Abstract
Iterative affinity selection procedures were used to isolate a number of single chain Fv (scFv) antibody fragment clones from naïve Tomlinson I + J phage display libraries that specifically recognize and bind a chemokine, monokine induced by interferon-gamma (MIG/CXCL9). MIG is an important transplant rejection/biology chemokine protein. ELISA-based affinity characterization results indicate that selectants preferentially bind to MIG in the presence of key biopanning component materials and closely related chemokine proteins. These novel antibody fragments may find utility as molecular affinity interface receptors in various electrochemical biosensor platforms to provide specific MIG binding capability with potential applications in transplant rejection monitoring, and other biomedical applications where detection of MIG level is important.
Introduction
Monokine induced by interferon-gamma (MIG/CXCL9) is a key protein in transplant rejection and the target analyte in transplant applications. Organ and tissue transplantation is an increasingly important treatment modality in multiple disease states (for example, heart/cardiac, kidney, liver and bone disease conditions), and will continue to become more prevalent as the American population ages. However, rejection of transplanted organs and tissues remains an obstacle to the efficacious long-term treatment of many of the disease conditions. A wide variety of complex immunological events play out during rejection of transplanted tissues. MIG has been clearly implicated as critical in transplant or allograft rejection (Watari et al., 2000). An essential aspect of transplant rejection is recruitment of specific immune cells to the graft site. A superfamily of about 50 small, chemo-attractive diffusible protein factors [chemokines, (Cascieri and Springer, 2000; Luter, 1998; Mantovani, 1999)] are involved in cellular trafficking to grafts. Key among chemokines responsible for rejection of kidney, skin and cardiac tissue is the chemokine MIG. MIG which binds the CXCR3 receptor on the surface of Th1 lymphocytes, is produced by IFN stimulated monocytes, macrophages and endothelial cells. Human MIG like other chemokines or chemo-attractant cytokines are basic glycosamino-glycan binding proteins that play an important role in the recruitment and activation of leukocytes (Luster, 1998). The human CXCL9 cDNA encodes a 125 amino acid residue precursor protein with a 22 amino acid residue signal peptide that is cleaved to yield a 103 amino acid residue mature protein, with a molecular weight of ~11.7 kDa. MIG is a potent chemo-attractant for the Th1 population (Cascieri and Springer, 2000; Bonecchi et al., 1998; Sallusto et al., 1998). MIG levels are positively correlated with rejection in murine skin allografts [2], and a number of other murine and human allografts (Miura et al., 2001; Miura et al., 2003; Fahmy et al., 2003; Zhao et al., 2002; Reiners et al., 2002). The range of MIG/CXCL9 concentration in normal and pathophysiological disease states (including allograft rejection episodes) in humans is ≈ 0.2–3 ng/mL and 10–400 ng/mL, respectively (Lauw et al., 2000; Yun et al., 2002).
We are involved in a systematic research program that utilize multiple phage display libraries (Scott and Smith, 1990; Burton, 1995; Hoess, 2001; Azzazy and Highsmith, 2002; Berry et al., 2003; Eteshola et al., 2005), and other modern antibody engineering/modification approaches to prepare a repertoire of novel interface biorecognition antibody fragment molecules for the rational design and fabrication of biochemically modified field effect transistor (BioFET) sensing channel or gate interfaces at the molecular level (Eteshola et al., 2008; Bergveld, 2003; Schöning and Poghossian, 2002). The prepared antibody fragment molecules will primarily provide specific MIG binding capability to the BioFET device (currently under research development in our group). The development of the BioFET sensor platform is aimed for minimally invasive detection of MIG as an early warning system in transplant rejection. We report here preliminary results of the isolation and sandwich ELISA characterization of scFv clones for MIG from a synthetic (Tomlinson I + J) naïve phage display libraries.
Materials and Methods
The Tomlinson I + J human single fold synthetic naïve phage display single chain antibody fragment libraries (in phagemid/scFv format - fused to the pIII minor coat protein of M13 bacteriophage), helper phage KM13, E. coli strains TG1 and HB2151 for selection of specific antibody clones and for production of soluble single chain Fvs, respectively, were obtained from The Medical Research Council (MRC), Cambridge, England (de Wildt et al., 2000; Torrance et al., 2006; Nissim et al., 1994; Hoogenboom et al., 1991). This scFv phagemid library contains synthetic V-gene (VH-VL) from lox library vector (Griffiths et al., 1994) recloned into the pHEN2 phagemid vector. The library size is 1.47 × 108 phagemid clones in E. coli TG1 cells, and has a high proportion of functional antibody fragments with approximately 96% of clones containing inserts. Biotinylated MIG was custom synthesized for us by Chemical Synthesis Services, Scotland, UK. Subsequently, biotinylated MIG was prepared in-house using Pierce EZ-Link biotinylation kit (Rockford, IL). NeutrAvidin coated magnetic particles were purchased from Spherotech, Inc., Libertyville, IL (USA).
