Abstract
To date, various G-quadruplex structures have been reported in human telomeric sequences. Human telomeric repeats can form many topological structures depending on conditions and on base modification; parallel, antiparallel, and hybrid forms. The effect of salts and some specific ligands on conformational switches between different conformers is known, but the influence of protruding sequences has rarely been discussed. In this paper, we analyze different quadruplex-forming oligomers derived from human telomeric sequences which contain 3′- and 5′-protruding nucleotides, not usually associated with the G-quadruplex motif. The study was performed using electrophoresis, CD, and UV spectroscopies. The major findings are (i) protruding nucleotides destabilize the G-quadruplex structure, and (ii) overhanging sequences influence the folding of the quadruplex.
1. Introduction
There is currently tremendous interest in understanding G-quadruplex formation from G-rich sequences. To date, more than 1600 scientific articles concerning the quadruplex problem have been published; more than 1200 of them are focused on quadruplexes which potentially occur in humans and approximately half of these are interested in quadruplex formation in telomeres. Telomeres are structures at the end of chromosomes which protect chromosomal DNA from degradation and recombination [1, 2]. Eukaryotic telomeres consist of tandem repeats of G-rich sequences, for example, (TTAGGG)n repeats in human. Several kilobases of that sequence are paired with a complementary strand to form duplex DNA, but approximately 100–200 nucleotides of the sequence remain unpaired and form a single-stranded overhang. These G-rich ends can fold into four-stranded G-quadruplex structures containing G-tetrads, paired by intra- or intermolecular Hoogsteen bonds; they are held together by π-π stacking interactions [3]. This noncanonical DNA secondary structure has been postulated as being involved in a variety of biological functions; it is suggested that they may also be important causal factors in cell aging and human diseases such as cancer [1, 2]; G-quadruplexes formed by these sequences represent attractive anticancer targets [4, 5]. There is supporting evidence that telomeres also serve as a biological clock, as telomere structures appear to shorten with each successive cell cycle. In immortalized cells and in cancer cells however, telomerase is activated to maintain the length of the telomere [6]. Specific proteins interact with the overhang and regulate telomerase activity. There is supporting evidence that quadruplex structures occur in vivo [7], and recent evidence suggests that quadruplexes form in telomeric DNA at specific times in the cell cycle [8]. However, G-rich DNA sequences can form a large number of structurally diverse G-quadruplex structures in the presence of monovalent ions, for example potassium and sodium [9]. Much still remains unclear about the condition and driving forces of quadruplex topology.
Repeats of the 5′-TTAGGG sequence spontaneously form intramolecular quadruplex structures in solution, with a variety of folded morphologies. Different conformations of human telomeric oligomers in water solution and in crystals have been reported by a number of authors [3, 10, 11]. In addition, one conformation of G-quadruplex can transform to another, for example, parallel-to-antiparallel form. Quadruplex folding depends on many factors; the DNA sequence, the presence of ions, the temperature, and the presence of various ligands [12, 13]. Recently, four-repeats of human telomere sequences have been shown to form two very similar intramolecular (3 + 1) G-quadruplexes in solutions containing K+ ion (hybrid form 1 and 2) [14–16]. Both structures contain the (3 + 1) G-tetrad core with one double-chain-reversal (DCR) and two edgewise loops (EW), but differ in the order of loop appearance within the G-quadruplex scaffold. In addition, a basket-type G-quadruplex with only two G-tetrad layers was detected where loops are successively edgewise, diagonal, and edgewise. Despite the presence of only two G-tetrads in the core, this structure is more stable than the three-G-tetrad intramolecular G-quadruplexes previously observed in human telomeric sequences in potassium solution. This novel structure highlights the conformational heterogeneity of human telomeric DNA [17]. Studies have also revealed a crystal structure of the human telomere sequence which has a unique symmetrical propeller-type structure with all parallel G-tetrads and three double-chain-reversal loops [3]. This structure does not seem to be the prevalent form in solution [11]. However, recent experiments show that the human telomeric repeats can form parallel G-quadruplex structures in crowding conditions simulated with polyethylene glycol and ethanol [18–20].
