Abstract
Third-generation (G3) poly(amidoamine) (PAMAM) dendrimers are simulated approaching 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayers with fully atomistic molecular dynamics, which enables the calculation of a free energy profile along the approach coordinate. Three different dendrimer terminations are examined: protonated primary amine, uncharged acetamide, and deprotonated carboxylic acid. As the dendrimer and lipids become closer, their attractive force increases (up to 240 pN) and the dendrimer becomes deformed as it interacts with the lipids. The total energy release upon binding of a G3–NH3+, G3–Ac, or G3–COO– dendrimer to a DMPC bilayer is, respectively, 36, 26, or 47 kcal/mol or, equivalently, 5.2, 3.2, or 4.7 × 10–3 kcal/g. These results are analyzed in terms of the dendrimers’ size, shape, and atomic distributions as well as proximity of individual lipid molecules and particular lipid atoms to the dendrimer. For example, an area of 9.6, 8.2, or 7.9 nm2 is covered on the bilayer for the G3–NH3+, G3–Ac, or G3–COO– dendrimers, respectively, while interacting strongly with 18–13 individual lipid molecules.
Introduction
Understanding the interaction of nanoparticles with biological membranes is of fundamental importance for both the design of effective nanodevices for medical applications and for avoiding unintended consequences from disruption of biological structures. Nanoparticles are chemically modified for precisely defined properties in attempts to create agents for gene therapy and targeted drug delivery. As nanodevices approach a living animal cell, the first interaction is with the cellular plasma membrane. Nanometer-sized particles are especially effective at penetrating the plasma membrane and altering the natural processes within the cell.1–8 Nanoparticles can be effective carriers of therapeutic cargos through the plasma membrane, likely due to one of two internalization mechanisms: physical rupturing and pore formation1–3 or receptor-mediated, energy-dependent cellular processes, such as endocytosis.6–8
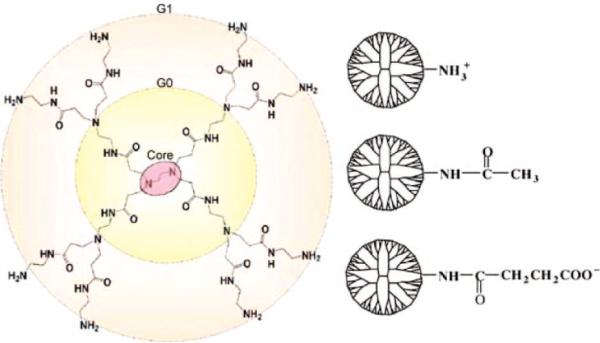
Synthetic polymer nanodevices have been developed from poly(lysine), poly(ethylenimine), and assorted dendrimers.9 Poly(amidoamine) (PAMAM) dendrimers are a widely studied synthetic polymer for both medical and basic science applications.10–12 Their structure incorporates the starburst addition of repeating units around the ethylenediamine core to the desired generation (G) (Figure 1). Dendrimers are particularly useful due to their high homogeneity (polydispersity index ≈1.01)2 and use as a framework in multifunctional nanodevices. PAMAM dendrimers are natively terminated with primary amines and are commonly modified by the covalent addition of small functional molecules (e.g., chromophores, chemotherapeutics, targeting moieties) and less reactive terminations (e.g., acetamide or hydroxyl) to create a nanodevice capable of targeting, binding, internalizing, labeling, and/or treating diseased cells.10,13–15
Figure 1.
Composition of a natively terminated first-generation (G1) PAMAM dendrimer and three possible terminations: protonated primary amine (–NH3+), neutral acetamide (–Ac), and deprotonated carboxylic acid (–COO–). A summary of dendrimer properties is given in Table 1.