Selection of anti-MIG scFvs clones
The supplied stock libraries I & J and stock helper phage KM13 were first expanded in order to obtain sufficient quantities for use in several rounds of biopanning. Each library stock was amplified and phage particles were rescued by superinfection with helper phage (Marks and Hoogenboom, 1991). Libraries I and J were biopanned separately to ensure selecting the most MIG antigen binding clones.
The biopanning procedure is essentially as described previously (Harrison et al., 1996; Chames et al., 2002), with some modifications. Briefly, ~1012 phage units of the library was mixed together with equal volume of 4% dried milk (containing 0.2% Tween 20) in PBS in polypropylene microcentrifuge tube. Biotinylated MIG (prepared in PBS) was added to a final specified concentration, and incubated with end-to-end rotation for 1 hr at room temperature. Aliquots of 150 μL of washed and blocked neutravidin-magnetic particles were added to the phage bound biotinylated MIG and incubated with end-over-end rotation for 15 min at room temperature. Tubes were placed in magnetic separators for ~3 minutes and supernatants carefully aspirated from tubes; particles in each tube were washed seven times with wash buffers. Finally, bound phages were eluted from the beads by resuspending in 20 mM dithiothreitol (DTT)-PBS and incubating tubes for 5 min with end-over-end mixing at room temperature. Particles were drawn to one side of each tube with magnetic separators and the supernatants were transferred to new microcentrifuge tubes. An aliquot of the eluted or output phages was used to infect fresh exponentially growing culture of E. coli TG1 cells, and incubated at 37 °C in a water bath (without shaking) for 30 min to allow optimal infection. Phage particles were rescued by superinfection with helper phage, amplified, and used for further rounds of panning as instructed in the Tomlinson (I + J) protocol. Four rounds of biopanning experiments were carried out for selection of scFv-phage clones with specific binding to MIG.
Phagemid DNA Sequencing, translation and alignment
Phagemid DNA sequencing of 44 randomly picked clones was performed using the QIAprep Spin M13 Kit (QIAGEN Inc., Valencia, CA). The phagemids were sequenced by the Plant-Microbe Genomics Facility, The Ohio State University, Columbus using the pHEN seq (5′ CTA TGC GGC CCC ATT CA 3′) or Link seq (5′ CGA CCC GCC ACC GCC GCT G 3′) sequence primers. Readily available web based tools were used for translation and alignment of DNA sequences.
Regular and competition monoclonal ELISA for fusion scFv-phage
Appropriately diluted glycerol stock of precipitated and purified phage/scFv from the fourth round of selection was used to infect log phase E. coli TG1 and plated on TYE/glucose/ampicilin plates. Individual colonies were picked and inoculated into 2xTY/glucose/ampicilin and grown shaking at 250 rpm until OD600 ~ 0.4. The culture was infected with KM13 helper phage, incubated for 30 minutes at 37 °C (without shaking), spun down at 4000 rpm for 10 minutes, resuspended in 2xTY/ampicilin/kanamycin/0.1 % glucose, and grown shaking overnight at 250 rpm and at 30 °C. Next day, culture aliquots were prepared in final glycerol concentration of 20% (v/v), mixed thoroughly and stored at −70 °C. The remainder of the culture was processed and purified according to standard protocol to obtain phage/scFv for subsequent use in monoclonal fusion phage-scFv ELISA. The ELISAs were performed essentially as described on the MRC website [www.geneservice.co.uk/products/proteomic/datasheets/tomlinsonIJ.pdf]. Briefly, phage displayed scFvs for each clone were added to Pierce (Rockford, IL, USA) high binding NeutrAvidin coated microtiter plates that have been previously reacted with biotinylated-MIG. Bound phage-scFv was detected using an anti-M13 antibody/HRP conjugate (GE Healthcare, Little Chalfont, UK) and 1-Step Ultra TMB-ELISA (Pierce).
For competion ELISA, non-biotinylated recombinant MIG in PBS was preincubated for 30 min with selected monoclonal purified phage/scFv clones supernatant suspension. This reaction mixture was added to Pierce neutravidin pre-coated/blocked microtiter plate wells that has been treated with biotin-MIG. Detection of bound phage-scFv was the same as indicated for monoclonal fusion phage ELISA above. The purpose of these competition experiments was to determine whether the presence of biotin is required by the isolated phage-scFv clones for specific recognition and binding to native MIG protein epitopes.
Small-scale expression and purification of soluble scFv fragments
For the production of soluble scFv antibody fragments from selected positive MIG binding clones, 1.0 mL inocula from glycerol stock of each phage/scFv clone already infected into E. coli HB2151 nonsuppressor strain (stored at −70 °C) was transferred into culture flasks containing 1000 mL 2xTY/ampicillin/0.1 % glucose. The culture was grown with shaking (250 rpm) at 37°C until the OD600 was approximately 0.9. At this stage, the transcription of scFv cassette driven with a lacZ promoter was then induced with isopropyl β-D-thiogalactopyranoside (IPTG), which was added to the culture at 1/8 volume in 2xTY containing 100 μg/mL ampicillin and 9 mM IPTG to a final concentration of 1 mM IPTG. Shaking was continued at 200 rpm, at 30 °C overnight to harvest antibody fragments secreted into culture supernatant and the E. coli periplasm by osmotic shock.