In this study, we analyze the human telomeric sequence 3′-G3(TTAG3)3-5′ containing various overhangs at 3′- and 5′-ends, Table 1. The effect of thymine tract length has previously been studied for different shorter sequences which form dimeric and tetrameric quadruplex molecules [21]. However, for the first time we focus on monomolecular quadruplexes containing longer protruding sequences of more than 7 nucleotides. It has been widely documented that changes of only one base in the sequence at the 3′- or 5′- end can drastically modify quadruplex topology [15]. Our goal was to choose sequences in which overhangs do not form any secondary structures, because additional secondary motifs can significantly affect CD spectra and can complicate spectra interpretation. Therefore, only one random protruding sequence was analyzed to avoid any unexpected interactions of overhanging guanines with parts of the sequence forming a core of G-quadruplex; other sequences in overhangs do not contain guanine and cytosine bases. Thus, d(T)n and d(TTA)m have been analyzed, where n = 7 and 11 and m = 1 and 6. The second goal was to use sequences which cannot form or associate to other quadruplex motifs, as for example, in the TEL2 sequence used here. Recent evidence suggests the existence of intramolecular quadruplex-quadruplex interactions between two contiguous quadruplex motifs in another quadruplex forming sequence, which can influence their overall structure [22]. These oligomeric sequences forming G-quadruplexes serve as a model of the arrangement of quadruplexes in telomeres in cellular conditions because G-rich islands are always associated with another part of telomeric DNA, mainly at 5′-end, where the telomeric sequence occurs as double-stranded DNA. Electrophoresis, CD and UV spectroscopies have been used for this purpose.
Table 1.
Oligodeoxynucleotides and their sequences used in this study.
| Name | N. nt. | Sequence 5′→ 3′ |
|---|---|---|
| TEL1 | 21 | GGGTTAGGGTTAGGGTTAGGG |
| TEL1-TTA | 24 | GGGTTAGGGTTAGGGTTAGGGTTA |
| TEL2 | 45 | GGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTA GGG TTAGGG |
| TEL1-5-T-7 | 28 | TTTTTTT-GGGTTAGGGTTAGGGTTAGGG |
| TEL1-3-T-7 | 28 | GGGTTAGGGTTAGGGTTAGGG-TTTTTTT |
| TEL1-5-A-7 | 28 | AAAAAAA- GGGTTAGGGTTAGGGTTAGGG |
| TEL1-3-A-7 | 28 | GGGTTAGGGTTAGGGTTAGGG-AAAAAAA |
| TEL1-5-T-11 | 32 | TTTTTTTTTTT-GGGTTAGGGTTAGGGTTAGGG |
| TEL1-3-T-11 | 32 | GGGTTAGGGTTAGGGTTAGGG-TTTTTTTTTTT |
| TEL1-3-TTA-11 | 32 | GGGTTAGGGTTAGGGTTAGGG-TTATTATTATT |
| TEL1-5-TTA-11 | 32 | TTATTATTATT GGGTTAGGGTTAGGGTTAGGG- |
| TEL1-3-5-T-7 | 35 | TTTTTTT-GGGTTAGGGTTAGGGTTAGGG-TTTTTTT |
| TEL1-3-T-18 | 39 | GGGTTAGGGTTAGGGTTAGGG-TTTTTTTTTTTTTTTTTT |
| TEL1-5-T-18 | 39 | TTTTTTTTTTTTTTTTTT-GGGTTAGGGTTAGGGTTAGGG |
| TEL1-3-5-A-7 | 39 | AAAAAAA- GGGTTAGGGTTAGGGTTAGGG-AAAAAAA |
| TEL1-5-TTA-18 | 39 | TTATTATTATTATTATTA-GGGTTAGGGTTAGGGTTAGGG |
| TEL1-3-5-TTA-18 | 57 | TTATTATTATTATTATTA-GGGTTAGGGTTAGGGTTA GGG-TTATTATTATTATTATTA |
| TEL1-ran | 39 | GATCCCAGATCTTC-GGGTTAGGGTTAGGGTTAGGG -CAGA |
2. Materials and Methods
2.1. Material and Equipments
All chemical and reagents were obtained from commercial sources. Acrylamide : bisacrylamide (19 : 1) solution and ammonium persulfate were purchased from Bio-Rad, polyethylene glycol—PEG 200 and N,N,N′,N′-tetramethylethylenediamine were purchased from Fisher Slovakia. DNA oligomers (sequences shown in Table 1) were obtained from Sigma Genosys and Biosearch Technologies, Inc. All DNA oligomers were PAGE purified and dissolved in double-distilled water before use. Single-strand DNA concentrations were determined by measuring the absorbance (260 nm) at high temperature. The concentration of DNA was determined by UV measurements carried out on a Varian Cary 100 UV-visible spectrophotometer (Amedis, Slovakia). Cells with optical path lengths of 10 mm were used, and the temperature of the cell holder was controlled with an external circulating water bath (Varian).