For example, acetylization of the PAMAM dendrimer's primary amine end groups can reduce nonspecific binding, enhance functional capabilities, and minimize nanoparticle toxicity.10,14–16 Positively charged nanoparticles are more likely to bind and internalize into cells than are the uncharged analogues.2,3 This strong relationship between nanoparticle termination and biological behavior demonstrates the importance of understanding the mechanisms of nanoparticle–membrane interactions for effective nanodevice design.
Atomic force microscopy (AFM) has been used to experimentally study the effects of PAMAM dendrimers on 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) lipid bilayers with nanometer-scale resolution.17–19 Drastic differences in the dendrimer–lipid interaction were observed for varying dendrimer termination. Charged G5 dendrimers at 20 nM caused membrane disruption via removal of membrane material via the formation of membrane holes and the expansion of preexisting defects. In contrast, neutral G5 dendrimers at 20 nM did not cause as much membrane degradation but predominantly accumulated around the edge of preexisting holes without removing lipid molecules.
Ginzburg and Balijepalli20 have utilized continuum thermodynamic models to demonstrate the effects nanoparticle surface charge density has on membrane binding. This model provides a phase diagram of nanoparticle–membrane structures, including regimes of nanoparticle binding within the bilayer's hydrophobic tails and enclosed within vesicles. Lee and Larson21 have looked at dendrimer–phospholipid binding in greater detail through coarse-grained simulations, providing qualitatively similar conclusions of nanoparticle charge and size dependence on membrane disruption.
The research reported in this paper presents a quantitative analysis of molecular dynamic simulations of the dendrimer–membrane binding process. To date, only a qualitative picture existed concerning the pore formation process by nanoparticles on lipid bilayers.20,21 Specifically, we examine the differences in energetics and forces of interaction between charged PAMAM dendrimers with zwitterionic DMPC bilayers. Three types of G3 PAMAM dendrimers were examined: positively charged primary amine (G3–NH3+), uncharged acetamide (G3–Ac), and negatively charged carboxyl (G3–COO–). The CHARMM27 force field is used for atomistic Langevin dynamics simulations along an interaction coordinate defined as the center-of-mass separation distance between the dendrimer and lipid bilayer. A distance-dependent dielectric function was used. A potential of mean force is calculated along the interaction coordinate, from which the forces of interaction are extracted. Further, the dendrimers’ structure (radii of gyration, asphericities, and atomic distributions) and binding morphology are examined as a function of their proximity to the lipid membrane.
Methods
Umbrella sampling22 and the weighted histogram analysis method (WHAM)23 were employed to extract the free energy of binding along the interaction coordinate defined as the dendrimer and lipid bilayer center-of-mass separation distance. The potential of mean force (PMF),24 or free energy, along the interaction coordinate (z) is extracted from the coordinate's distribution function, averaged over all degrees of freedom as represented in the configurational vector (X):
| (1) |
with Boltzmann constant (kB), the temperature (T), and the CHARMM25 potential (V) with the protein parameter set 22.26
The PMF along particular (slow) coordinates is a central concept in the statistical mechanical representation of molecular systemsandhasbeenwidelyusedincomputationalapplications27–30 The PMF reveals the equilibrium conformation (i.e., PMF minimum) and free energy changes. The negative gradient of the PMF is exactly the force averaged over the conformational ensemble.24 In the umbrella sampling calculation performed here long-time events along the interaction coordinate are encouraged to happen within the nanosecond time scale by stepwise moving of the umbrella sampling window offset along the interaction coordinate.