The induced bacterial culture was centrifuged at 30,000 × g at 4 °C for 30 min and the supernatant containing secreted antibody fragments was collected, and clarified by filtration through disposable 0.45 μm filters. The resulting supernatant was concentrated by ammonium sulfate precipitation followed by dialysis [10 kDa cut dialysis system] into binding/loading buffer at 4°C overnight. The scFv antibody fragments was purified using Protein L immobilized on agarose resin (according to manufacturer’s [Pierce] instructions).
To extract scFv secreted into the E. coli cell periplasm, the bacterial pellet from the above centrifugation step was resuspended in 1/50 the original culture volume of ice-cold 50 mM Tris-HCl, pH 8.0, containing 20% sucrose, 1 mM EDTA (with sufficient volume of Halt Protease Inhibitor sinlge-use cocktail EDTA-free (10 μL vial content/1 mL culture) to produce a 1X final concentration. The suspension was agitated gently for 20 min in an ice bath or cold room, centrifuged for 30 min at 30,000 × g at 4 °C and the supernatant collected (periplasmic fraction of scFv protein) into a new polypropylene tube maintained at 4 °C. To obtain the osmotic shock fraction, the resulting pellet from the previous scFv periplasm extraction step was again resuspended in the same volume [1/50] of ice-cold 5 mM MgSO4, 1 mM EDTA, and sufficient volume of Halt Protease Inhibitor. The suspension was agitated gently for 15 min in an ice bath or cold room, centrifuged for 30 min at 30,000 × g at 4 °C, and the supernatant containing the osmotic shock fraction of scFv proteins was collected. Supernatants from both the periplasmic and osmotic shock fractions containing scFv proteins were pooled, filtered through disposable 0.22 or 0.45 μm filters and dialyzed (10 kDa cut dialysis system) into binding/loading buffer. Soluble scFvs enriched in the periplasmic/osmotic fractions were then purified on ImmunoPure® immobilized protein L columns (Pierce).
Finally, all eluted protein fractions (supernatant, periplasmic and osmotic shock) were pooled and dialyzed to PBS buffer, pH 7.4, and simultaneously concentrated to 1.0–1.5 mL using iCON concentrator (Pierce). Purity of expressed and extracted soluble scFv was evaluated by 12% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). The concentration of the purified scFv was determined by Pierce micro BCA technique. The expressed soluble and purified protein from selected clones were then re-tested in regular and competition ELISA as well as cross-reactivity ELISA.
Regular, competition and functional cross-reactivity ELISA tests for soluble & purified scFv
Purified soluble scFvs at specified concentration were added to Pierce HisGrab copper coated microtiter plate wells (exploiting the His-tag on the scFv to achieve immobilization). The wells were subsequently blocked with 3% BSA, 0.05% Tween 20 in PBS, and biotinylated MIG added. Finally, the plates were treated with Sigma ultrasensitive streptavidin-HRP conjugate prepared in PBS-0.05% Tween 20 (1:2000 dilution). Plate wells were washed at appropriate steps. For competition ELISA, non-biotinylated recombinant MIG in PBS was preincubated for 60 min with specified amount of purified soluble scFv clones prepared in PBS-0.05% Tween 20. This reaction mixture was then added to Pierce HisGrab copper coated microtiter plate wells. For detection, the wells were treated as described for regular ELISA experiments above. The purpose of these ELISA experiments was to determine whether the soluble scFv proteins would still recognize and bind to MIG protein epitopes after expression and purification (that is, in the absence of the phage particles to which they were fused during initial biopanning and monoclonal fusion phage-scFv ELISA experiments). The binding specificity of expressed, soluble and purified scFvs for MIG was further probed using functional cross-reactivity ELISA. Three other structurally or biochemically related and most widely studied chemokines subfamilies [CXC α- and CC β-], namely: gamma-interferon inducible protein 10 (IP-10, CXCL10), neutrophil activating protein (NAP, CXCL8), and eotaxin (CCL11) were used. For these experiments, ELISA plate wells were coated with a given soluble, purified scFv fragment at specified concentration in carbonate buffer, blocked with 3% (w/v) BSA in PBS. This was followed by addition of cold recombinant IP-10, IL-8 or eotaxin preincubated with the same soluble purified scFv. Biotinylated MIG was then added to the wells; detection of MIG was the same as indicated for soluble and purified scFv ELISA above. The purpose of these experiments was to ascertain the possible level of binding of the soluble purified scFv clones to chemokines that are biochemically and structurally similar to MIG.