2.2. Circular Dichroism Spectroscopy
CD spectra were recorded on a Jasco J-810 spectropolarimeter (Easton, MD, USA) equipped with a PTC-423L temperature controller using a quartz cell of 1 mm optical path length in a reaction volume of 300 μl and an instrument scanning speed of 100 nm/min, 1 nm pitch, and 1 nm bandwidth, with a response time of 2 s, over a wavelength range of 220–320 nm. The scan of the buffer was subtracted from the average scan for each sample. All DNA samples were dissolved and diluted in suitable buffers containing appropriate concentrations of ions. The amount of DNA oligomers used in the experiment was kept close to ~2.5 μM. The normalized CD spectra were compared. DNA samples were annealed at 95°C for 5 minutes then allowed to cool down to the initial temperature as at the beginning of the experiment for ~2 hours. CD data represents three averaged scans taken at a temperature range of 20°C. The Britton-Robinson buffer was used in all experiments: 25 mM H3PO4, 25 mM Boric acid, 25 mM acetic acid, and supplemented by 50 mM of KCl, pH was adjusted by Tris to the final value of 7.0.
2.3. Melting Curves
The CD melting profiles were collected at 265 and at 295 nm. The thermal stability of different antiparallel quadruplexes was also measured by recording UV absorbance and the CD ellipticity at 295 nm as a function of temperature, by a method similar to that published previously [23, 24]. The temperature ranged from 20 to 100°C, the heating rate was 0.25°C per minute. The melting temperature (T m) was defined as the temperature of the midtransition point. This T m value was used as an initial parameter of van't Hoff analysis [24].
2.4. Electrophoresis
Native polyacrylamide gel electrophoresis (PAGE) was run in a temperature-controlled vertical electrophoretic apparatus (Z375039-1EA; Sigma-Aldrich, San Francisco, CA). Gel concentration was 16% (19 : 1 monomer to bis ratio, Applichem, Darmstadt). About two micrograms of DNA (~1/5 of DNA as used in the CD experiments) was loaded on 14 × 16 × 0.1 cm gels. Electrophoresis was run at 20°C for 4 hours at 126 V (~8 V·cm−1). DNA oligomers were visualized with silver after the electrophoresis, and the electrophoretic record was photographed by an Olympus Camedia 3000 camera [25].
3. Results and Discussion
3.1. CD Spectroscopy
Although CD spectra are routinely used to assign DNA folds [11], the interpretation of optical properties such as hypochromicity or the shape and sign of CD bands can be controversial [26]. G-tetrad stacking and the polarity of DNA strands are determining factors in the intensity and shape of the CD spectrum, and specifically the rotation angle between the stacks. It is well known that parallel G-quadruplex structures give a positive band ~265 nm and a negative band ~240 nm, while antiparallel G-quadruplex structures, such as the basket and chair forms, show two positive bands ~295 and ~245 nm and a negative band ~260 nm [26]. These spectral features are mainly attributed to the specific guanine stacking in various G-quadruplex structures [24, 27].
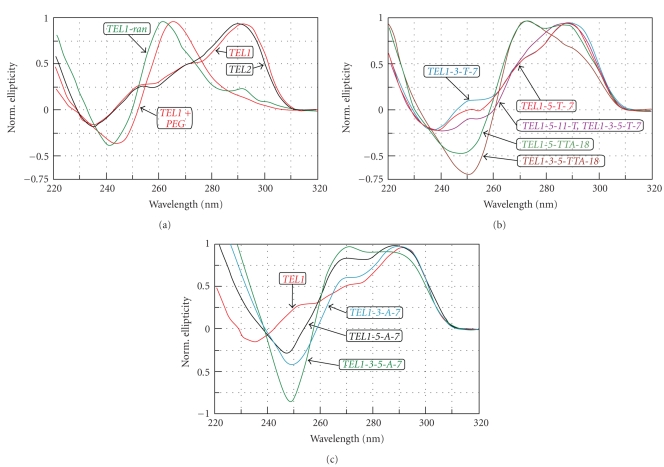
Figure 1 shows CD spectra of oligomers, the sequences of which are summarized in Table 1. CD measurements were performed in a Britton-Robinson buffer containing 50 mM of KCl. These measurements clearly show that protruding nucleotides have a considerable impact on the profile of spectra; longer protrusions have an even more significant influence. The positive CD band of TEL1 and TEL2 at 293 nm associated with a ~265 nm positive shoulder (red and black lines in Figure 1(a)), correspond to the antiparallel hybrid forms in K+ solution [28]. TEL1 in the presence of 50% PEG 200 folds into a parallel conformer, which the CD spectrum shows as no peak at ~295 nm, red-dashed line in Figure 1(a). The spectra of TEL1-3-5-T-7 and TEL1-5-T-11 are in principle the same which is why only one spectrum is presented in Figure 1(b). Prolongation of the quadruplex overhanging sequence increases the negative and positive signals at ~250 nm and 265 nm, respectively, Figures 1(b) and 1(c). These results indicate that tails of G-quadruplexes can influence the quadruplex topology. Our data also supports the suggestions made by other authors that even small changes to flanking sequences can disturb the equilibrium between different coexisting G-quadruplex forms. In a recent study, two separated bands of comparable magnitude were observed at ~270 and ~295 nm for A3G3(T2AG3)3A2 oligomer [20]. Similar features of spectra were observed to that of TEL1-3-5-TTA-18, TEL1-5-TTA-18, and TEL1-3-5-A-7 in K+ solution. In addition, a study of A3G3(T2AG3)3A2 using NMR confirmed a hybrid 2 of quadruplex structure [16]. The flanking nucleotides resulted in a shoulder on the short wavelength side of the 295 nm CD band. The height of this shoulder increased with the number of flanking nucleotides. A similar CD spectrum was observed for G3(T2AG3)4, which contains one redundant T2AG3 repeat [18].