The interaction coordinate (z), defined as the dendrimer–lipid center-of-mass separation distance, was examined from values of 2.8–6.9 nm. Sampling dynamics occurred in equally spaced windows along the interaction coordinate with 0.05 nm step size and a harmonic confinement of the interaction coordinate to the window center by 500 kcal mol–1 nm–2. To start the simulations, the dendrimer and lipid bilayer were equilibrated with a harmonic confinement to z = 6 nm for 500 ps. The result of this equilibration was used as the initial conditions for the production dynamics in the z = 6 nm window as well as the starting configuration for the equilibration at z = 5.95 nm, which lasted another 100 ps. The result of the equilibration at 5.95 nm was used to begin the z = 5.9 nm window equilibration and so forth until all windows from 6 to 2.8 nm were recursively equilibrated. Once each window was equilibrated, production runs were calculated in parallel for 4 ns per window with atomic configurations saved every 2 ps. To confirm that z > 6 nm was outside the interaction range of the dendrimer with the lipid bilayers, further sampling windows were run for 6.05 nm ≤ z ≤ 6.9 nm with identical recursive equilibration procedures starting from the z = 6 nm configuration and recursively stepping to larger z.
The interaction coordinate versus time was recorded during all production runs and later used with WHAM to calculate the PMF.23,24 The recursive calculation of the PMF via WHAM was performed until a fitting threshold of 0.001 kcal/mol was achieved.27 WHAM results for the PMF were demonstrated to be dependent on the threshold such that more stringent thresholds resulted in a difference smaller than that between the PMF at z = 6 nm and z = 6.9 nm. Since the PMF at this extended range is flat and a sufficiently low threshold value was too costly to compute, a linear fit to the PMF was calculated for z = 6.0–6.9 nm and subtracted for the entirety of the PMF.
The initial structure for the DMPC bilayer originated from a preequilibrated patch of 32 lipid molecules replicated 16 times to create a single patch with 512 lipid molecules, from which a 10 nm diameter circular disk was extracted. Lipid equilibration occurred at every step during the creation of the final 10 nm patch for greater than 200 ps each. The equilibrated DMPC disk consisted of 263 phospholipid molecules at a lipid density (60 Å2/lipid) and bilayer thickness (4.3 nm) in agreement with experimental measurements of DMPC bilayers.31,32
The focus of this paper is the examination of binding of dendrimers to a membrane with biological relevance. The primary contribution to the initial binding of dendrimers to membranes was thought to be the interaction between the charged dendrimer moieties with the polar lipid head groups.33–35
Subsequently, the hydrophobic dendrimer moieties and lipid tails are likely playing an important factor in the later pore formation and penetration of the dendrimer into the bilayer. This manuscript focuses on the initial binding of dendrimers to the lipid bilayer surface. As such, in the interest of computational efficiency, the lipid tail atoms have been fixed in location. Indeed, the PMF computations were not feasible allowing full lipid mobility. This approximation limits the lateral and vertical motion of the individual lipid molecules and prevents lipid tail rearrangement from being incorporated into the resulting energetic calculations. This lipid tail fixation is not expected to significantly affect the binding for large dendrimer–lipid separations (z ≥ 4 nm) but does affect the binding at smaller separations. The effects of this approximation, and the influence of lipid tail mobility in general, is the subject of a companion article.36
Three different PAMAM dendrimers of varying surface chemistry are simulated with DMPC bilayers using the CHARMM27 parameters for lipids37 and, for dendrimers, parameters from set 22, within the CHARMM c32b2 academic software package.37,38 The G3 dendrimers natively contain 32 terminal groups, a net zero charge, and 1092 atoms before modifying the terminal chemistry. The terminations are modified to yield the G3–NH3+, G3–Ac, and G3–COO– dendrimers (Figure 1), to make the dendrimers representative of a physiological pH 7 environment.39 Dendrimer properties are given in Table 1 with each protonated primary amine terminal group having a +e charge and each deprotonated carboxylic acid terminal group having a –e charge. This yields three different dendrimers terminations giving the dendrimers a net charge of +32e, 0, or –32e. The initial structure for the dendrimer was generated by a recursive script in CHARMM38 and equilibrated for over 400 ps in the absence of lipids. At this point the temperature, potential energy, and radius of gyration were stabilized. All images of the molecular structure within this manuscript were created with the software VMD.40
TABLE 1.