Results and Discussion
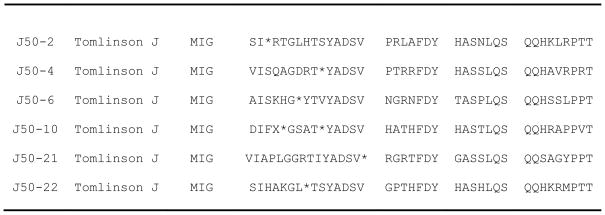
Binding efficiency is measured as the fraction of input phage that bind the target molecule, MIG. In these experiments, phage was quantified by titering after each round of the four biopanning experiments. The concentration input phage used for each round of selection was kept constant at ~1012 particles/mL. The results obtained showed that the relative eluate yield/output of binding phage particles was increased approximately 1000-fold and 100-fold from the first to the fourth round of affinity selection for libraries J (4 × 106 – 5 × 109) and I (1.1 × 106 – 3.5 × 108), respectively. During successful biopanning experiments, phage yields increase from one round to another round of selection enrichment (Eteshola et al., 2005; Eteshola et al., 2006). The results clearly indicates that isolation and enrichment of MIG binding phages was accomplished. The phagemid DNA sequences were examined by inspection to ensure that the selected MIG-binding scFv clones have full length VH and Vκ inserts. Of the 44 phage clones randomly picked for sequencing, 14 clones from both Tomlinson I + J libraries had distinct amino acid sequences (Fig. 1). The translation of the DNA sequences into amino acid sequences and subsequent protein alignment were accomplished using readily available free web based tools: (www.bioinformatics.org/SMS/rev_comp.html; ca.expasy.org/tools/dna.html and www.ebi.ac.uk/Tools/clustalw2/index.html). The results show we succeeded in selecting clones incorporating complete sequences of scFvs. Only scFv clones with complete nucleotide sequences or amino acid inserts were used for further studies. However, in this report, only the four clones isolated from the Tomlinson I library (Fig. 1) are expressed in soluble form for further ELISA experiments (see later discussion of the soluble expression results below).
Fig. 1.
Deduced amino acid sequences (including depiction of CDR and His-tag sections) of scFv fragments with binding specificity to MIG isolated from the Tomlinson I + J libraries. The random amber stop codons present in the sequence of the J clones is represented by an asterisk (*). The numbers on the right indicate positions of amino acids.
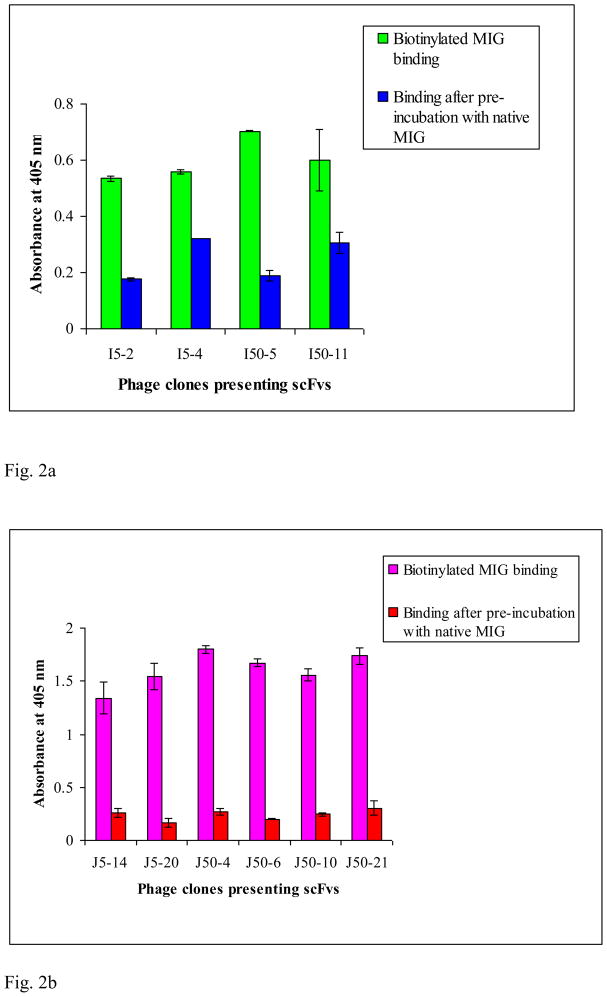
To demonstrate that the individually picked and sequenced phage clones were indeed MIG binders, some of these were grown up and the ability to bind MIG was verified by monoclonal fusion phage/scFv ELISA. Results of the monoclonal phage/scFv ELISA (not shown) clearly indicate that there is significant difference in the absorbance reading in all wells with the MIG antigen versus control wells (no MIG, with OD readings of only ~0.05–0.3). Thus, the results of the monoclonal phage/scFv ELISA show that positive clones with specific binding to biotinylated MIG have been selected from both Tomlinson I + J libraries. Furthermore, competition ELISA experiments in which unbiotinylated native MIG was pre-incubated with selected monoclonal phage-scFv clones from both Tomlinson I + J libraries prior to incubation with biotinylated MIG resulted in OD reading values comparable to control experiments (Fig. 2a & 2b). These results indicate that the selected clones recognize actual epitopes of native MIG, and do not require those epitopes to be biotinylated to be recognized. Additionally, various ELISA control experiments in which the isolated clones were exposed to key material components [biotin, neutravidin, milk in PBS (MPBS)] present in the biopanning experiments indicate that the isolated scFv clones were indeed MIG binders (results not shown) and that the presence of these key components did not significantly influence the binding affinity of the isolated clones for MIG.