Figure 1.
Normalized CD spectra of G-quadruplex-forming sequences in Britton-Robinson buffer containing 50 mM KCl, pH 7.0 distinguished by different color: TEL1, TEL2, and TEL1 in the presence of 50% PEG-200 and TEL1-ran in (a); TEL1-3-T-7, TEL1-5-T-7, TEL1-3-5-T-7, TEL1-5-T-11, TEL1-5-TTA-18, TEL1-3-5-TTA-18 in (b), and TEL1-3-A-7, TEL1-5-A-7, TEL1-3-5-A-7, and TEL1 in (c).
The presence of multiple G-quadruplex conformations in K+ solution makes structural interpretation difficult under conditions of NMR measurements [15]. To date, only two hybrid conformers have been clearly confirmed by NMR. However, Phan et al. have detected a small population of other (3 + 1) hybrids containing two DCR loops [29]. Our hypotheses explaining the relative increase and decrease of peaks at ~270 nm and ~295 nm, respectively, is as follows; increasing the number of protruding nucleotides shifts the equilibrium from a hybrid with one DCR towards the (3 + 1) arrangement containing two DCR loops. The subsequent introduction of crowding conditions causes a propeller-like parallel arrangement of G-quadruplex to develop
| (1) |
However, our hypothesis has to be verified with NMR at corresponding condition. Interestingly, the CD pattern of TEL1-ran (green line) shows a maximum of ellipticity at ~262 nm associated with ~295 nm positive shoulder. This oligomer supports and confirms our previous suggestions. However, when a protruding sequence is able to form an additional quadruplex, as for example, the TEL2 oligomer used in this study, then two tandemic quadruplexes are more likely to be formed. The same spectral features are to be observed in both TEL1 and TEL2 [26, 30, 31]. Nevertheless, a significant destabilization of TEL2 quadruplex structure can also be observed, Table 2. These results are in agreement with previous studies [26].
Table 2.
Thermodynamic parameters obtained from CD and UV melting curves. UV melting curves were obtained at ~293 nm in the presence of 50 mM KCl. Values in which a two-state mechanism was clearly confirmed by dual-wavelength parametric test are highlighted by bold font.
| Oligomer | Protrusion [nt.] | CD | UV | ||||||
|---|---|---|---|---|---|---|---|---|---|
| T m | ΔH | ΔS | ΔG 37 o | T m | ΔH | ΔS | ΔG 37 o | ||
| [°C] | [kcal · mol−1] | [cal · mol−1 · K−1] | [kcal · mol−1] | [°C] | [kcal · mol−1] | [cal · mol−1 · K−1] | [kcal·mol−1] | ||
| TEL1 | 0–0 | 64.7 | 59.3 | 207 | 4.9 | 65.3 | 58.8 | 205 | 4.9 |
| TEL2 | 0-3-0 | 55.9 | 39.5 | 135 | 2.3 | 55.3 | 34.0 | 116 | 1.9 |
| 5′-overhang | |||||||||
|
| |||||||||
| TEL1-5-TTA | 3–0 | 57.6 | 57.9 | 198 | 3.6 | 57.8 | 58.7 | 201 | 3.7 |
| TEL1-5-T-7 | 7–0 | 53.6 | 51.4 | 174 | 2.6 | 50.6 | 39.2 | 132 | 1.6 |
| TEL1-5-A-7 | 7–0 | 54.8 | 39.0 | 133 | 2.1 | 56.0 | 41.8 | 143 | 2.4 |
| TEL1-5-T-11 | 11–0 | 55.1 | 51.6 | 175 | 2.8 | 54.9 | 60.7 | 206 | 3.3 |
| TEL1-5-TTA-11 | 11–0 | 54.8 | 50.6 | 172 | 2.7 | 53.9 | 58.1 | 197 | 3.0 |
| TEL1-5-T-18 | 18–0 | 51.2 | 43.4 | 143 | 1.9 | nd | nd | nd | nd |
| TEL1-5-TTA-18 | |||||||||
| 293 nm | 18–0 | 51.0 | 42.4 | 137 | 1.8 | nd | nd | nd | nd |
| 263 nm | 52.3 | 42.8 | 144 | 2.0 | nd | nd | nd | nd | |
|
| |||||||||
| 3′-overhang | |||||||||
|
| |||||||||
| TEL1-3-TTA | 0–3 | 59.6 | 55.9 | 193 | 3.8 | 58.9 | 61.6 | 212 | 4.1 |
| TEL1-3-T-7 | 0–7 | 56.8 | 52.8 | 181 | 3.2 | 55.4 | 59.1 | 201 | 3.3 |
| TEL1-3-A-7 | 0–7 | 59.8 | 50.5 | 174 | 3.5 | 59.3 | 50.7 | 175 | 3.4 |
| TEL1-3-T-11 | 0–11 | 57.9 | 57.5 | 197 | 3.6 | 56.1 | 26.8 | 92 | 1.6 |
