctoral Fellowship (Grant 0815382D). Shanen M. Sherrer was supported by a Predoctoral Fellowship from the National Institutes of Health Chemistry-Biology Interface Program at The Ohio State University (Grant 5 T32 GM008512-13). Brown and Zhang contributed equally to this work.
Abbreviations
- BSA:
Bovine serum albumin
- dNTP:
2′-deoxynucleoside 5′-triphosphate
- Dpo4:
Sulfolobus solfataricus P2 DNA polymerase IV
- dPTP:
Pyrene 5′-triphosphate
- EMSA:
Electrophoretic mobility shift assay
- HPolγ:
Human mitochondrial DNA polymerase gamma
- PCNA:
Proliferating cell nuclear antigen
- PIP:
PCNA-interacting peptide
- PolB1:
Exonuclease-deficient DNA polymerase B1 from
sulfolobus solfataricus P2
- Polι:
DNA polymerase iota
- rPolβ:
Rat DNA polymerase beta
- TBE
Tris/boric acid/EDTA
- UBZ:
Ubiquitin-binding zinc finger
- YPolη:
Saccharomyces cerevisiae DNA polymerase eta.
References
- 1.Fowler JD, Suo Z. Biochemical, structural, and physiological characterization of terminal deoxynucleotidyl transferase. Chemical Reviews. 2006;106(6):2092–2110. doi: 10.1021/cr040445w. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Diaz M, Bebenek K. Multiple functions of DNA polymerases. Critical Reviews in Plant Sciences. 2007;26(2):105–122. doi: 10.1080/07352680701252817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burgers PMJ, Koonin EV, Bruford E, et al. Eukaryotic DNA polymerases: proposal for a revised nomenclature. Journal of Biological Chemistry. 2001;276(47):43487–43490. doi: 10.1074/jbc.R100056200. [DOI] [PubMed] [Google Scholar]
- 4.Steitz TA. DNA- and RNA-dependent DNA polymerases. Current Opinion in Structural Biology. 1993;3(1):31–38. doi: 10.1016/s0959-440x(99)00058-5. [DOI] [PubMed] [Google Scholar]
- 5.Fowler JD, Brown JA, Kvaratskhelia M, Suo Z. Probing conformational changes of human DNA polymerase lambda using mass spectrometry-based protein footprinting. Journal of Molecular Biology. 2009;390(3):368–379. doi: 10.1016/j.jmb.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro C, Smidansky ED, Arnold JJ, et al. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nature Structural and Molecular Biology. 2009;16(2):212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AA, Fiala KA, Suo Z. DNA polymerases and their interactions with DNA and nucleotides. In: Vaghefi MM, editor. Nucleoside Triphosphates and Their Analogs: Chemistry, Biotechnology, and Biological Applications. Boca Raton, Fla, USA: Taylor & Francis; 2005. pp. 133–168. [Google Scholar]
- 8.Steitz TA. DNA polymerases: structural diversity and common mechanisms. Journal of Biological Chemistry. 1999;274(25):17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 9.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43(45):14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 10.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Research. 2008;18(1):148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107(1):91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 12.Mizukami S, Kim TW, Helquist SA, Kool ET. Varying DNA base-pair size in subangstrom increments: evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry. 2006;45(9):2772–2778. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]
- 13.Xu C, Maxwell BA, Brown JA, Zhang L, Suo Z. Global conformational dynamics of a Y-family DNA polymerase during catalysis. PLoS Biology. 2009;7 doi: 10.1371/journal.pbio.1000225. Article ID e1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohmori H, Friedberg EC, Fuchs RPP, et al. The Y-family of DNA Polymerases. Molecular Cell. 2001;8(1):7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 15.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147(4):1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. Journal of Biological Chemistry. 1999;274(23):15975–15977. doi: 10.1074/jbc.274.23.15975. [DOI] [PubMed] [Google Scholar]
- 17.Roush AA, Suarez M, Friedberg EC, Radman M, Siede W. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Molecular and General Genetics. 1998;257(6):686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη . Science. 1999;283(5404):1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 19.Yu S-L, Johnson RE, Prakash S, Prakash L. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Molecular and Cellular Biology. 2001;21(1):185–188. doi: 10.1128/MCB.21.1.185-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masutani C, Kusumoto R, Yamada A, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η . Nature. 1999;399(6737):700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 21.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Molecular Cell. 2001;8(2):417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 22.Alt A, Lammens K, Chiocchini C, et al. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase η . Science. 2007;318(5852):967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 23.Fiala KA, Suo Z. Mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43(7):2116–2125. doi: 10.1021/bi035746z. [DOI] [PubMed] [Google Scholar]
- 24.Cannistraro VJ, Taylor J-S. DNA-thumb interactions and processivity of T7 DNA polymerase in comparison to yeast polymerase η . Journal of Biological Chemistry. 2004;279(18):18288–18295. doi: 10.1074/jbc.M400282200. [DOI] [PubMed] [Google Scholar]
