Abstract
Targeted therapeutic and imaging agents are becoming more prevalent, and are used to treat increasingly smaller segments of the patient population. This has lead to dramatic increases in the costs for clinical trials. Biomarkers have great potential to reduce the numbers of patients needed to test novel targeted agents by predicting or identifying non-response early-on and thus enriching the clinical trial population with patients more likely to respond. Biomarkers are characteristics that are objectively measured and evaluated as indicators of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention. Biomarkers can be used to predict response to specific therapies, predict response regardless of therapy, or to monitor response once a therapy has begun. In terms of drug development, predictive biomarkers have the greatest impact, as they can be used as inclusion criteria for patient segmentation. Prognostic markers are used routinely in clinical practice but do not provide direction for the use of targeted therapies. Imaging biomarkers have distinct advantages over those that require a biopsy sample in that they are “non-invasive” and can be monitored longitudinally at multiple time points in the same patient. This review will examine the role of functional and molecular imaging in predicting response to specific therapies; will explore the advantages and disadvantages of targeting intracellular or extracellular markers; and will discuss the attributes of useful targets and methods for target identification and validation.
Keywords: Biomarker, targeted therapy, intracellular, extracellular, target
1. Introduction
In cancer treatment, there is an increasing reliance on therapeutic agents that are specifically targeted to gene products, pathways or physiologies. These can be matched, in a personalized fashion, to the presence of that target in an individual patient. It is axiomatic that targeted agents will impact increasingly smaller segments of the patient population. This invariably leads to staggering increases in the cost of clinical trials for drugs with an intended use by a small fraction of the patient population. In 2002, the average cost to bring a new drug to market was approximately $1.8 Billion and, unless there is a paradigm shift, this will increase with newer, more targeted therapies. The most egregious example comes from the trials the receptor tyrosine kinase (RTK) inhibitor, gefitinib (Iressa) in non small cell lung cancer (NSCLC). The response rates to this EGFR inhibitor are around 7% [1–4]. To get this approved, over 20,000 patients were enrolled and more than $2 Billion was spent worldwide before it was determined that the responders bore a specific EGFR polymorphism [5, 6]. Thus, if this trial had been enriched in this population to begin with, it would have been more efficient and less expensive. Admittedly, this is hindsight, as it can be argued that the presence of the EGFR polymorphism could not have been anticipated. However, high-resolution quantitative CT volume measurements taken 3 weeks after initiation of gefitinib therapy were capable of discriminating a population that was highly enriched in patients with the polymorphism [7]. Thus, without knowledge of the molecular mechanisms underlying sensitivity or resistance, quantitative imaging could have been used to enrich a clinical trial population by excluding non-responding patients early in the trial process.
There is a consensus belief that an appropriate biomarker could have significantly increased the efficiency of these trials, either by predicting which patients would respond to RTK inhibition by either pre-identifying patients who harbored the mutation, or by monitoring the patients who had an immediate response following therapy initiation. A biomarker is a “characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.” [8]. In the last few years, biomarkers have been incorporated into, and explicitly used to affect the course of clinical trials [9].
In therapeutics, biomarkers can be used to predict response to specific therapies, predict response regardless of therapy, or to monitor response once a therapy has begun (Box 1). In terms of drug development, Predictive Biomarkers have the greatest impact, as they can be used as inclusion criteria for patient segmentation. In other words, does the patient express the drug target? For example, if 10% of the patient population is 'target-positive', 159 patients would be needed to obtain 90% power, whereas only 37 patients would be needed if the target-positive fraction was 45%.1 Prognostic markers are used routinely in clinical practice but do not provide direction for the use of targeted therapies. Response biomarkers, especially for imaging, are finding increased application in trials, as described below.
Box 1. Types of therapy biomarkers
Response: Used to measure a quantitative change in response to therapy.
Prognostic: Used to predict patient outcome regardless of therapy.
Predictive: Used to predict response to a specific therapy.
A number of targeted therapies have been developed and approved in recent years, e.g. trastuzumab and lapatinib for treatment of HER-2 positive metastatic breast cancer [10], and there are many more currently in development. Hence, biomarkers have great potential for use as companion diagnostics through the identification of patients likely to respond to a specific targeted therapeutic agent.
Serum-based biomarkers are clearly and reasonably a major focus of discovery, as samples can be readily obtained and tested, and are therefore generally cost-effective [11, 12]. Many putative blood derived biomarkers have been identified, but few are currently validated and used routinely in the clinic. The best example of a validated serum-based biomarker is prostate serum antigen (PSA) for prostate cancer. However the utility of PSA has recently been questioned due to low sensitivity and specificity for diagnosis [13]. Validation and development of serum–based biomarkers is therefore of great importance. Most validated cancer biomarkers in use today are either tumor gene expression or immunohistochemically based [14–17]. Unfortunately, these approaches require biopsies and thus are inappropriate to be used to monitor therapy response. Modern imaging biomarkers have distinct advantages over these non-serum based biomarkers in that they are generally considered to be “non-invasive” and hence, can be monitored longitudinally at multiple time points in the same patient.
In this review, we will examine the role of functional and molecular imaging in predicting response to specific therapies. The use of imaging biomarkers to monitor response to targeted therapy has recently been reviewed by us [18].
2. Imaging Biomarkers
Response Evaluation Criteria in Solid Tumors (RECIST) are MR or CT based anatomic unidimensional measurements that are commonplace in evaluating “objective response” clinical trials [19, 20]. Notably, changes in tumor size following therapy do not always indicate a better clinical outcome, which is necessary to qualify these as “surrogate” markers of response [21]. This can be expected with cytostatic and targeted therapies, so there has been a push to develop more sensitive imaging biomarkers for measurement of tumor function or molecular expression patterns. Some of these approaches are listed in Box 2.
Box 2. Imaging biomarkers
Anatomic: RECIST, volumetrics (CT, MRI).
Functional: Dynamic Contrast Enhanced-MRI, Diffusion MRI, Magnetization Transfer MRI, and Macromolecule contrast.
Molecular: PET (Receptors, metabolites, hypoxia, pH), CT/US/MR Nanoparticles, and MR (Spectroscopy, pH, hypoxia, chemical exchange saturation transfer, hyperpolarization).
Functional Imaging measures physical aspects of tumor physiology (e.g. perfusion, hypoxia, pH) that may be targets for therapy. For example, blood flow and vascular permeability can be measured by dynamic contrast enhanced (DCE-) MRI and this has increasingly been used as a predictive and response biomarker for anti-angiogenic therapies [22]. Notably, intratumoral heterogeneity of response may be a more powerful predictor than the average measurement, which is a clear advantage of imaging that can sample the entire tumor volume [23]. Diffusion MRI is a measure of cell density, which can also be prognostic and does change with successful therapy [24]. In select cases tumor hypoxia can be predicted with Blood-oxygen-level-dependent (BOLD) MRI, which is sensitive to levels of deoxyhemoglobin [25]. Hypoxia can also be quantitatively imaged with PET tracers, such as F-misonidazole [26]. A low insertion peptide tracer has recently been developed to be sensitive to tumor pH [27], which can also be used to target some new nanoparticle based therapies.