Properties of the G3 PAMAM Dendrimers
| terminal groupa | no. of atoms per dendrimer | mass (kDa) | charge (e) | |
|---|---|---|---|---|
| G3–NH3+ | –NH3+ | 1124 | 6.94 | +32 |
| G3–Ac | –NHCOCH3 | 1252 | 8.25 | 0 |
| G3–COO– | –NHCOCH2CH2COO– | 1412 | 10.1 | –32 |
All 32 primary amines of each G3 PAMAM dendrimer are converted to one of these three terminal groups, giving the dendrimers a +32e, 0, or –32e net charge.
Electrostatic interactions are modeled with spatially dependent screening incorporated by an r-dependent dielectric function of the type ε(r) = 4r.41 This model accounts for the enthalpic electrostatic screening effects of the solvent and counterions but does not incorporate solvent or counterion entropic effects. Nonbonded interactions are cut off for atom–atom separation distances greater than 1.3 nm and switched at 0.8 nm.
The accuracy of the distance-dependent dielectric function on the dendrimer structure has been previously compared to molecular dynamics in explicit water with counterions with satisfactory results.17,41–44 Although the 4r-dependent dielectric solvent model is an approximation relative to explicit water solvation, the size of these simulations, if surrounded by appropriate explicit solvent molecules, would have created a system with a volume of 3600 nm3 and 260 000 atoms. Already, simulation of the system with implicit solvent is computationally challenging, especially for the highly demanding process of developing a PMF, as is performed here. Similar calculations using explicit water are currently too computationally demanding. The distance-dependent dielectric simulations we perform provide new, meaningful details of the energetics and morphology of dendrimer–phospholipid binding, while avoiding drawbacks that explicit solvent simulations may incorporate, such as excessive computation time impeding the achievement of ergodicity, slow water equilibration, or insufficient averaging of counterion distribution.
As an example of current difficulties using explicit solvent, recently Changand Violi45 have performed similar nanoparticle–lipid bilayer molecular dynamics on a considerably smaller system and had difficulty achieving adequate equilibration of the explicit solvent. Their study of lipid–nanoparticle interactions includes a carbon nanoparticle ( the size of the G3 dendrimer) and demonstrates hysteresis effects over the course of the simulations due to the initial water molecule positions and slow water equilibration times. Therefore, the use of implicit solvent will be necessary for computational studies of the dynamics of nanoparticles and membranes for the time being, especially nanoparticles of the relatively large size and internal complexity of dendrimers.
An alternative to the 4r dielectric function is the use of generalized Born solvation models.46–48 For example, implicit solvent models using the analytical continuum electrostatics (ACE) or generalized Born using molecular volume (GBMV) analysis49,50 have been effectively utilized with the CHARMM force field to study protein folding/unfolding.51,52 ACE and GBMV and other models53 have demonstrated, for proteins, good agreement with the thermal unfolding properties reported by experiments and MD simulations with explicit water (with exceptions such as differences in solvent dynamics leading to a dewetting transition.)54–56 However, our tentative utilization of generalized Born implicit solvent models within simulations of dendrimers and lipids yielded results that were unsatisfactory because of the lack of well-established parameters for solvation of lipids in water.
Although an explicit atomic representation of water molecules is the most accurate means to account for the effect of the solvent on molecular kinetics and thermodynamics,55,56 we already have presented the computational limitations regarding the simulation for 129 windows for >35 000 dendrimer and lipid atoms in addition to explicit solvent atoms. Additionally the implicit solvent model enhances computational time due through instantaneous (adiabatic) equilibration of implicit water around the solute. If water would be explicitly represented, it would be out of equilibrium for this duration and the objective of simulating representative dendrimer dynamics in equilibrated water would not be obtained. Implicit representation is not necessarily a poorer choice in this regard because it maintains the time scale separation of water dynamics and dendrimer dynamics by instantaneous relaxation akin to an adiabatic separation. Given these considerations, in combination with the necessity of many trajectories to sample even the most probable conformations reasonably well, we have opted for an implicit water representation.