- 25.Garg P, Stith CM, Majka J, Burgers PMJ. Proliferating cell nuclear antigen promotes translesion synthesis by DNA polymerase ζ . Journal of Biological Chemistry. 2005;280(25):23446–23450. doi: 10.1074/jbc.C500173200. [DOI] [PubMed] [Google Scholar]
- 26.Fiala KA, Brown JA, Ling H, et al. Mechanism of template-independent nucleotide incorporation catalyzed by a template-dependent DNA polymerase. Journal of Molecular Biology. 2007;365(3):590–602. doi: 10.1016/j.jmb.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiala KA, Hypes CD, Suo Z. Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. Journal of Biological Chemistry. 2007;282(11):8188–8198. doi: 10.1074/jbc.M610718200. [DOI] [PubMed] [Google Scholar]
- 28.Washington MT, Prakash L, Prakash S. Yeast DNA polymerase η utilizes an induced-fit mechanism of nucleotide incorporation. Cell. 2001;107(7):917–927. doi: 10.1016/s0092-8674(01)00613-4. [DOI] [PubMed] [Google Scholar]
- 29.Brown JA, Suo Z. Elucidating the kinetic mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase B1. Biochemistry. 2009;48(31):7502–7511. doi: 10.1021/bi9005336. [DOI] [PubMed] [Google Scholar]
- 30.Fiala KA, Sherrer SM, Brown JA, Suo Z. Mechanistic consequences of temperature on DNA polymerization catalyzed by a Y-family DNA polymerase. Nucleic Acids Research. 2008;36(6):1990–2001. doi: 10.1093/nar/gkn004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai Y-C, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45(32):9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson KA. Transient-state kinetic analysis of enzyme reaction pathways. Enzymes. 1992;20:1–61. [Google Scholar]
- 33.Guckian KM, Schweitzer BA, Ren RX-F, Sheils CJ, Tahmassebi DC, Kool ET. Factors contributing to aromatic stacking in water: evaluation in the context of DNA. Journal of the American Chemical Society. 2000;122(10):2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fiala KA, Duym WW, Zhang J, Suo Z. Up-regulation of the fidelity of human DNA polymerase λ by its non-enzymatic proline-rich domain. Journal of Biological Chemistry. 2006;281(28):19038–19044. doi: 10.1074/jbc.M601178200. [DOI] [PubMed] [Google Scholar]
- 35.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Molecular Cell. 2001;8(2):407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 36.Acharya N, Brahma A, Haracska L, Prakash L, Prakash S. Mutations in the ubiquitin binding UBZ motif of DNA polymerase η do not impair its function in translesion synthesis during replication. Molecular and Cellular Biology. 2007;27(20):7266–7272. doi: 10.1128/MCB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Polη and Polδ by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(14):5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Washington MT, Johnson RE, Prakash S, Prakash L. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η . Journal of Biological Chemistry. 1999;274(52):36835–36838. doi: 10.1074/jbc.274.52.36835. [DOI] [PubMed] [Google Scholar]
- 39.Matsuda T, Bebenek K, Masultanl C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-η . Nature. 2000;404(6781):1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 40.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η . Journal of Biological Chemistry. 2000;275(11):7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Yuan F, Xin H, et al. Human DNA polymerase κ synthesizes DNA with extraordinarily low fidelity. Nucleic Acids Research. 2000;28(21):4147–4156. doi: 10.1093/nar/28.21.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohashi E, Bebenek K, Matsuda T, et al. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. Journal of Biological Chemistry. 2000;275(50):39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 43.Tissier A, McDonald JP, Frank EG, Woodgate R. polι, a remarkably error-prone human DNA polymerase. Genes and Development. 2000;14(13):1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 44.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dp04): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic polη . Nucleic Acids Research. 2001;29(22):4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kokoska RJ, Bebenek K, Boudsocq F, Woodgate R, Kunkel TA. Low fidelity DNA synthesis by a Y family DNA polymerase due to misalignment in the active site. Journal of Biological Chemistry. 2002;277(22):19633–19638. doi: 10.1074/jbc.M202021200. [DOI] [PubMed] [Google Scholar]