| TEL1-3-TTA-11 | 0–11 | 57.7 | 55.5 | 190 | 3.5 | 57.1 | 46.8 | 160 | 2.8 |
| TEL1-3-T-18 | 0–18 | 53.4 | 45.8 | 155 | 2.3 | nd | Nd | nd | Nd |
|
| |||||||||
| 3′- and 5′-overhangs | |||||||||
|
| |||||||||
| TEL1-3-5-T-7 | 7–7 | 47.5 | 55.9 | 186 | 1.8 | 48.3 | 39.5 | 132 | 1.4 |
| TEL1-3-5-A-7 | 7–7 | 51.3 | 46.8 | 158 | 2.1 | 52.6 | 52.4 | 179 | 3.1 |
| TEL1-ran 263 nm | 14–4 | 53.5 | 37.5 | 127 | 1.9 | nd | Nd | nd | nd |
| TEL1-3-5-TTA-18 | |||||||||
| 290 nm | 18–18 | 40.5 | 49.5 | 161 | 0.5 | nd | nd | nd | nd |
| 265 nm | 40.9 | 37.6 | 129 | 0.5 | nd | nd | nd | nd | |
3.2. Electrophoretic Analysis
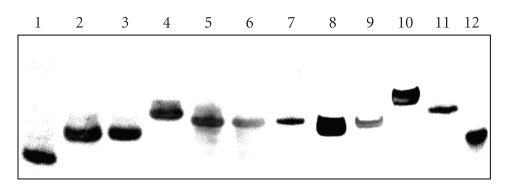
The overhanging sequences significantly influence the mobility of DNA samples. Frequently, the unusually small value of mobility of the parallel conformers which does not correspond to their size can be explained by a higher molecularity. It has been suggested that the mobility of antiparallel and parallel conformers depends only on the number of molecules associated with the formation of its structure [26]. However, in a nondenaturing condition the mobility depends not only on the molecular mass and the charge of macromolecules, but is also strongly determined by topology [24]. Figure 2 displays electrophoretic records of native 16% polyacrylamide gels illustrating the relative mobilities of the oligomers in the presence of 50 mM KCl. The electrophoretic mobilities of TEL1 and their derivatives are clearly visible.
Figure 2.
Electrophoresis at 20°C in 16% polyacrylamide gels in 25 mM Britton-Robinson buffer (pH 7.0) containing 50 mM KCl. Each well contains ~0.5 μM of oligomer. In lines 1–10 there were loaded TEL1, TEL1-3-T-7, TEL1-5-T-7, TEL1-3-5-T-7, TEL1-3-T-11, TEL1-5-T-11, TEL1-ran, TEL2, TEL1-5-TTA-18, and TEL1-3-5-TTA-18 oligomers. Random 20- and 40-mer are loaded in lines 11 and 12.
Random 20-mer and 40-mer were used as DNA standards. The fastest electrophoretic mobility is exhibited by the TEL1 oligomer (line 1); it is consistent with the mobility of monomolecularly folded G-quadruplexes [24, 26, 30]. The mobility retardation caused by the overhanging nucleotides of TEL1 oligomer is evident. The electrophoresis did not confirm any anomalous mobility of oligomers; sequences with the same length move equally and longer oligomers more slowly. For example, the mobilities of TEL1-ran and TEL1-5-TTA-18 sequences are comparable in the given conditions due to similarities in their lengths. The CD spectra of oligomers TEL1, TEL1-3-T-7, TEL1-5-T-7, TEL1-3-T-11, TEL1-5-T-11, and TEL2 show similar profiles, suggesting that all these sequences form a topologically equivalent core of quadruplex, and then their mobilities depend on the length of protruding sequences and not on DNA topology. Though TEL1-ran, TEL1-5-TTA-18, and TEL1-3-5-TTA-18 oligomers show the most intensive peaks at range of 262–272 nm, their mobility is not anomalous to the oligomers discussed above. Therefore, we suggest that the topology of the (3 + 1) hybrid arrangement did not have to be disturbed. However, we cannot decide based on CD and electrophoretic experiment unambiguously whether the arrangement of TEL1-ran is still a hybrid or parallel form. Nevertheless, the TEL2 consists of 45 nucleotides, but this oligomer moves faster than other “tailed” but shorter oligomers; TEL1-3-5-T-7 (35 nt), TEL1-5-T-11, and TEL1-3-T-11 (32 nt). Thus, we suggest that based on CD spectra and electrophoresis, this oligomer forms two quadruplexes within a (3 + 1) hybrid topology ordered in tandem [26].