Molecular Imaging measures specifically targeted expression or activity of gene products. These are intimately coupled to predicting effects of targeted therapies so will be the subject of the rest of this review. PET is commonly referred to as a molecular imaging modality, because its sensitivity is orders of magnitude higher than that of CT or MR. The most common PET tracer is fluoro-deoxy glucose (FDG), which has been used to measure the activity of glucose transporters in over a million patients worldwide [28]. Although development of PET tracers is not trivial, over 100 are available for research use and thus, are too many to enumerate here. Readers are pointed to recent reviews [29, 30]. Despite low sensitivity, MR probes for highly expressed receptors (e.g. HER-2) have been developed [31]. A unique aspect of MR is the ability to spatially resolve metabolites with different spectral resonance frequencies, called magnetic resonance spectroscopic imaging, MRSI. This is generally limited to cholines, creatine, and lactate, but has been used effectively to diagnose and prognose breast, prostate and brain cancers in humans. This has also been recently reviewed by us [32]. A new technology involves the hyperpolarization of MR-active stable nuclei, which can result in 10–50,000 fold increases in sensitivity and the ability to image metabolism in vivo [33]. Although early work focused on the measurement of 13C enriched pyruvate, more recent studies have been interrogating other 13C labeled substrates, as well as tracers using other MR active nuclei [34–40].
Nanoparticles themselves deserve special mention as they can be theragnostic to deliver therapy, as well as imaging contrast [41, 42]. This is a rapidly changing field and a recent review has identified over 100 different formulations for using nanoparticles in imaging and therapy [43]. Notably, many of these agents are responsive, and thus will be activated by specific cancer expression patterns or environments, such as pH [44].
3. Intracellular vs. Extracellular Targets?
In developing imaging agents, as well as targeted therapeutics, the most important decision is whether the target of the therapy (or imaging agent) will be intra-or extra-cellular. There are clear advantages and disadvantages for each location. Intracellular targets are advantageous in that there are many more available targets, compared to extracellular targets, and many of these are the focus of anti-cancer drug discovery efforts. However, intracellular agents must cross the plasma membrane barrier either by carrier mediated transport or by passive diffusion. Hence the design criteria for intracellular agents are much more stringent in terms of hydrophilicity, chemical nature and size. While extracellular markers may be limited in number, they have been successfully targeted by both therapy and imaging. An added benefit of using cell-surface markers for targeting is that the agent can act independently of the molecular function of the target, i.e. the target can act as a landing pad for delivery of an imaging or therapeutic agent without the necessity for a complete understanding of target biology. This allows selection of obscure targets that were recently identified by the human genome project where little is yet known about the cellular function. The remainder of this review will illustrate examples for each type of target, and end by considering criteria in evaluating what it takes to validate a target.
4. Intracellular Targets
The following examples demonstrate molecular imaging of intracellular targets using PET, diffusion-weighted MRI and optical imaging modalities:
4.1 Alpha methyl tryptophan (AMT)
Tryptophan is an essential amino acid required for biosynthesis of proteins, serotonin, and nicotinamide adenine dinucleotide (NAD/NADP). Recent studies indicate that tryptophan oxidation via the enzyme indoleamine 2,3-dioxygenase (IDO) can modulate immunoresistance of cancers [45, 46]. Most human tumors constitutively express IDO, and IDO products (i.e. kynurenine) in tumors may also exert a potent immunosuppressive effect by blocking T-lymphocyte proliferation, thus diminishing T cell mediated tumor rejection [47–52]. Thus, manipulation of tryptophan metabolism via the kynurenine pathway may have important implications in tumor immunotherapy. Recent studies have consistently shown expression of IDO in a variety of resected human extra-cerebral tumors, including lung cancer [53, 54], colorectal cancer [46, 55], breast cancer [56], hepatocellular carcinoma [57], prostate cancer [46], pancreatic carcinoma [46], and ovarian cancer [58]. Several of these studies demonstrated that high expression of IDO was associated with a reduced survival [53, 55, 58]. These findings suggest that IDO could be a powerful prognostic marker in a variety of extra-cerebral tumors.
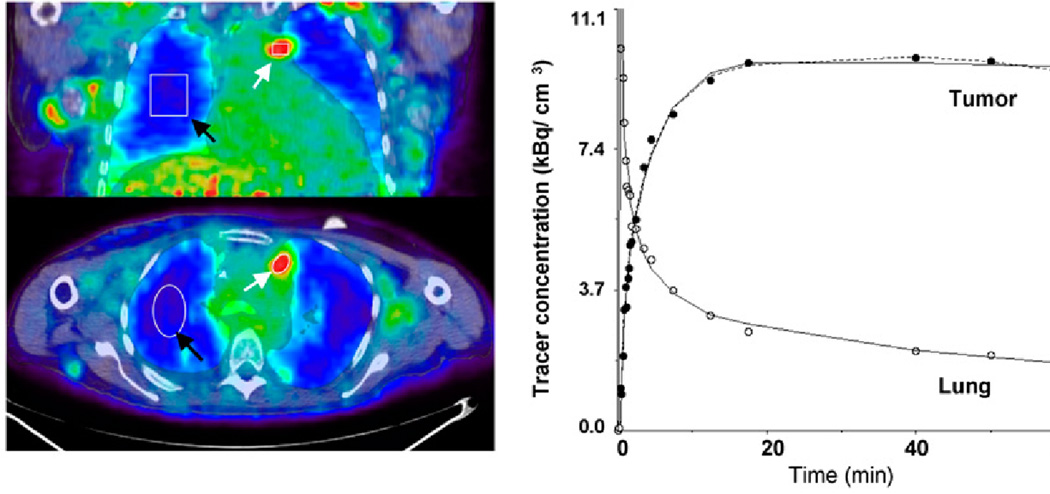
In vivo detection of IDO may facilitate therapeutic measures by identifying patients likely to benefit and to quantitatively monitor IDO inhibition in vivo. The positron emission tomography (PET) radiotracer alpha-[11C]methyl-L-tryptophan (AMT) is metabolized by IDO to yield kynurenine pathway products. In preliminary PET studies of children and adults with brain tumors, increased AMT uptake was found in a variety of human gliomas and even glioneuronal tumors [59]. AMT PET was able to differentiate between tumors and non-cancerous lesions. A peculiar feature of AMT was its accumulation in both high- and low-grade tumors, but with different kinetics. A second study investigated AMT PET imaging in ten NSCLC patients [60]. It found that the majority of those tumors demonstrated prolonged AMT uptake suggesting increased IDO activity (Figure 1).
Figure 1.
Images of AMT tracer uptake between 40 and 60 min after injection (left) in patient with multiple local NSCLC metastases. Images show high accumulation in tumor tissue and excellent contrast between tumor and lung tissue. ROIs were defined for tumor nodules (white arrows) and lung tissue (black arrows). Corresponding tumor time–activity curve (right) indicates rapid initial uptake of AMT, followed by slight decrease at late time points. Curve fit applying reduced compartmental model (k4 5 0) is shown as solid line, and full compartmental model is displayed as broken line. In addition, 2-compartmental model fit is shown for lung tissue. Image from reference 60.
4.2 Misonidazoles
One of the more exciting probes in this arena is 18F-misonidazole, F-MISO [61]. As with other 2-nitroimidazoles, F-MISO is specifically targeted to viable hypoxic cells through the action of oneelectron reductases, such as cytochrome P450 reductase. In the absence of molecular oxygen, a highly reactive hydroxylamine is formed which covalently attaches to cellular proteins [62]. Hence, this tracer can be used to identify hypoxic tumor regions, which are negatively prognostic for radiotherapy and can be positive predictors of response to hypoxia activated prodrugs, such as tirapazamine or TH-302 [63].