- 46.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. Journal of Biological Chemistry. 2002;277(18):15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 47.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO Journal. 2005;24(17):2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Wu X, Rechkoblit O, Geacintov NE, Taylor J-S, Wang Z. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Research. 2002;30(7):1630–1638. doi: 10.1093/nar/30.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Washington MT, Johnson RE, Prakash L, Prakash S. The mechanism of nucleotide incorporation by human DNA polymerase η differs from that of the yeast enzyme. Molecular and Cellular Biology. 2003;23(22):8316–8322. doi: 10.1128/MCB.23.22.8316-8322.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fiala KA, Suo Z. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43(7):2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 51.Washington MT, Johnson RE, Prakash L, Prakash S. Human DNA polymerase ι utilizes different nucleotide incorporation mechanisms dependent upon the template base. Molecular and Cellular Biology. 2004;24(2):936–943. doi: 10.1128/MCB.24.2.936-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howell CA, Prakash S, Washington MT. Pre-steady-state kinetic studies of protein-template-directed nucleotide incorporation by the yeast Rev1 protein. Biochemistry. 2007;46(46):13451–13459. doi: 10.1021/bi701429v. [DOI] [PubMed] [Google Scholar]
- 53.Sherrer SM, Brown JA, Pack LR, et al. Mechanistic studies of the bypass of a bulky single-base lesion catalyzed by a Y-family DNA polymerase. Journal of Biological Chemistry. 2009;284(10):6379–6388. doi: 10.1074/jbc.M808161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potapova O, Grindley NDF, Joyce CM. The mutational specificity of the Dbh lesion bypass polymerase and its implications. Journal of Biological Chemistry. 2002;277(31):28157–28166. doi: 10.1074/jbc.M202607200. [DOI] [PubMed] [Google Scholar]
- 55.Boudsocq F, Kokoska RJ, Plosky BB, et al. Investigating the role of the little finger domain of Y-family DNA polymerases in low fidelity synthesis and translesion replication. Journal of Biological Chemistry. 2004;279(31):32932–32940. doi: 10.1074/jbc.M405249200. [DOI] [PubMed] [Google Scholar]
- 56.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285(5425):263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 57.Washington MT, Prakash L, Prakash S. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase η . Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):12093–12098. doi: 10.1073/pnas.2134223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haracska L, Yu S-L, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η . Nature Genetics. 2000;25(4):458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 59.Carlson KD, Washington MT. Mechanism of efficient and accurate nucleotide incorporation opposite 7,8-dihydro-8-oxoguanine by Saccharomyces cerevisiae DNA polymerase η . Molecular and Cellular Biology. 2005;25(6):2169–2176. doi: 10.1128/MCB.25.6.2169-2176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η . Biochemistry. 2000;39(16):4575–4580. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 61.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η . EMBO Journal. 2000;19(12):3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassett E, Vaisman A, Tropea KA, et al. Frameshifts and deletions during in vitro translesion synthesis past Pt-DNA adducts by DNA polymerases β and η . DNA Repair. 2002;1(12):1003–1016. doi: 10.1016/s1568-7864(02)00150-7. [DOI] [PubMed] [Google Scholar]
- 63.Bassett E, Vaisman A, Havener JM, Masutani C, Hanaoka F, Chaney SG. Efficiency of extension of mismatched primer termini across from cisplatin and oxaliplatin adducts by human DNA polymerases β and η in vitro. Biochemistry. 2003;42(48):14197–14206. doi: 10.1021/bi035359p. [DOI] [PubMed] [Google Scholar]
- 64.Choi J-Y, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N 2-guanine DNA lesions by human DNA polymerase η . Journal of Molecular Biology. 2005;352(1):72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 65.Choi J-Y, Zang H, Angel KC, et al. Translesion synthesis across 1,N2-ethenoguanine by human DNA polymerases. Chemical Research in Toxicology. 2006;19(6):879–886. doi: 10.1021/tx060051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahn J, Kraynov VS, Zhong X, Werneburg BG, Tsai M-D. DNA polymerase β: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity and conformational changes. Biochemical Journal. 1998;331, part 1:79–87. doi: 10.1042/bj3310079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, Brown JA, Newmister SA, Suo Z. Polymerization fidelity of a replicative DNA polymerase from the hyperthermophilic archaeon Sulfolobus solfataricus P2. Biochemistry. 2009;48(31):7492–7501. doi: 10.1021/bi900532w. [DOI] [PubMed] [Google Scholar]
- 68.Lee HR, Johnson KA. Fidelity of the human mitochondrial DNA polymerase. Journal of Biological Chemistry. 2006;281(47):36236–36240. doi: 10.1074/jbc.M607964200. [DOI] [PubMed] [Google Scholar]
- 69.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-ι occurs by Hoogsteen base-pairing. Nature. 2004;430(6997):377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 70.Washington MT, Helquist SA, Kool ET, Prakash L, Prakash S. Requirement of Watson-Crick hydrogen bonding for DNA synthesis by yeast DNA polymerase η . Molecular and Cellular Biology. 2003;23(14):5107–5112. doi: 10.1128/MCB.23.14.5107-5112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong JH, Fiala KA, Suo Z, Ling H. Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. Journal of Molecular Biology. 2008;379(2):317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 72.Petruska J, Goodman MF, Boosalis MS, Sowers LC, Cheong C, Tinoco I., Jr. Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(17):6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]