However, we cannot exclude the possibility of a mixed population of parallel and antiparallel conformers in a solution containing potassium, because both corresponding peaks/shoulders (265 and 295 nm) were observed in some sequences. Electrophoresis excludes a mix of topologically different conformers because only one clear band is detected, although thermodynamically transient conformers can cause smears in certain lines during the electrophoretic separation [24]. The silver staining procedure is highly sensitive in DNA visualization; the advantage is that it allows us to detect even very small populations of structural forms [25].
3.3. Analysis of Melting Curves
Melting curve analysis can help to clarify some other aspects concerning quadruplex stability. Nonsigmoidal shapes of the melting curve have been reviewed by many authors, in particular those concerning quadruplex evaluation [32]. Although van't Hoff's evaluation of melting curves can offer important information, inappropriate use of the application can also lead to incorrect interpretations. This analysis can only offer relevant results for two-state mechanisms, as described by the following scheme: F↔U, where F and N represent the folded and unfolded states of molecule, respectively. Chaires have critically reviewed a routine application of van't Hoff analysis to a number of melting curves [33]. The following methods should be used to verify a two-state mechanism; (i) dual-wavelength parametric test [34], (ii) detection of whole-scale spectra at each temperature, for example, detection of isoelliptic and isosbestic points [12, 23, 33], and (iii) so-called singular value decomposition (SVD) analysis which allows us to detect the presence of kinetically stable intermediate species in the process of quadruplex formation [12, 35]. The dual-wavelength parametric test and detection of isoelliptic and isosbestic points gave ambiguous results concerning a two-state mechanism of quadruplex unfolding (not shown). Only the TEL1-ran, TEL1-3-5-TTA-18, and TEL-5-A-7 oligomers used in this work show signs of two-state melting behavior, therefore the result of van't Hoff enthalpy, entropy, and Gibbs energy change, summarized in Table 2, are provided for information purposes and might be verified by microcalorimetry.
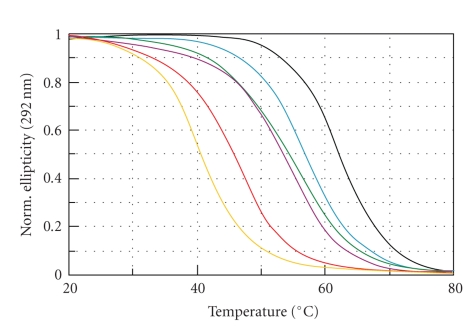
The effect of quadruplex destabilization by tails could be partially explained with analogy to a negatively charged head and a whip. Local thermal fluctuations cause a different movement of the tail and of the head of quadruplex in solution, which can result in a destabilization of the proximal G-tetrad, Figure 3. It appears that 5′-tail destabilizes TEL1 slightly more than 3′-overhanging nucleotides, Table 2. This fact confirms that the folding topology of human quadruplexes, the arrangements of DCR and EW loops in (3 + 1) hybrid conformers, governs a structural stability of G-quadruplex. It seems that the melting temperature and other thermodynamic parameters are affected by protruding nucleotides. However, it is impossible to describe the destabilization effect using simple formulae, because the effect depends on at least two factors: (i) the sequence of overhanging sequence and (ii) the length of DNA overhanging. The random overhanging sequence of TEL-ran presented here provides a noteworthy effect. Its melting temperature is comparable with TEL1-5-TTA-18 oligomer of the same nucleotide size, but the shift of the main CD peak towards lower wavelengths is more evident in comparison with other sequences, Figure 1. The TEL2 oligomer also shows destabilization, although the CD spectrum is in principle equivalent to TEL1. The CD spectrum of TEL2 agrees with the spectrum obtained by Petraccone at al. for (TTAG3)8TT [31], where the CD spectrum of the dimer closely matches the sum of the CD spectra of hybrid 1 and hybrid 2 monomers. According to the analysis of human telomeric quadruplex multimers by molecular dynamics, it has been shown that quadruplexes behave as two independent subunits connected by a TTA linker. The TTA linker loop also loses all of its original conformational features and becomes extended [36]. Therefore, we suggest here that when two thermodynamically equivalent quadruplex units of TEL1 in TEL2 connected by a TTA loop are exhibited to thermal fluctuations in solution, the proximal guanines in G-tetrads might be preferentially destabilized in comparison with TEL1 oligomer. Our suggestion is also supported by previous studies where there is shown that the number of TTAG3 repeats influences the quadruplex thermostability and enthalpy [26]. 5′-protruding nucleotides partially model the end of telomere. We can extrapolate our results and suggest that the 5′-overhanging sequence forces contiguous G-quadruplex to fold into conformers where the positive CD signal at 265–270 nm, originally only being a shoulder in TEL1, is dominant. This can be compared to the “mild” crowding condition, in which DNA oligomer is forced to fold into similar conformers at the presence of potassium and certain concentration of PEG [30, 37].