4.3 Diffusion MRI
Diffusion weighted MRI is used to calculate the apparent diffusion coefficient (ADC) of water inside the tissue being imaged. Increased water mobility is associated with decreased cellularity within the tissue and, thus, increased ADC values. DW-MRI is being evaluated as method for diagnosis, grading and staging and of a number of cancers. Recent reviews outline the efficacy and use of DW-MRI for characterization of breast cancer [64] and head and neck cancers [65]. ADC is being evaluated as a prognostic indicator and as a predictor of therapeutic efficacy. For pancreatic cancer, lower ADC values were predictive of tumor progression, poor prognosis, in patients imaged 3 or 6 months after treatment [66]. For glioblastoma multiforme (GBM), it was hypothesized that bevacizumab treatment would be most effective for highly necrotic tumors. Analysis of recurrent GBM tumors demonstrated that tumor ADC can be used to predict the efficacy of bevacizumab treatment prior to initiation of therapy [67].
4.4 Reporter genes
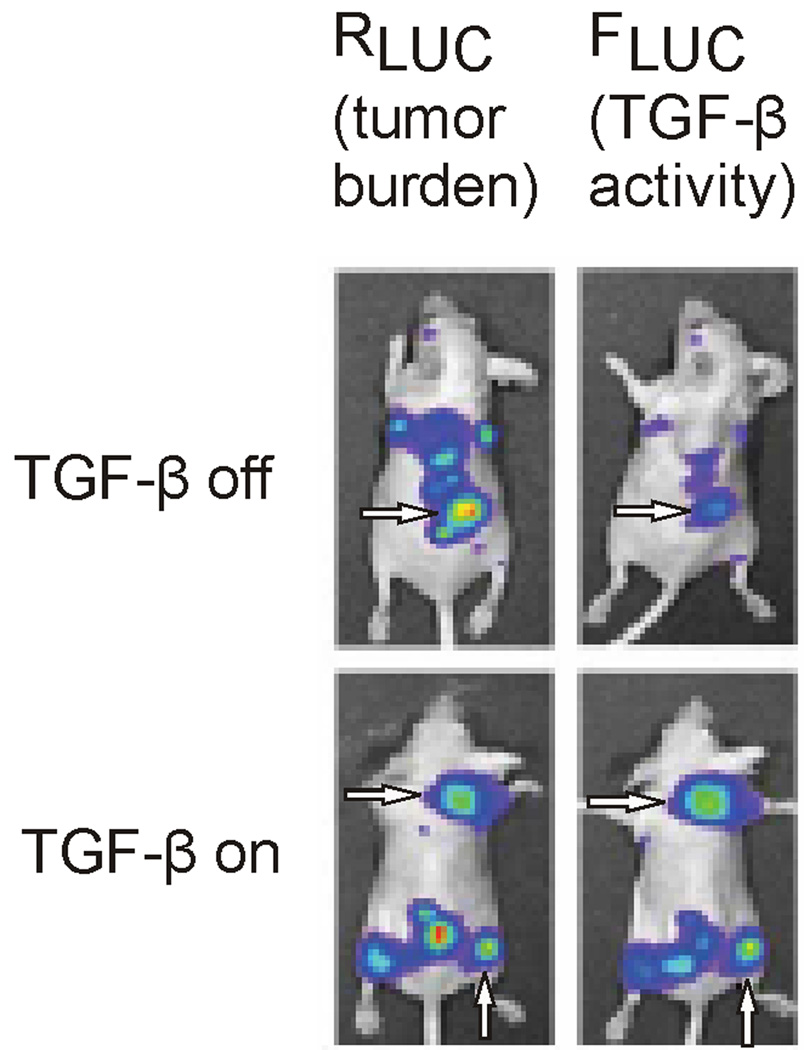
Although only useful for preclinical studies at this time, a multitude of reporter-constructs are commercially available for imaging activation of specific pathways. Cellular signaling is detected by expression of luciferase or fluorescence protein, e.g. GFP, under the control of a promoter sequence that is activated by a transcription factor associated with the signaling pathway of interest. Bioluminescence in cells carrying these reporter constructs is related to activation of the pathway being studied. While these constructs are commonly used in cell-based assays, xenografts have been used to study the effect of modulating signaling pathways in vivo. Recently, the dependence of TGF-β-SMAD signaling on growth of osteolytic bone metastases was studied in vivo using a dual reporter system in mice [68]. Cells used to form the bone metastases constitutively expressed Renilla luciferase (RLUC) to determine tumor burden, and expressed firefly luciferase (FLUC) upon activation of the TGF-β pathway. Luminescence originating from the two luciferases can be distinguished by their respective peak emission wavelengths. Administration of TGF-β and inhibitors was used to study pathway activation and pathway dependent tumor growth (Figure 2). Infrared fluorescent protein (IFP) was recently expressed in mammalian cells with excitation and emission maxima of 684 and 708 nm respectively [69]. Because these wavelengths penetrate tissue with minimal autofluorescence, reporter constructs using IFP may allow for the in vivo study of pathways in small animals without the need for injection of the luciferase substrate luciferin.
Figure 2.
In vivo chemiluminescence imaging of a dual expression system in mouse bone metastases, with RLUC expression (left panels), under control of the constitutive CMV promoter, representing tumor burden and FLUC expression (right panels), under control of the multiple SMAD binding element (SBE) promoter which is activated by the TGF-β receptor mediated pathway. Note that when the TGF-β pathway is off (top panels) the FLUC signal (top right, arrow) is greatly reduced relative to the corresponding RLUC expression (top left, arrow) which is representative of the tumor burden; and that when the TGF-β pathway is on (bottom panels) the FLUC signal (bottom right, arrows) is comparable to the corresponding RLUC expression (bottom left, arrows). Image from reference 68.
5. Extracellular targets
As mentioned above, the design criteria for extracellular targeted agents are significantly less stringent than those for intracellular targets. However, there are significant sequelae to large hydrophilic agents that must be considered. First the ADME (absorption, distribution, metabolism and excretion) pharmacokinetics can be complex, especially for large extracellular agents. Particles up to 100 nm can extravasate from neovasculature and can thus be retained in tumors by Enhanced Permeability and Retention (EPR), even without targeting [70, 71]. They may also have very long plasma half-lives if they are too large (> 50 KDa) for renal clearance and are not substrates for reticuloendothelial (RES) clearance. This can have significance to imaging agents, which rely on clearance to increase conspicuity and can have off target toxicities if the pharmacokinetic AUC is large, such as Gadolinium-induced nephrogenic systemic fibrosis, NSF [42, 72]. Although there is a temptation to combine imaging and therapeutic agents to target receptors, this may not always be appropriate as therapeutics are often antagonists with very high affinities, yet imaging agents are often better as agonists, as they can induce receptor recycling and hence, amplification of signal [73]. The following are examples of ligands, antibodies, nanoparticles and substrates that have been successfully used to target extracellular proteins.