The constant temperature ensemble was generated using Langevin dynamics to simulate the thermal fluctuations within an implicit solvent by incorporating random fluctuations, (Ri(t)), and a damping constant (γ) into Newton's equations, as shown in eq 2. xi represents the position of the ith atom of mass mi in CHARMM potential V:
| (2) |
The friction coefficient γ is related to Ri(t) by fluctuation–dissipation; 〈Ri(0) · Ri(t)〉 = 2mikBTγδ(t). For bulk water, γ = 91 ps–1. During equilibration, γ was decreased to 10 ps–1 because lower γ values accelerate molecular kinetics while yielding identical equilibrium structures. During dynamics, we set γ = 50 ps–1 to mimic the decreased viscosity within the dendrimer. During both equilibration and production runs, the temperature was set to 300 K.
Results and Discussion
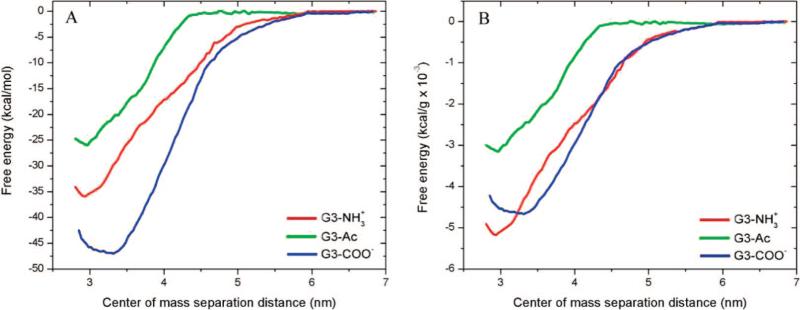
The binding of G3–NH3+, G3–Ac, and G3–COO– to DMPC bilayers was simulated (Figure 2). The energetics, macromolecular structure, and mechanisms of interaction are examined with atomistic detail. The PMF has been calculated along the interaction coordinate defined as the center-of-mass separation for the dendrimer–lipid system from 2.8 to 6.9 nm (Figure 3). The charged dendrimers (G3–NH3+ and G3–COO–3) are more strongly attracted to the lipids than the uncharged dendrimer (G3–Ac). As listed in Table 2, the total free energy released upon interaction with the bilayer is 36, 26, and 47 kcal/ mol for G3–NH3+, G3–Ac, and G3–COO–, respectively. The majority of the released energy occurs at smaller separation distances, where the dendrimer and lipid have made contact and the attractive force is large. Within these simulations, G3–COO– released the most energy per molecule upon binding to the DMPC bilayer; however, G3–NH3+ released the most energy per mass. As shown in Figure 3B and Table 2 with the mass-weighted energy release of binding per dendrimer is 5.2, 3.2, and 4.7 × 10–3 kcal/g for G3–NH3+, G3–Ac, and G3–COO–, respectively. In both the per-molecule and per-mass analyses, G3–Ac released the least energy upon binding.
Figure 2.