Figure 3.
Normalized melting curves of TEL1 (black), TEL2 (green), TEL1-3-T-7 (blue), TEL1-5-T-11 (magenta), TEL1-3-5-7-T (red), and TEL1-3-5-TTA-18 (orange) at pH 7.0 obtained by CD spectroscopy collected at 293 nm.
Interestingly, the van't Hoff enthalpies for TEL1-ran and TEL1-3-5-TTA-18 agree with the calorimetric measurements obtained for TEL1 [35]. However, Antonacci et al. reported an unfolding process involving a three-state mechanism for the sequence (TTAGGG)4 with a total enthalpy change of two transitions ranging from 32.1 to 36.3 kcal·mol−1 depending on salt concentration; two clear transitions were clearly distinguished. The difference between melting temperatures of both transitions is about 12°C. One of the possible explanations why we observe a process that fits a two-state mechanism is as follows. Based on CD melting experiments at different wavelengths we are suggesting that unpaired overhangs decrease the melting temperature difference which does not allow distinguishing between the transitions. It could mean that in solution there either occur two topological isoforms with the same mobilities before temperature unfolding and similar unfolding energies or a transition of one topological structure is governed by a three-state mechanism but with proximal melting temperatures. Recently we have discussed in detail a similar effect observed for intermolecular and intramolecular G-quadruplexes formed from various G-rich repetitive sequences, two melting curves at different wavelengths and at least two electrophoretic bands [24]. Therefore, we presume that “tailed” monomolecular quadruplexes might occur in two different structural forms with the same electrophoretic mobility, and the melting process in each of them is governed by a two-state mechanism as illustrated by the example of TEL1-3-5-TTA-18 where two different enthalpy values of 49.5 and 37.6 are reported in Table 2.
4. Conclusions
The human overhanging sequence on the 3′-end consists of 100–200 nucleotides forming G-quadruplexes [2, 3]. 21 nucleotides are consumed in the formation of one quadruplex subunit and three additional nucleotides are necessary for the connection of two neighboring quadruplexes. Therefore, the end of telomere can contain no more than 4–8 tandem quadruplexes.
The favorable formation of a G-quartet stack is enthalpy-driven, and this favorable enthalpy is compensated by an unfavorable entropy contribution, consistent with the immobilization of counterions [37]. A decrease in degree of freedom is normally associated with the entropy changes. Overhanging sequences at 3′- and/or 5′-end cause a decrease in the degree of freedom of DNA folding; this effect is the most evident for longer quadruplex protrusions and can be compared with the TEL2 oligomer, however, in case the protruding sequence again consists of telomeric repeats able to form an additional quadruplex structure [26]. Our results confirm that the 3′- and 5′-overhangs in human telomeric quadruplexes are not equivalent due to the arrangement of DCR and EW loops.
Acknowledgments
This study was supported by grants from the Slovak Grant Agency (1/0153/09), the Science and Technology Assistance Agency (APVT-20-006604), and COST action MP0802. The authers would like to thank L. Sieber and G. W. Cowper for a critical paper proofreading.
Abbreviations
- DCR:
Double-chain-reversal loop
- EW:
Edgewise loops
- nt:
Nucleotide
- PEG:
Polyethylene glycol.