5.1 Ligands
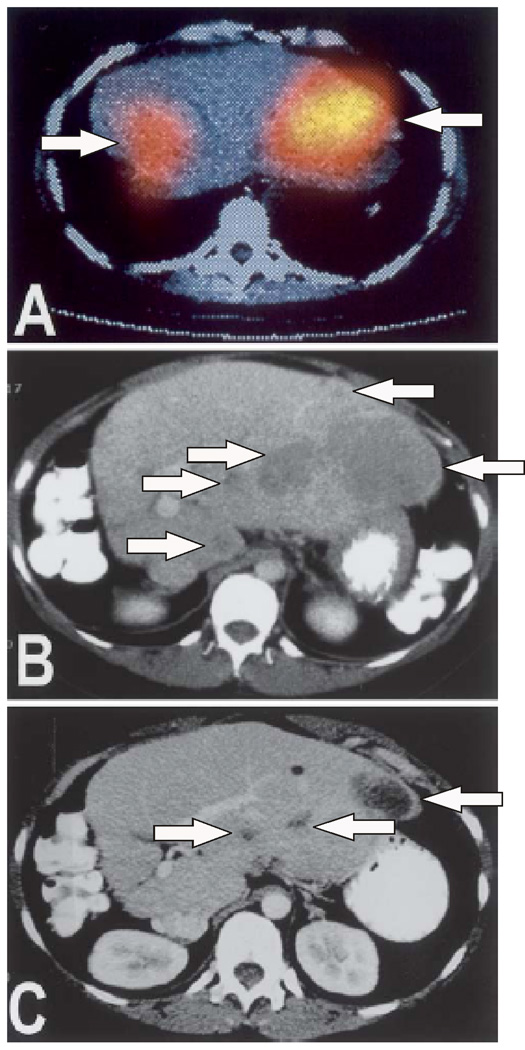
Small molecule ligands for receptors have significant advantages over larger agents, in terms of more rapid clearance, and higher tumor-to-background values due to lower EPR effect. Illustrative examples include peptide and peptidomimetic ligands for integrins, and receptors for somatostatin and folic acid. Integrins are hetero bivalent cell surface receptors that mediate cell-cell and cell-martix interactions [74]. Notably, integrins of different classes are known to be aberrantly expressed in growing cancers. For example, αvβ3 and αvβ5 integrins are expressed in angiogenic vasculature, which occurs in tumors and in wound healing. With the discovery that an arginine-glycine-aspartate (RGD) motif will selectively target αvβ3 and αvβ5, this motif has been used in numerous platforms (e.g. PET, MR, optical) to image angiogenesis in vivo [75–79] and to mediate the delivery of therapeutic agents [80, 81]. Recently, non-peptide ligands for αvβ3 and αvβ5 have been developed and these have been incorporated into optical dyes that are commercially available [82]. Additionally, the somatostatin receptor, SSTR2, is known to be over-expressed in a number of neuroendocrine tumors. Radiolabeled ligands for SSTR2 are readily available and are used in patients both for in vivo molecular diagnosis but also for delivery of radiotherapy (Figure 3) [83]. Similarly, the folic acid receptor is known to be upregulated in a number of cancers, and has also been developed as a target for multimodal imaging agents [44, 84–86].
Figure 3.
Liver metastases from pancreatic insulinoma before and after radiolabeled therapy targeting the somatostatin receptor (SSTR2). A) SSTR2 targeted molecular imaging agent (OctreoScan) imaging prior to treatment, arrows indicate areas of signal related to the agent; B) CT prior to treatment, arrows indicate liver metastases; and C) CT image showing reduction in liver lesions 8 months post therapy, arrows indicate location of reduced metastases. Image from reference 83.
5.2 Antibodies
Over the past decade, antibodies have been developed to successfully target cancers. Antibodies can work by inhibiting their targets or for delivery of radiotherapy. Examples include bevacizumab (a.k.a. avastin) inhibits VEGF, trastuzumab (a.k.a. herceptin) inhibits Her-2/neu and rituximab (a.k.a. rituxan) inhibits CD20+ cells. These can also be modified to carry therapy, such as radiotherapy, in the case of anti-CD20 with Bexxar. Examples of the latter include anti-CD20 and Her-2/neu antibodies.
5.3 Substrates
Enzymes can also be used to activate contrast agents. This has distinct advantages in that there can be tremendous amplification of signals in a localized area. There are many fine examples of this approach, including MRI of myeloperoxidase [87, 88] or hyaluronidase [89], as well as protease activation of optical agents, which are commercially available [82, 90].
6. What Makes a “Good” Target?
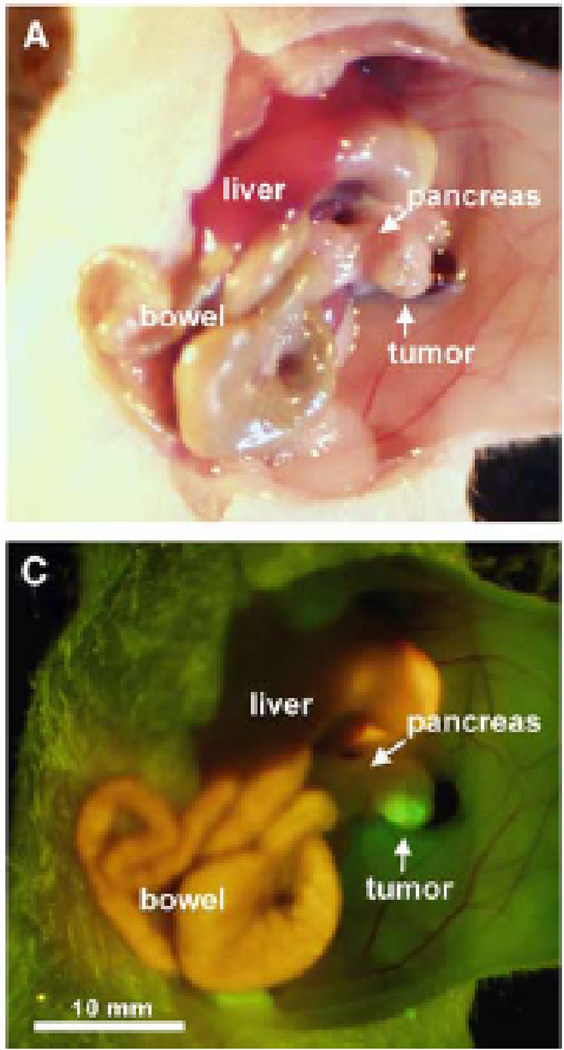
Ideally a cellular marker for targeted imaging or therapy is expressed in the target tissue of a given pathology, but is not expressed in any other normal, unaffected tissue. In this case a drug could be conjugated to a ligand or substrate specific for the marker, administered systemically and be readily localized at high concentration to the affected tissue, while maintaining low systemic concentrations and, hence, low off-target toxicities. In effect increasing the therapeutic window, where tolerable systemic dosages of the drug result in highly elevated and effective levels at the target site. Similarly in the case of an imaging agent, although there may be low toxicity associated with the agent, ideal targets are not expressed in normal tissues in order to avoid false-positive and confounding image results. Unfortunately very few, if any, cellular markers have been identified that have this property of non-expression in unaffected tissues. Because of this, it is important to evaluate target expression in unaffected tissues while considering the intended use of the agent to be developed. For example, an imaging agent designed to distinguish a primary pancreatic tumor for the purpose of intra-operative surgical guidance by determining tumor margins (Figure 4) [91], should be highly and broadly expressed amongst all pancreatic tumor types, but not expressed in normal pancreas. Other than that, expression in other tissues may be tolerable as long as there are no serious associated toxicities or difficulty in distinguishing a lesion due to expression in a tissue of close proximity. To avoid toxicity, it is best if markers are not expressed in vital organs or organs involved in clearance, e.g. heart, lung, liver and kidney. Such an imaging agent may also be used for molecular characterization of the disease, as markers may be related to specific prognoses and may be associated with effectiveness of a specific therapeutic regimen.
Figure 4.
Intravital imaging of pancreatic cancer following orthotopic implantation in mouse pancreas using a tumor targeted agent, fluorophore conjugated CA19-9 antibody. CA19-19 is a carbohydrate tumor-associated antigen found in up to 94% of pancreatic adenocarcinomas. A) Brightfield image of CA 19-9 treated subject and C) fluorescence image of treated subject with bright tumor associated fluorescence. Image from reference 91.