Fig. 2.
Fig. 2a. Competition ELISA determined binding properties of MIG recognizing purified monoclonal phage-scFv clones selected from Tomlinson I library. MIG analyte concentrations used in biopanning were 5 nM (I5-2, I5-4) and 50 nM (I50-5, I50-11). The data represent mean ± standard deviation from duplicate measurements.
Fig. 2b. Competition ELISA determined binding properties of MIG recognizing purified monoclonal phage-scFv clones selected from Tomlinson J library. MIG analyte concentrations used in biopanning were 5 nM (J5-14, J5-20) and 50 nM (J50-4, J50-6, J50-10, J50-21). The data represent mean ± standard deviation from duplicate measurements.
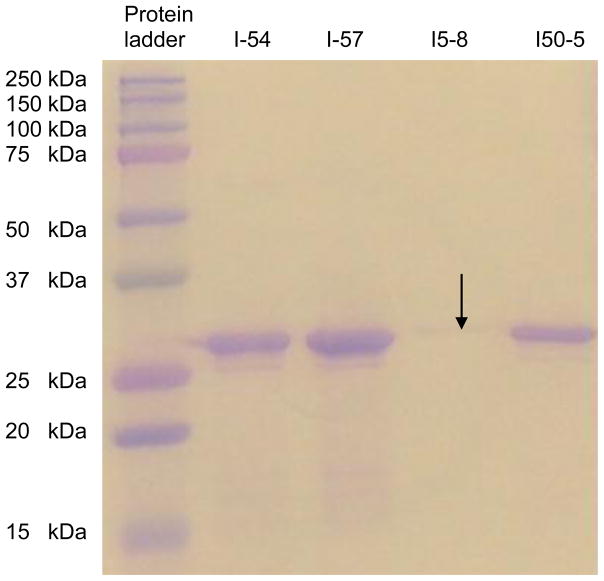
Based on the above results, we selected some of the positive MIG binding clones for small scale production of soluble scFv antibody fragments in the nonsuppressor HB2151 E. coli strain according to the Tomlinson library protocol. However, we encountered difficulties in the ability of the HB2151 E. coli strain expressing soluble scFv for the clones selected from the J library due to the presence of amber stop codons in the antibody fragment genes for all the promising scFv clones selected for further studies from the J library (Marcus et al., 2006). These amber stop codons, while still allowing phage/scFv production in TG1 cells, prevent the expression of full-length scFvs in the nonsuppressor HB2151 E. coli strain. We are currently using molecular biology techniques (Wu et al., 2007) to repair these clones containing randomly generated amber codons. However, while work is ongoing to repair the amber codons in the sequences of the J library clones, we proceeded to produce soluble scFv fragments using selected clones isolated from the I library with no amber stop codons. Soluble scFv fragments protein expression levels for these clones were in the range of 0.044 to 1.8 mg/liter of culture. These yields are in general agreement with published yields in the range of 0.1 to 5.0 mg per liter of E. coli culture for clones isolated from the Tomlinson libraries (Kennel et al., 2004; Wu et al., 2007; Lobova et al., 2008). SDS-PAGE analysis for these four clones show bands corresponding approximately to 26–28 kD molecular weight when compared to the 25 kD band of the protein standard ladder run in parallel with the samples (Fig. 3). The molecular weights of scFvs are generally indicated to be between 25–30 kD; thus, the results would imply that we succeeded in expressing the soluble scFv fragments for the Tomlinson I library clones in the HB2151 E. coli strain. The band for clone I5-8 in the SDS-PAGE gel picture appears quite faint because equal volumes of soluble scFv stock for each clone was mixed with equal volume of sample buffer for loading into gel well for the experiment. Following successful soluble expression and purification, we then proceeded to further test these scFv fragments in competition and cross-reactivity ELISA tests.
Fig. 3.
SDS-PAGE (12% Tris glycine-HCl gel) analysis of purified single chain antibody fragments (scFvs) from clones (I5-4, I5-7, I5-8 and I50-5) isolated from Tomlinson I library. The position of the bands appears to indicate that the molecular weight of the expressed scFv fragments is approximately 26–28 kD.