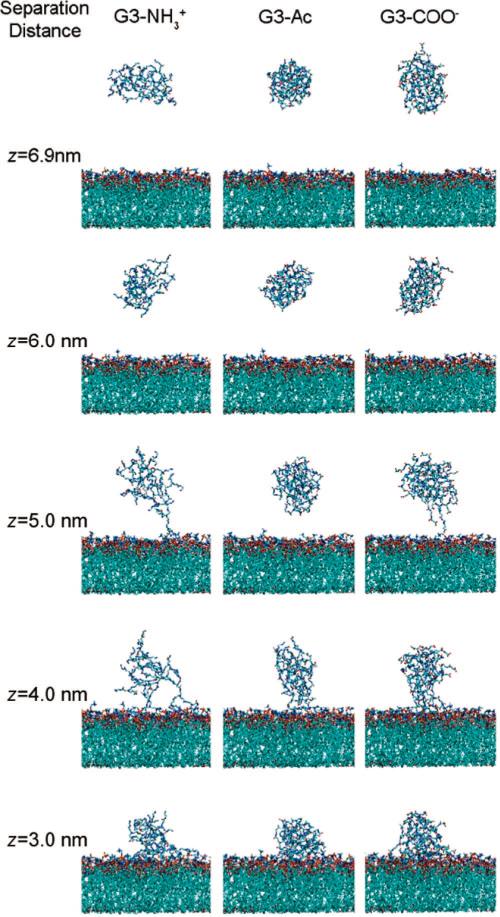
Images of G3–NH3+, G3–Ac, and G3–COO– at center-of-mass separation distances of 6.9, 6.0, 5.0, 4.0, and 3.0 nm from the DMPC bilayer. For z > 6 nm, there is no interaction between the dendrimers and lipids. At z = 3 nm the dendrimers are near the equilibrium separation, as determined by the potential of mean force (PMF) (Figure 3). Notice the variation in dendrimers’ size, shape, and density, as quantified in Figures 5 and 6. Animations of dendrimers binding to the DMPC bilayer are found in the Supporting Information.
Figure 3.
Potential of mean forces for dendrimers, of varying termination, binding to DMPC bilayers. (A) shows the energy per molecule binding to the bilayers while (B) shows the energy per mass. The total energy release for the binding is 36, 26, and 47 kcal/mol and 5.2, 3.2, 4.7 × 10–3 kcal/g for the G3–NH3+, G3–Ac, and G3–COO–, respectively.
TABLE 2.
Simulation Results of G3 PAMAM Dendrimers Binding to DMPC Bilayers
| energy released upon binding |
maximum attractive force (pN) | maximum radius of gyration (nm) | maximum asphericity (10–2) | ||
|---|---|---|---|---|---|
| (kcal/mol) | (10–3 kcal/g) | ||||
| G3–NH3+ | 36 | 5.2 | 170 | 1.55 | 9 |
| G3–Ac | 26 | 3.2 | 200 | 1.3 | 7 |
| G3–COO– | 47 | 4.7 | 240 | 1.4 | 6 |
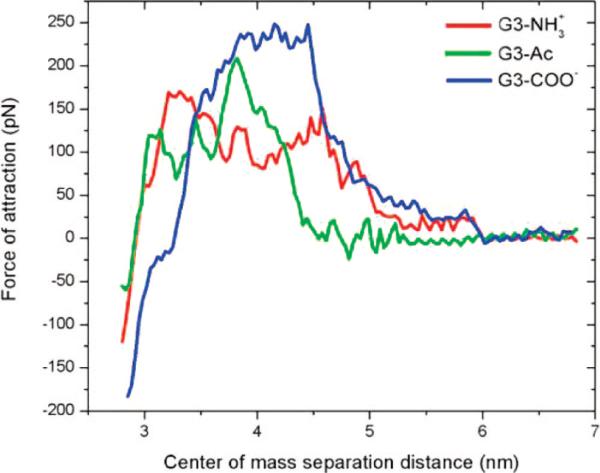
The derivative of the PMF as a function of the interaction coordinate is the mean force between the dendrimer and lipid bilayer (Figure 4). The attractive force between the dendrimers and lipid bilayers ranged from 0 to 240 pN (Table 2), depending on dendrimer termination and dendrimer–lipid separation distance. The attractive force increases as contact is made between the dendrimer and lipids and decreases to zero as the dendrimer and lipid approach their equilibrium separation distances of z ≈ 3 nm. The attractive force before contact is made (z > 4.5 nm) seems to depend only on the magnitude of dendrimer charge and not on the sign of the charge; G3–NH3+ and G3–COO– have similar forces of attraction (≈35 pN) in this range. At distances z < 4.5 nm, variables such as number of atoms per dendrimer and terminal group size contribute to the differences between the dendrimers. The maximum attractive force is greatest for the carboxyl dendrimer, the largest of the three dendrimers examined.