6.1 Target Identification
Many targets are reported in the literature as being specific for various pathologies. It is important to understand how a target was identified and whether or not expression in normal tissues was also considered in the study. In the case of cancer, few targets have been identified that cover all types of cancer from a given site of origin, e.g. breast or pancreas. Hence, it is important that a large number of samples are used for screening that represent the full range of disease types. Understanding the coverage of a target amongst the disease subtypes, can allow for development of a cocktail of agents that cover the entire range of disease and can provide for the molecular characterization of disease subtype in an individual patient.
Gene expression profiling of DNA microarray data generated from patient and normal “unaffected” tissue samples is an effective approach for target identification. DNA microarray is an established and relatively inexpensive technology with high-throughput capability for determination of mRNA expression in virtually every gene in the human genome. Expression of mRNA is a prerequisite for protein expression, but due to post-transcriptional and post-translational processing, detection of mRNA expression does not necessarily correlate with the presence of the corresponding protein product. Therefore, once a target is identified as being expressed as mRNA, it is essential that protein expression be validated. Alternatively, proteomics methods are being developed that can determine protein expression in tissue samples with high-throughput [92]. Proteomics allows for the immediate and quantitative determination of target protein.
6.2 Target Validation
Prior to undertaking the significant effort and expense of developing a targeted agent, it is necessary that expression and sub-cellular localization of the target protein be evaluated for expression amongst a broad range of diseased and unaffected tissues. Targets with a known ligand or substrate, or known structure activity relationships (SAR) are given priority as this will aid in ligand and agent development following validation. Differential expression, expression levels, and breadth of coverage amongst subtypes of the disease may also be considered when ranking identified targets for further validation. As most potential targets are expressed in a subset of normal, unaffected tissues, it is critical that the expression profile of each target be fully validated in a wide range, if not all, of normal tissues. As discussed above, this is necessary to select targets that are appropriate for a given imaging or therapeutic application. If target expression is not identified via a large-scale proteomics screen as described above, e.g. by mRNA expression profiling or otherwise, validation of protein expression may be performed via mass spectrometry-based proteomics using cell lines or tissue biopsy samples, or using immunohistochemistry (IHC) of diseased and unaffected tissues in tissue microarrays. IHC requires the availability of a highly specific antibody for the target, but, in the case of cancer, provides the added benefit of distinguishing expression in tumor cells, versus infiltrating vascular or immune system cells, different cell types within a given normal tissue, or normal cells adjacent to the pathology. In some cases, IHC can distinguish subcellular localization, e.g. cell-surface versus cytoplasmic or nuclear staining.
It is best if multiple validations are performed, e.g. for cancer targets, validation in established tumor cell lines allows for their use in pre-clinical evaluation of targeted agents by whole cell assays, or by generation of orthotopic tumor xenografts in small animals for in vivo testing. Additional validations include the use of publicly available data, such as in the Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/projects/geo/index.cgi) for mRNA expression, or at the Human Protein Atlas (http://www.proteinatlas.org) for IHC protein expression, and in the scientific literature. These validations are a useful adjunct to the direct validation of protein expression in patient samples by IHC or proteomics.
6.3 Tissue vs. Cell Lines
Although determining expression in established cell lines is recommended for tertiary validations, the use of tissue samples for the initial target identification screen and initial validation of protein expression is superior to identification of targets solely from established tumor lines. Cell lines are cultured in artificial conditions that do not fully recapitulate the expression patterns found in the tumor microenvironment, e.g. gene expression can be regionally altered or induced by microenvironmental conditions within a tumor such as hypoxia and pH, as is seen with regional expression of hypoxia response genes [93]. However, using tumor tissues involves the analysis of a mixture of cell types, tumor and host, and may have a confounding effect on the analysis, e.g. tumor vasculature, or, in the case of pancreatic cancer, the tumor microenvironment includes a pronounced stromal component [94, 95]. This effect can be reduced by micro- or macro-dissection of the tissue fortifying for cancer cells. However, targeting the tumor stroma may have a therapeutic advantage [96] and should be considered during agent design, as stromal interactions may potentially influence the efficacy of a targeted agent. Care should also be taken when using xenograft tumor tissue as a surrogate for patient tissue, as the microevironmental makeup of xenografts likely does not reflect the corresponding microenvironment in a patient tumor.
7. Conclusion
Molecular imaging and therapeutic agents are being developed for intracellular and extracellular targets. These new agents are useful for molecular characterization of disease, and may be used as predictive biomarkers for therapeutic response, as diagnostic determiners of disease state, and as prognostic indicators of patient outcomes. Such agents hold great promise for reducing the cost of specialized drug development and for improving patient outcomes by directing therapy and non-invasively providing diagnoses. Understanding the expression profile of a putative target protein in a broad range of affected and unaffected tissues is essential for development of targeted imaging and therapeutic agents. Ideal targets that exhibit high expression in a broad range of a given pathology but low or non-expression in unaffected tissues are scarce. However, markers may be identified that are useful for targeting sub-populations of disease, and may be used for the molecular characterization of disease and as predictive or prognostic indicators. Also, while considering the intended use of the agent to be developed, target expression in some normal, unaffected tissues may be tolerated if expression is relatively lower than in disease, or if the expression is not in a vital tissue associated with clearance or toxicity, or is not proximal to the organ site of interest for imaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Assuming ‘target-positive’ has a response rate of 70%; a 'Target-negative' patient has response rate of 5% and a 5% spontaneous regression rate, the (α=0.05 with two-sided Fisher's Exact test).
Contributor Information
David L. Morse, Email: David.Morse@moffitt.org.
Robert J. Gillies, Email: Robert.Gillies@moffitt.org.