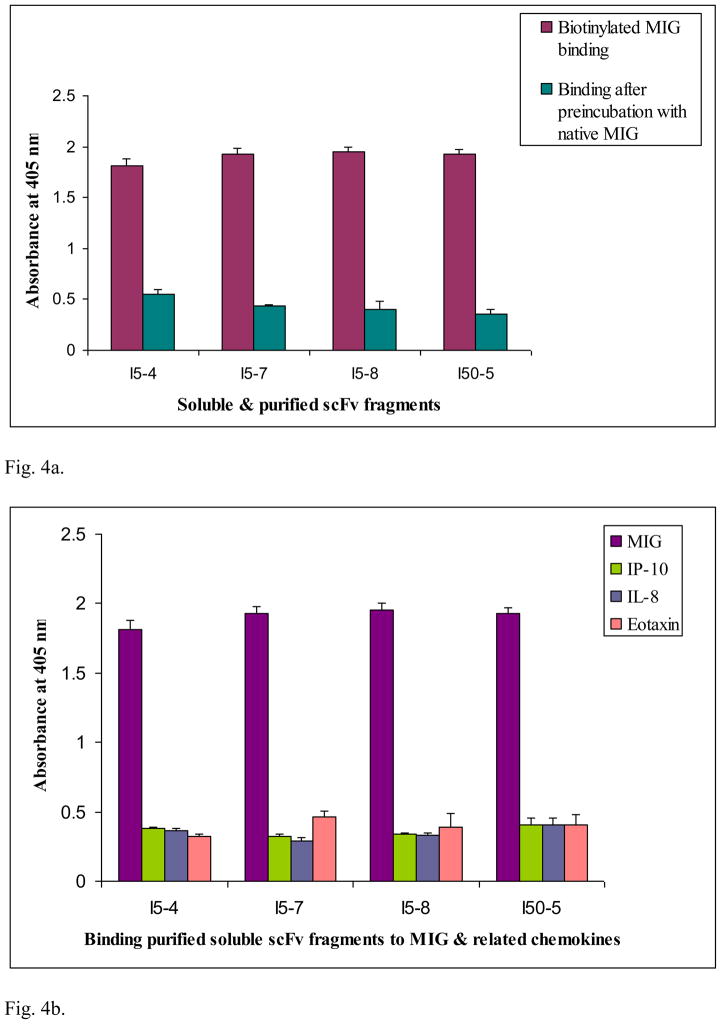
Results of competition ELISA experiments (Fig. 4a) in which unbiotinylated native MIG was pre-incubated with soluble and purified scFv prior to incubation with biotinylated MIG resulted in OD reading values comparable to control experiments. These results indicate that the selected clones recognize actual epitopes of native MIG after soluble expression and purification, and do not require those epitopes to be biotinylated or fused to phage particle to recognize and bind to MIG. Functional cross-reactivity ELISA results (Fig. 4b) indicates that the soluble and purified scFv fragments bound MIG in preference to closely related chemokines IP-10, NAP and eotaxin. Some measure of cross-reactivity of the selected soluble scFv fragments from the I library to chemokines closely related to MIG in the α- and β-chemokine subfamilies is observed. However, these results are not entirely surprising - in view of the high level of homology in amino acid sequences (generally 20 to 70%) within the CXC and CC chemokine subfamilies. In addition, all known α-chemokines share 25 to 90% sequence homology - thus, these are highly homologous chemokines (Luster, 1998). Moreover, some research suggests that 90% of all monoclonal antibodies (well known for their high antigen binding specificity) can recognize other antigens when screened across several antigens (Kay, 2006). Thus, the results obtained here would indicate the necessity to affinity mature some of the selected scFv clones - with the aim of generating scFv clones possessing higher specificity and binding affinity characteristics for use in the bio-functionalization of the sensing channel interface of the BioFET sensor device under development in our group.
Fig. 4.
Fig. 4a. Competition ELISA determined binding properties of MIG recognizing soluble and purified scFv fragments (5 μg/mL) synthesized from selected clones isolated from Tomlinson I library. Each well was coated with a given soluble, purified scFv fragment (5 μg/mL or 1.7 × 10−7 M) in carbonate buffer; blocked with 3% (w/v) BSA in PBS, followed by addition of cold recombinant MIG (3.5 × 10−6 M) pre-incubated with the same soluble purified scFv; then biotinylated MIG (6.6 × 10−7 M) was added. The data represent mean ± standard deviation from duplicate measurements.
Fig. 4b. Differential binding to MIG and structurally related chemokines IP-10, NAP and Eotaxin by soluble and purified scFv fragments synthesized from selected clones (I5-4, I5-7, I5-8 and I50-5) isolated from Tomlinson I library. Each well was coated with a given soluble, purified scFv fragment (5 μg/mL or 1.7 × 10−7 M) in carbonate buffer; blocked with 3% (w/v) BSA in PBS, followed by addition of cold recombinant IP-10 or IL-8 (1.2 × 10−6 M) and cold recombinant Eotaxin (1.0 × 10−6 M), pre-incubated with the same soluble purified scFv, and then biotinylated MIG (6.6 × 10−7 M) was added; see text for further explanation of the results. The data represent mean ± standard deviation from duplicate measurements.