References
- 1.Grandin N, Charbonneau M. Protection against chromosome degradation at the telomeres. Biochimie. 2008;90(1):41–59. doi: 10.1016/j.biochi.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 2.McEachern MJ, Krauskopf A, Blackburn EH. Telomeres and their control. Annual Review of Genetics. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417(6891):876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 4.Sun D, Thompson B, Cathers BE, et al. Inhibition of human telomerase by a G-quadruplex-interactive compound. Journal of Medicinal Chemistry. 1997;40(14):2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 5.Mergny J-L, Hélène C. G-quadruplex DNA: a target for drug design. Nature Medicine. 1998;4(12):1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 6.Londoño-Vallejo JA. Telomere instability and cancer. Biochimie. 2008;90(1):73–82. doi: 10.1016/j.biochi.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Granotier C, Pennarun G, Riou L, et al. Preferential binding of a G-quadruplex ligand to human chromosome ends. Nucleic Acids Research. 2005;33(13):4182–4190. doi: 10.1093/nar/gki722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-β facilitates G-quadruplex DNA unfolding in ciliates. Nature Structural and Molecular Biology. 2008;15(6):598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 9.Smargiasso N, Rosu F, Hsia W, et al. G-quadruplex DNA assemblies: loop length, cation identity, and multimer formation. Journal of the American Chemical Society. 2008;130(31):10208–10216. doi: 10.1021/ja801535e. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1(4):263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Research. 2005;33(14):4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray RD, Li J, Chaires JB. Energetics and kinetics of a conformational switch in G-quadruplex DNA. Journal of Physical Chemistry B. 2009;113(9):2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Research. 2008;36(12):4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Research. 2007;35(19):6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Research. 2006;34(9):2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation. Nucleic Acids Research. 2007;35(7):2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim KW, Amrane S, Bouaziz S, et al. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. Journal of the American Chemical Society. 2009;131(12):4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue Y, Kan Z-Y, Wang Q, et al. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. Journal of the American Chemical Society. 2007;129(36):11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 19.Zheng KW, Chen Z, Hao YH, Tan Z. Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA. Nucleic Acids Research. 2009;38(1):327–338. doi: 10.1093/nar/gkp898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renčiuk D, Kejnovská I, Školáková P, Bednářová K, Motlová J, Vorlíčková M. Arrangements of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Research. 2009;37(19):6625–6634. doi: 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Q, Lu M, Kallenbach NR. Effect of thymine tract length on the structure and stability of model telomeric sequences. Biochemistry. 1993;32(14):3596–3603. doi: 10.1021/bi00065a010. [DOI] [PubMed] [Google Scholar]
- 22.Schonhoft JD, Bajracharya R, Dhakal S, Yu ZB, Mao HB, Basu S. Direct experimental evidence for quadruplex-quadruplex interaction within the human ILPR. Nucleic Acids Research. 2009;37(10):3310–3320. doi: 10.1093/nar/gkp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mergny J-L, Phan A-T, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Letters. 1998;435(1):74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 24.Víglaský V, Bauer L, Tlučková K. Structural features of intra- and intermolecular G-quadruplexes derived from telomeric repeats. Biochemistry. 2010;49(10):2110–2120. doi: 10.1021/bi902099u. [DOI] [PubMed] [Google Scholar]
- 25.Bassam BJ, Gresshoff PM. Silver staining DNA in polyacrylamide gels. Nature Protocols. 2007;2(11):2649–2654. doi: 10.1038/nprot.2007.330. [DOI] [PubMed] [Google Scholar]
- 26.Vorlíčková M, Chládková J, Kejnovská I, Fialová M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Research. 2005;33(18):5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray DM, Wen J-D, Gray CW, et al. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008;20(3-4):431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 28.Kypr J, Kejnovská I, Renčiuk D, Vorlíčková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Research. 2009;37(6):1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Research. 2006;34(19):5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyoshi D, Nakao A, Sugimoto N. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry. 2002;41(50):15017–15024. doi: 10.1021/bi020412f. [DOI] [PubMed] [Google Scholar]
- 31.Petraccone L, Trent JO, Chaires JB. The tail of the telomere. Journal of the American Chemical Society. 2008;130(49):16530–16532. doi: 10.1021/ja8075567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Research. 2008;36(17):5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaires JB. Human telomeric G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS Journal. 2010;277(5):1098–1106. doi: 10.1111/j.1742-4658.2009.07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallimann P, Kennedy RJ, Miller JS, Shalongo W, Kemp DS. Dual wavelength parametric test of two-state models for circular dichroism spectra of helical polypeptides: anomalous dichroic properties of alanine-rich peptides. Journal of the American Chemical Society. 2003;125(5):1203–1220. doi: 10.1021/ja0275360. [DOI] [PubMed] [Google Scholar]
- 35.Antonacci C, Chaires JB, Sheardy RD. Biophysical characterization of the human telomeric (TTAGGG)4 repeat in a potassium solution. Biochemistry. 2007;46(15):4654–4660. doi: 10.1021/bi602511p. [DOI] [PubMed] [Google Scholar]
- 36.Yu H-Q, Miyoshi D, Sugimoto N. Characterization of structure and stability of long telomeric DNA G-quadruplexes. Journal of the American Chemical Society. 2006;128(48):15461–15468. doi: 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- 37.Olsen CM, Gmeiner WH, Marky LA. Unfolding of G-quadruplexes: energetic, and ion and water contributions of G-quartet stacking. Journal of Physical Chemistry B. 2006;110(13):6962–6969. doi: 10.1021/jp0574697. [DOI] [PubMed] [Google Scholar]