References
- 1.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 2.Kris MG, Natale RB, Herbst RS, Lynch TJ, Jr, Prager D, Belani CP, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. Jama. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- 3.Giaccone G, Herbst RS, Manegold C, Scagliotti G, Rosell R, Miller V, et al. Gefitinib in combination with gemcitabine and cisplatin in advanced non-small-cell lung cancer: a phase III trial--INTACT 1. J Clin Oncol. 2004;22:777–784. doi: 10.1200/JCO.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Giaccone G, Schiller JH, Natale RB, Miller V, Manegold C, et al. Gefitinib in combination with paclitaxel and carboplatin in advanced non-small-cell lung cancer: a phase III trial--INTACT 2. J Clin Oncol. 2004;22:785–794. doi: 10.1200/JCO.2004.07.215. [DOI] [PubMed] [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science (New York, NY. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Zhao B, Schwartz LH, Moskowitz CS, Ginsberg MS, Rizvi NA, Kris MG. Lung cancer: computerized quantification of tumor response--initial results. Radiology. 2006;241:892–898. doi: 10.1148/radiol.2413051887. [DOI] [PubMed] [Google Scholar]
- 8.Arthur J, Atkinson J, MD, Wayne A, Colburn P, Victor G, DeGruttola S, David L, DeMets P, Gregory J, Downing D, PhD, Daniel F, Hoth M, et al. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clinical pharmacology and therapeutics. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 9.Tan DSW, Thomas GV, Garrett MD, Banerji U, de Bono JS, Kaye SB, et al. Biomarker-driven early clinical trials in oncology: a paradigm shift in drug development. Cancer Journal. 2009;15:406–420. doi: 10.1097/PPO.0b013e3181bd0445. [DOI] [PubMed] [Google Scholar]
- 10.Tagliabue E, Balsari A, Campiglio M, Pupa SM. HER2 as a target for breast cancer therapy. Expert Opin Biol Ther. doi: 10.1517/14712591003689972. [DOI] [PubMed] [Google Scholar]
- 11.Wei B-R, Hoover SB, Ross MM, Zhou W, Meani F, Edwards JB, et al. Serum S100A6 concentration predicts peritoneal tumor burden in mice with epithelial ovarian cancer and is associated with advanced stage in patients. PLoS ONE [Electronic Resource] 2009;4:e7670. doi: 10.1371/journal.pone.0007670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lausted C, Hu Z, Hood L. Quantitative serum proteomics from surface plasmon resonance imaging. Molecular & Cellular Proteomics. 2008;7:2464–2474. doi: 10.1074/mcp.M800121-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Leman ES, Getzenberg RH. Biomarkers for prostate cancer. J Cell Biochem. 2009;108:3–9. doi: 10.1002/jcb.22227. [DOI] [PubMed] [Google Scholar]
- 14.Rubin MA. Using molecular markers to predict outcome. Journal of Urology. 2004;172:S18–S21. doi: 10.1097/01.ju.0000142448.58831.d9. discussion S-2. [DOI] [PubMed] [Google Scholar]
- 15.Ross JS, Schenkein DP, Kashala O, Linette GP, Stec J, Symmans WF, et al. Pharmacogenomics. Advances in Anatomic Pathology. 2004;11:211–220. doi: 10.1097/01.pap.0000131825.77317.ee. [DOI] [PubMed] [Google Scholar]
- 16.Wagner JA. Overview of biomarkers and surrogate endpoints in drug development. Disease markers. 2002;18:41–46. doi: 10.1155/2002/929274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lesko LJ, Atkinson AJ., Jr Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: criteria, validation, strategies. Annual review of pharmacology and toxicology. 2001;41:347–366. doi: 10.1146/annurev.pharmtox.41.1.347. [DOI] [PubMed] [Google Scholar]
- 18.Stephen RM, Gillies RJ. Promise and progress for functional and molecular imaging of response to targeted therapies. Pharmaceutical Research. 2007;24:1172–1185. doi: 10.1007/s11095-007-9250-3. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz LH, Bogaerts J, Ford R, Shankar L, Therasse P, Gwyther S, et al. Evaluation of lymph nodes with RECIST 1.1. Eur J Cancer. 2009;45:261–267. doi: 10.1016/j.ejca.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 20.Curran SD, Muellner AU, Schwartz LH. Imaging response assessment in oncology. Cancer Imaging. 2006;6:S126–S130. doi: 10.1102/1470-7330.2006.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birchard KR, Hoang JK, Herndon JE, Jr, Patz EF., Jr Early changes in tumor size in patients treated for advanced stage nonsmall cell lung cancer do not correlate with survival. Cancer. 2009;115:581–586. doi: 10.1002/cncr.24060. [DOI] [PubMed] [Google Scholar]
- 22.Wilmes LJ, Pallavicini MG, Fleming LM, Gibbs J, Wang D, Li K-L, et al. AG-013736, a novel inhibitor of VEGF receptor tyrosine kinases, inhibits breast cancer growth and decreases vascular permeability as detected by dynamic contrast-enhanced magnetic resonance imaging. Magnetic Resonance Imaging. 2007;25:319–327. doi: 10.1016/j.mri.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 23.Rose CJ, Mills SJ, O'Connor JPB, Buonaccorsi GA, Roberts C, Watson Y, et al. Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magnetic Resonance in Medicine. 2009;62:488–499. doi: 10.1002/mrm.22003. [DOI] [PubMed] [Google Scholar]
- 24.Patterson DM, Padhani AR, Collins DJ. Technology insight: water diffusion MRI--a potential new biomarker of response to cancer therapy. Nature Clinical Practice Oncology. 2008;5:220–233. doi: 10.1038/ncponc1073. [DOI] [PubMed] [Google Scholar]
- 25.O'Connor JPB, Naish JH, Parker GJM, Waterton JC, Watson Y, Jayson GC, et al. Preliminary study of oxygen-enhanced longitudinal relaxation in MRI: a potential novel biomarker of oxygenation changes in solid tumors. International Journal of Radiation Oncology, Biology, Physics. 2009;75:1209–1215. doi: 10.1016/j.ijrobp.2008.12.040. [DOI] [PubMed] [Google Scholar]
- 26.Padhani AR, Krohn KA, Lewis JS, Alber M. Imaging oxygenation of human tumours. European radiology. 2007;17:861–872. doi: 10.1007/s00330-006-0431-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vavere AL, Biddlecombe GB, Spees WM, Garbow JR, Wijesinghe D, Andreev OA, et al. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Cancer research. 2009;69:4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gambhir SS, Czernin J, Schwimmer J, Silverman DH, Coleman RE, Phelps ME. A tabulated summary of the FDG PET literature. J Nucl Med. 2001;42:1S–93S. [PubMed] [Google Scholar]
- 29.Margolis DJ, Hoffman JM, Herfkens RJ, Jeffrey RB, Quon A, Gambhir SS. Molecular imaging techniques in body imaging. Radiology. 2007;245:333–356. doi: 10.1148/radiol.2452061117. [DOI] [PubMed] [Google Scholar]
- 30.Alford R, Ogawa M, Choyke PL, Kobayashi H. Molecular probes for the in vivo imaging of cancer. Molecular bioSystems. 2009;5:1279–1291. doi: 10.1039/b911307j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu W, Okollie B, Bhujwalla ZM, Artemov D. PAMAM dendrimer-based contrast agents for MR imaging of Her-2/neu receptors by a three-step pretargeting approach. Magn Reson Med. 2008;59:679–685. doi: 10.1002/mrm.21508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillies RJ, Morse DL. In vivo magnetic resonance spectroscopy in cancer. Annual review of biomedical engineering. 2005;7:287–326. doi: 10.1146/annurev.bioeng.7.060804.100411. [DOI] [PubMed] [Google Scholar]