Concluding remarks
We have successfully isolated a number of novel scFv clone variants from two naïve or single-pot libraries with specific binding affinity to actual epitopes of native MIG protein. The results indicate that some of these isolated scFv clones would have potential to serve as molecular affinity interface receptors in a MIG sensitive BioFET sensor device. To our knowledge, these scFvs represent the first recombinant antibodies to this very important chemokine protein with high relevance to human health. Currently, more detailed studies of affinity/kinetics of binding of selected clones for MIG using functional BIAcore techniques are ongoing. These studies would enable us to differentiate between variants that differ in affinity or the kinetics of binding for MIG, and as well determine which of the selected scFv fragments to affinity mature to possess optimal or higher specificity and binding affinity characteristics for MIG.
Acknowledgments
This work was supported by the NIH/NIBIB Career Development grant number EB004960. I thank John Shapiro for technical assistance and Dr. SC Lee for helpful discussions and technical advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azzazy HM, Highsmith WE., Jr Phage display technology: clinical applications and recent innovations. Clin Biochem. 2002;35(6):425. doi: 10.1016/s0009-9120(02)00343-0. [DOI] [PubMed] [Google Scholar]
- Bergveld P. Thirty years of ISFETOLOGY: What happened in the past 30 years and what may happen in the next 30 years. Sensors & Actuators. 2003;B88:1. [Google Scholar]
- Berry JD, Popkov M, Gubbins M, Mandeville R. Recent innovations and analytical applications of phage display libraries. Anal Lett. 2003;36:3227. [Google Scholar]
- Bonecchi R, Bianchi G, Bordignon PP, D’Ambrosio D, Lang R, Borsatti A, Sozzani S, Allavena P, Gray PA, Mantovani A, Sinigaglia F. Differential Expression of Chemokine Receptors and Chemotactic Responsiveness of Type 1 T Helper Cells (Th1s) and Th2s. J Exp Med. 1998;187:129. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR. Phage display. Immunotechnol. 1995;1:87. doi: 10.1016/1380-2933(95)00013-5. [DOI] [PubMed] [Google Scholar]
- Cascieri MA, Springer MS. The chemokine/chemokine-receptor family: potential and progress for therapeutic intervention. Curr Opin Chem Biol. 2000;4(4):420. doi: 10.1016/s1367-5931(00)00113-7. [DOI] [PubMed] [Google Scholar]
- Chames P, Hoogenboom HR, Henderikx P. Selection of antibodies against biotinylated antigens. Methods Mol Biol. 2002;178:147. doi: 10.1385/1-59259-240-6:147. [DOI] [PubMed] [Google Scholar]
- Eteshola E, Brillson LJ, Lee SC. Selection and characteristics of peptides that bind thermally grown silicon dioxide films. Biomol Eng. 2005;22(5–6):201. doi: 10.1016/j.bioeng.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Eteshola E, van Valkenburgh CD, Merlin S, Rowold E, Adams J, Ibdah R, Pegg LE, Donelly A, Klover J, Lee SC. Screening Libraries of Circularly Permuted Proteins by Phage Display to Manipulate Protein Topographies. Proc IMechE, Part N: J Nanoengineering and Nanosystems. 2006;219(N2):45. [Google Scholar]
- Eteshola E, Keener MT, Elias M, Shapiro J, Brillson LJ, Bhushan B, Lee SC. Engineering functional protein interfaces for immunologically modified field effect transistor (ImmunoFET) by molecular genetic means. J R Soc Interface. 2008;5(18):123. doi: 10.1098/rsif.2007.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths AD, Williams SC, Hartley O, Tomlinson IM, Waterhouse P, Crosby WL, Kontermann RE, Jones PT, Low NM, Allison TJ, Prospero TD, Hoogenboom HR, Nissim A, Jonathan PL, Cox JPL, Harrison JL, Zaccolo M, Gherardi E, Winter G. Isolation of high affinity human antibodies directly from large synthetic repertoires. EMBO J. 1994;13(14):3245. doi: 10.1002/j.1460-2075.1994.tb06626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JL, Williams SC, Winter G, Nissim A. Screening of phage antibody libraries. Methods Enzymol. 1996;267:83. doi: 10.1016/s0076-6879(96)67007-4. [DOI] [PubMed] [Google Scholar]
- Hoess RH. Protein design and phage display. Chem Rev. 2001;101:3205. doi: 10.1021/cr000056b. [DOI] [PubMed] [Google Scholar]
- Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19(15):4133. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay B. September 26–27, 2006; The Future of Biodetection Systems Workshop Proceedings; Santa Fe, NM. Weblink: www.lanl.gov/orgs/b/Biodetection_Proceedings/Biodetection_Futures_Analysis.pdf {pg. 15} [Google Scholar]
- Kennel SJ, Lankford T, Foote L, Wall M, Davern S. Phage Display Selection of scFv to Murine Endothelial Cell Membranes. Hybridoma and Hybridomics. 2004;23(4):205. doi: 10.1089/1536859041651295. [DOI] [PubMed] [Google Scholar]