- 33.Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer research. 2006;66:10855–10860. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- 34.Witney TH, Kettunen MI, Day SE, Hu DE, Neves AA, Gallagher FA, et al. A comparison between radiolabeled fluorodeoxyglucose uptake and hyperpolarized (13)C-labeled pyruvate utilization as methods for detecting tumor response to treatment. Neoplasia (New York, NY. 2009;11:574–582. doi: 10.1593/neo.09254. 1 p following 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto S, Yasui H, Batra S, Kinoshita Y, Bernardo M, Munasinghe JP, et al. Simultaneous imaging of tumor oxygenation and microvascular permeability using Overhauser enhanced MRI. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17898–17903. doi: 10.1073/pnas.0908447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keshari KR, Wilson DM, Chen AP, Bok R, Larson PE, Hu S, et al. Hyperpolarized [2-13C]-fructose: a hemiketal DNP substrate for in vivo metabolic imaging. Journal of the American Chemical Society. 2009;131:17591–17596. doi: 10.1021/ja9049355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlsson M, Jensen PR, In 't Zandt R, Gisselsson A, Hansson G, Duus JO, et al. Imaging of branched chain amino acid metabolism in tumors with hyperpolarized (13)C ketoisocaproate. International journal of cancer. 2009 doi: 10.1002/ijc.25072. [DOI] [PubMed] [Google Scholar]
- 38.Gallagher FA, Kettunen MI, Hu DE, Jensen PR, Zandt RI, Karlsson M, et al. Production of hyperpolarized [1,4-13C2]malate from [1,4-13C2]fumarate is a marker of cell necrosis and treatment response in tumors. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19801–19806. doi: 10.1073/pnas.0911447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallagher FA, Kettunen MI, Day SE, Hu DE, Ardenkjaer-Larsen JH, Zandt R, et al. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature. 2008;453:940–943. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- 40.Gallagher FA, Kettunen MI, Day SE, Lerche M, Brindle KM. 13C MR spectroscopy measurements of glutaminase activity in human hepatocellular carcinoma cells using hyperpolarized 13C-labeled glutamine. Magn Reson Med. 2008;60:253–257. doi: 10.1002/mrm.21650. [DOI] [PubMed] [Google Scholar]
- 41.Banerjee HN, Verma M. Application of nanotechnology in cancer. Technology in Cancer Research & Treatment. 2008;7:149–154. doi: 10.1177/153303460800700208. [DOI] [PubMed] [Google Scholar]
- 42.Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine (London, England) 2008;3:703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Louie A. Mulimodal Imaging Probes. Chemical Reviews. 2010 doi: 10.1021/cr9003538. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bae Y, Nishiyama N, Kataoka K. In vivo antitumor activity of the folate-conjugated pH-sensitive polymeric micelle selectively releasing adriamycin in the intracellular acidic compartments. Bioconjugate chemistry. 2007;18:1131–1139. doi: 10.1021/bc060401p. [DOI] [PubMed] [Google Scholar]
- 45.Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, et al. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science (New York, NY. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 46.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nature medicine. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 47.Friberg M, Jennings R, Alsarraj M, Dessureault S, Cantor A, Extermann M, et al. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. International journal of cancer. 2002;101:151–155. doi: 10.1002/ijc.10645. [DOI] [PubMed] [Google Scholar]
- 48.Mellor AL, Keskin DB, Johnson T, Chandler P, Munn DH. Cells expressing indoleamine 2,3-dioxygenase inhibit T cell responses. J Immunol. 2002;168:3771–3776. doi: 10.4049/jimmunol.168.8.3771. [DOI] [PubMed] [Google Scholar]
- 49.Mellor AL, Munn D, Chandler P, Keskin D, Johnson T, Marshall B, et al. Tryptophan catabolism and T cell responses. Advances in experimental medicine and biology. 2003;527:27–35. doi: 10.1007/978-1-4615-0135-0_3. [DOI] [PubMed] [Google Scholar]
- 50.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nature medicine. 2005;11:312–319. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 51.Munn DH. Indoleamine 2,3-dioxygenase, tumor-induced tolerance and counter-regulation. Current opinion in immunology. 2006;18:220–225. doi: 10.1016/j.coi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. The Journal of experimental medicine. 1999;189:1363–1372. doi: 10.1084/jem.189.9.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Astigiano S, Morandi B, Costa R, Mastracci L, D'Agostino A, Ratto GB, et al. Eosinophil granulocytes account for indoleamine 2,3-dioxygenase-mediated immune escape in human non-small cell lung cancer. Neoplasia (New York, NY. 2005;7:390–396. doi: 10.1593/neo.04658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yasui H, Takai K, Yoshida R, Hayaishi O. Interferon enhances tryptophan metabolism by inducing pulmonary indoleamine 2,3-dioxygenase: its possible occurrence in cancer patients. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:6622–6626. doi: 10.1073/pnas.83.17.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brandacher G, Perathoner A, Ladurner R, Schneeberger S, Obrist P, Winkler C, et al. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: effect on tumor-infiltrating T cells. Clin Cancer Res. 2006;12:1144–1151. doi: 10.1158/1078-0432.CCR-05-1966. [DOI] [PubMed] [Google Scholar]
- 56.Sakurai K, Amano S, Enomoto K, Kashio M, Saito Y, Sakamoto A, et al. Study of indoleamine 2,3-dioxygenase expression in patients with breast cancer. Gan to kagaku ryoho. 2005;32:1546–1549. [PubMed] [Google Scholar]
- 57.Ishio T, Goto S, Tahara K, Tone S, Kawano K, Kitano S. Immunoactivative role of indoleamine 2,3-dioxygenase in human hepatocellular carcinoma. Journal of gastroenterology and hepatology. 2004;19:319–326. doi: 10.1111/j.1440-1746.2003.03259.x. [DOI] [PubMed] [Google Scholar]
- 58.Okamoto A, Nikaido T, Ochiai K, Takakura S, Saito M, Aoki Y, et al. Indoleamine 2,3-dioxygenase serves as a marker of poor prognosis in gene expression profiles of serous ovarian cancer cells. Clin Cancer Res. 2005;11:6030–6039. doi: 10.1158/1078-0432.CCR-04-2671. [DOI] [PubMed] [Google Scholar]
- 59.Juhasz C, Chugani DC, Muzik O, Wu D, Sloan AE, Barger G, et al. In vivo uptake and metabolism of alpha-[11C]methyl-L-tryptophan in human brain tumors. J Cereb Blood Flow Metab. 2006;26:345–357. doi: 10.1038/sj.jcbfm.9600199. [DOI] [PubMed] [Google Scholar]
- 60.Juhasz C, Muzik O, Lu X, Jahania MS, Soubani AO, Khalaf M, et al. Quantification of tryptophan transport and metabolism in lung tumors using PET. J Nucl Med. 2009;50:356–363. doi: 10.2967/jnumed.108.058776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rasey JS, Koh WJ, Grierson JR, Grunbaum Z, Krohn KA. Radiolabelled fluoromisonidazole as an imaging agent for tumor hypoxia. International Journal of Radiation Oncology, Biology, Physics. 1989;17:985–991. doi: 10.1016/0360-3016(89)90146-6. [DOI] [PubMed] [Google Scholar]
- 62.Lee ST, Scott AM. Hypoxia positron emission tomography imaging with 18f-fluoromisonidazole. Seminars in Nuclear Medicine. 2007;37:451–461. doi: 10.1053/j.semnuclmed.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 63.Rischin D, Hicks RJ, Fisher R, Binns D, Corry J, Porceddu S, et al. Prognostic significance of [18F]-misonidazole positron emission tomography-detected tumor hypoxia in patients with advanced head and neck cancer randomly assigned to chemoradiation with or without tirapazamine: a substudy of Trans-Tasman Radiation Oncology Group Study 98.02. J Clin Oncol. 2006;24:2098–2104. doi: 10.1200/JCO.2005.05.2878. [DOI] [PubMed] [Google Scholar]
- 64.Abdel Razek AA, Gaballa G, Denewer A, Tawakol I. Diffusion weighted MR imaging of the breast. Acad Radiol. 17:382–386. doi: 10.1016/j.acra.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 65.Chawla S, Kim S, Wang S, Poptani H. Diffusion-weighted imaging in head and neck cancers. Future Oncol. 2009;5:959–975. doi: 10.2217/fon.09.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Niwa T, Ueno M, Ohkawa S, Yoshida T, Doiuchi T, Ito K, et al. Advanced pancreatic cancer: the use of the apparent diffusion coefficient to predict response to chemotherapy. Br J Radiol. 2009;82:28–34. doi: 10.1259/bjr/43911400. [DOI] [PubMed] [Google Scholar]