- Lauw FN, Simpson AJ, Prins JM, van Deventer SJ, Chaowagul W, White NJ, van der Poll T. The CXC chemokines gamma interferon (IFN-gamma)-inducible protein 10 and monokine induced by IFN-gamma are released during severe melioidosis. Infect Immun. 2000;68(7):3888. doi: 10.1128/iai.68.7.3888-3893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobova D, Čizek A, Celer V. The Selection of Single-Chain Fv Antibody Fragments Specific to Bhlp 29.7 Protein of Brachyspira hyodysenteriae. Folia Microbiol. 2008;53(6):517. doi: 10.1007/s12223-008-0081-3. [DOI] [PubMed] [Google Scholar]
- Luster AD. Chemokines--chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338(7):436. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- Mantovani A. Chemokines - introduction and overview. Chem Immunol. 1999;72:1. [PubMed] [Google Scholar]
- Marcus WD, Lindsey SM, Sierks MR. Identification and repair of positive binding antibodies containing randomly generated amber codons from synthetic phage display libraries. Biotechnol Prog. 2006;22:919. doi: 10.1021/bp050420y. [DOI] [PubMed] [Google Scholar]
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222(3):581. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- Miura M, Morita K, Kobayashi H, Hamilton TA, Burdick MD, Strieter RM, Fairchild RL. Monokine Induced by IFN-γ Is a Dominant Factor Directing T Cells into Murine Cardiac Allografts During Acute Rejection. J Immunol. 2001;167:3494. doi: 10.4049/jimmunol.167.6.3494. [DOI] [PubMed] [Google Scholar]
- Miura M, Morita K, Koyanagi T, Fairchild RL. Neutralization of monokine induced by interferon-γ during the early posttransplantation period prevents development of chronic allograft vasculopathy and graft fibrosis. Transplant Proceed. 2003;35:875. doi: 10.1016/s0041-1345(02)04036-8. [DOI] [PubMed] [Google Scholar]
- Nissim A, Hoogenboom HR, Tomlinson IM, Flynn G, Midgley C, Lane D, Winter G. Antibody fragments from a ‘single pot’ phage display library as immunochemical reagents. EMBO J. 1994;13(3):692. doi: 10.1002/j.1460-2075.1994.tb06308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiners J, Henne T, Offner G, von Schmakenburg C, Strehlau J, Latta K, Ehrich JHH, Melter M. Mig, IP-10, and CXCR3 gene expression is predictive for the individual response of children with chronic allograft nephropathy to mycophenolate mofetil. Transplant Proceed. 2002;34:2217. doi: 10.1016/s0041-1345(02)03209-8. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible Programs of Chemokine Receptor Expression on Human Polarized T Helper 1 and 2 Lymphocytes. J Exp Med. 1998;187:875. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning MJ, Poghossian A. Recent advances in biologically sensitive field-effect transistors (BioFETs) Analyst. 2002;127:1137. doi: 10.1039/b204444g. [DOI] [PubMed] [Google Scholar]
- Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Sci. 1990;249:386. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Torrance L, Ziegler A, Pittman H, Paterson M, Toth R, Eggleston I. Oriented immobilisation of engineered single-chain antibodies to develop biosensors for virus detection. J Virol Methods. 2006;134(1–2):164. doi: 10.1016/j.jviromet.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Watarai Y, Koga S, Paolone DR, Engeman TM, Tannenbaum C, Hamilton TA, Fairchild RL. Intraallograft chemokine RNA and protein during rejection of MHC-matched/multiple minor histocompatibility-disparate skin grafts. J Immunol. 2000;164(11):6027. doi: 10.4049/jimmunol.164.11.6027. [DOI] [PubMed] [Google Scholar]
- de Wildt RM, Mundy CR, Gorick BD, Tomlinson IM. Antibody arrays for high-throughput screening of antibody-antigen interactions. Nat Biotechnol. 2000;18(9):989. doi: 10.1038/79494. [DOI] [PubMed] [Google Scholar]
- Wu S, Ke A, Doudna JA. A fast and efficient procedure to produce scFvs specific for large macromolecular complexes. J Immunol Methods. 2007;318:95. doi: 10.1016/j.jim.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun JJ, Fischbein MP, Whiting D, Irie Y, Fishbein MC, Burdick MD, Belperio J, Strieter RM, Laks H, Berliner JA, Ardehali A. The role of MIG/CXCL9 in cardiac allograft vasculopathy. Am J Pathol. 2002;161(4):1307. doi: 10.1016/S0002-9440(10)64407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DXM, Hu Y, Miller GG, Luster AD, Mitchell RN, Libby P. Differential Expression of the IFN-γ-Inducible CXCR3-Binding Chemokines, IFN-Inducible Protein 10, Monokine Induced by IFN, and IFN-Inducible T Cell Chemoattractant in Human Cardiac Allografts: Association with Cardiac Allograft Vasculopathy and Acute Rejection. J Immunol. 2002;169:1556. doi: 10.4049/jimmunol.169.3.1556. [DOI] [PubMed] [Google Scholar]