- 67.Pope WB, Kim HJ, Huo J, Alger J, Brown MS, Gjertson D, et al. Recurrent glioblastoma multiforme: ADC histogram analysis predicts response to bevacizumab treatment. Radiology. 2009;252:182–189. doi: 10.1148/radiol.2521081534. [DOI] [PubMed] [Google Scholar]
- 68.Korpal M, Yan J, Lu X, Xu S, Lerit DA, Kang Y. Imaging transforming growth factor-beta signaling dynamics and therapeutic response in breast cancer bone metastasis. Nature medicine. 2009;15:960–966. doi: 10.1038/nm.1943. [DOI] [PubMed] [Google Scholar]
- 69.Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science (New York, NY. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maeda H, Bharate GY, Daruwalla J. Polymeric drugs for efficient tumor-targeted drug delivery based on EPR-effect. Eur J Pharm Biopharm. 2009;71:409–419. doi: 10.1016/j.ejpb.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 71.Gullotti E, Yeo Y. Extracellularly activated nanocarriers: a new paradigm of tumor targeted drug delivery. Molecular pharmaceutics. 2009;6:1041–1051. doi: 10.1021/mp900090z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leiner T, Kucharczyk W. Special issue: nephrogenic systemic fibrosis. J Magn Reson Imaging. 2009;30:1233–1235. doi: 10.1002/jmri.21985. [DOI] [PubMed] [Google Scholar]
- 73.Aime S, Barge A, Cabella C, Crich SG, Gianolio E. Targeting cells with MR imaging probes based on paramagnetic Gd(III) chelates. Current pharmaceutical biotechnology. 2004;5:509–518. doi: 10.2174/1389201043376580. [DOI] [PubMed] [Google Scholar]
- 74.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nature reviews. 10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schnell O, Krebs B, Carlsen J, Miederer I, Goetz C, Goldbrunner RH, et al. Imaging of integrin alpha(v)beta(3) expression in patients with malignant glioma by [18F] Galacto-RGD positron emission tomography. Neuro-oncology. 2009;11:861–870. doi: 10.1215/15228517-2009-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu Z, Niu G, Shi J, Liu S, Wang F, Liu S, et al. (68)Ga-labeled cyclic RGD dimers with Gly3 and PEG4 linkers: promising agents for tumor integrin alphavbeta3 PET imaging. European journal of nuclear medicine and molecular imaging. 2009;36:947–957. doi: 10.1007/s00259-008-1045-1. [DOI] [PubMed] [Google Scholar]
- 77.Edwards WB, Akers WJ, Ye Y, Cheney PP, Bloch S, Xu B, et al. Multimodal imaging of integrin receptor-positive tumors by bioluminescence, fluorescence, gamma scintigraphy, and single-photon emission computed tomography using a cyclic RGD peptide labeled with a near-infrared fluorescent dye and a radionuclide. Mol Imaging. 2009;8:101–110. [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Y, Cai W, Chen X. Near-infrared fluorescence imaging of tumor integrin alpha v beta 3 expression with Cy7-labeled RGD multimers. Mol Imaging Biol. 2006;8:226–236. doi: 10.1007/s11307-006-0041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huo T, Du X, Zhang S, Liu X, Li X. Gd-EDDA/HYNIC-RGD as an MR molecular probe imaging integrin alphanubeta3 receptor-expressed tumor-MR molecular imaging of angiogenesis. European journal of radiology. 73:420–427. doi: 10.1016/j.ejrad.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y, Wang X, Zhang Y, Yang S, Wang J, Zhang X, et al. RGD-modified polymeric micelles as potential carriers for targeted delivery to integrin-overexpressing tumor vasculature and tumor cells. Journal of drug targeting. 2009;17:459–467. doi: 10.1080/10611860902974085. [DOI] [PubMed] [Google Scholar]
- 81.Chen X, Plasencia C, Hou Y, Neamati N. Synthesis and biological evaluation of dimeric RGD peptide-paclitaxel conjugate as a model for integrin-targeted drug delivery. Journal of medicinal chemistry. 2005;48:1098–1106. doi: 10.1021/jm049165z. [DOI] [PubMed] [Google Scholar]
- 82.Kossodo S, Pickarski M, Lin SA, Gleason A, Gaspar R, Buono C, et al. Dual In Vivo Quantification of Integrin-targeted and Protease-activated Agents in Cancer Using Fluorescence Molecular Tomography (FMT) Mol Imaging Biol. 2009 doi: 10.1007/s11307-009-0279-z. [DOI] [PubMed] [Google Scholar]
- 83.Bodie L, Cremonesi M, Zoboli S, Grana C, Bartolomei M, Rocca P, et al. Receptor-mediated radionuclide therapy with 90Y-DOTATOC in association with amino acid infusion: a phase I study. Eur J Nucl Med. 2003;30:207–216. doi: 10.1007/s00259-002-1023-y. [DOI] [PubMed] [Google Scholar]
- 84.Corot C, Robert P, Lancelot E, Prigent P, Ballet S, Guilbert I, et al. Tumor imaging using P866, a high-relaxivity gadolinium chelate designed for folate receptor targeting. Magn Reson Med. 2008;60:1337–1346. doi: 10.1002/mrm.21773. [DOI] [PubMed] [Google Scholar]
- 85.Sega EI, Low PS. Tumor detection using folate receptor-targeted imaging agents. Cancer metastasis reviews. 2008;27:655–664. doi: 10.1007/s10555-008-9155-6. [DOI] [PubMed] [Google Scholar]
- 86.Bettio A, Honer M, Muller C, Bruhlmeier M, Muller U, Schibli R, et al. Synthesis and preclinical evaluation of a folic acid derivative labeled with 18F for PET imaging of folate receptor-positive tumors. J Nucl Med. 2006;47:1153–1160. [PubMed] [Google Scholar]
- 87.Ronald JA, Chen JW, Chen Y, Hamilton AM, Rodriguez E, Reynolds F, et al. Enzyme-sensitive magnetic resonance imaging targeting myeloperoxidase identifies active inflammation in experimental rabbit atherosclerotic plaques. Circulation. 2009;120:592–599. doi: 10.1161/CIRCULATIONAHA.108.813998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Querol M, Chen JW, Bogdanov AA., Jr A paramagnetic contrast agent with myeloperoxidase-sensing properties. Organic & biomolecular chemistry. 2006;4:1887–1895. doi: 10.1039/b601540a. [DOI] [PubMed] [Google Scholar]
- 89.Shiftan L, Neeman M. Kinetic analysis of hyaluronidase activity using a bioactive MRI contrast agent. Contrast media & molecular imaging. 2006;1:106–112. doi: 10.1002/cmmi.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weissleder R, Tung CH, Mahmood U, Bogdanov A., Jr In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nature biotechnology. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 91.McElroy M, Kaushal S, Luiken GA, Talamini MA, Moossa AR, Hoffman RM, et al. Imaging of primary and metastatic pancreatic cancer using a fluorophore-conjugated anti-CA19-9 antibody for surgical navigation. World J Surg. 2008;32:1057–1066. doi: 10.1007/s00268-007-9452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fang X, Balgley BM, Lee CS. Recent advances in capillary electrophoresis-based proteomic techniques for biomarker discovery. Electrophoresis. 2009;30:3998–4007. doi: 10.1002/elps.200900219. [DOI] [PubMed] [Google Scholar]
- 93.Robey IF, Stephen RM, Brown KS, Baggett BK, Gatenby RA, Gillies RJ. Regulation of the Warburg effect in early-passage breast cancer cells. Neoplasia (New York, NY. 2008;10:745–756. doi: 10.1593/neo.07724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Korc M. Pancreatic cancer-associated stroma production. Am J Surg. 2007;194:S84–S86. doi: 10.1016/j.amjsurg.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mahadevan D, Von Hoff DD. Tumor-stroma interactions in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2007;6:1186–1197. doi: 10.1158/1535-7163.MCT-06-0686. [DOI] [PubMed] [Google Scholar]
- 96.Zischek C, Niess H, Ischenko I, Conrad C, Huss R, Jauch KW, et al. Targeting tumor stroma using engineered mesenchymal stem cells reduces the growth of pancreatic carcinoma. Ann Surg. 2009;250:747–753. doi: 10.1097/SLA.0b013e3181bd62d0. [DOI] [PubMed] [Google Scholar]