Figure 4.
Attractive force between dendrimers of varying termination and a DMPC bilayer. There is no force at larger separation distances (z > 6 nm), which increases to 170, 200, and 240 pN for G3–NH3+, G3–Ac, and G3–COO–, respectively.
The dendrimer structure is affected by the lipids, as quantified by the dendrimers’ radii of gyration within each umbrella sampling window. A dendrimer's radius of gyration (RG) represents the magnitude by which the dendrimer atoms are stretched out away from their close-packed state. RG is calculated according to eq 3, incorporating the dendrimer's mass (M) and center of mass (x0), in addition to each dendrimer atom's mass (mi) and position (xi):
| (3) |
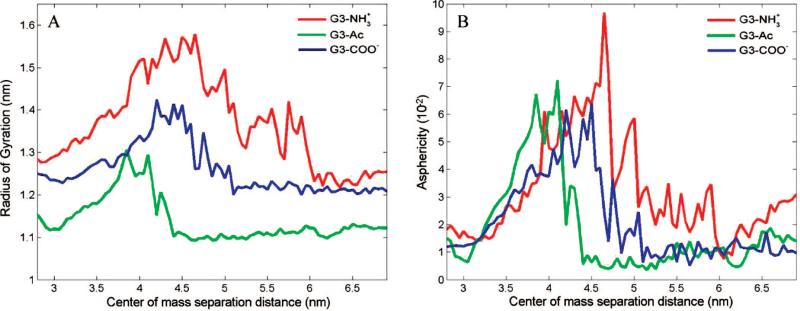
The radius of gyration increases, by 19% on average, as the dendrimer approaches the lipids (Figure 5A). As the system becomes close to the equilibrium separation (z < 3.7 nm) the radius of gyration decreases to approximately the initial, isolated values. The dendrimer elongates in response to the interactions with the lipid and settles into an equilibrated size similar to that without the influence of lipids. Throughout the entire range of interaction coordinate, the size of the three dendrimers is ordered as G3-NH3+ > G3–COO–3 > G3–Ac, with the smallest mass dendrimer demonstrating the largest RG.
Figure 5.
Radius of gyration and asphericity vs interaction coordinate for the three different dendrimers. The dendrimers become influenced by the lipids for z < 6 nm, and the radii of gyration and asphericities increase. Both the radii of gyration and asphericities decreases as the dendrimers approach their equilibrium bound states at z ≈ 3 nm. These results can be qualitatively observed in the images of the simulation shown in Figure 2.
The asphericity (A) is a quantifiable description of how deformed from a reference spherical shape the dendrimer has become (Figure 5B). The asphericity has been calculated according to eq 4 by comparing the eigenvectors Ix,y,z of the dendrimer's moment of inertia matrix:57
| (4) |
The three dendrimers in this study have similar asphericity (A = 0.014) at z ≈ 6 nm, which demonstrates the slight asymmetry in the PAMAM dendrimer core. As the dendrimer and lipids become closer, the asphericities of the dendrimers increase to A ≥ 0.06 at z ≈ 4.2 nm, a 4.3-fold increase. With further decrease in separation, the dendrimer returns to a near spherical shape and settles into the equilibrium interaction coordinate, z ≈ 3 nm, with A = 0.015.
Upon decreasing dendrimer–lipid separation distance the dendrimer stretches normal to the bilayer for greater contact with the lipids. This transition to larger asphericity occurs simultaneously with the increasing dendrimer radius of gyration, increased attractive force to the bilayer, and initial contact between the dendrimer and lipids. The charged dendrimers contact the bilayer at z = 5.1 nm, whereas the uncharged dendrimer does not contact the bilayer until z = 4.5 nm (Figure 2).
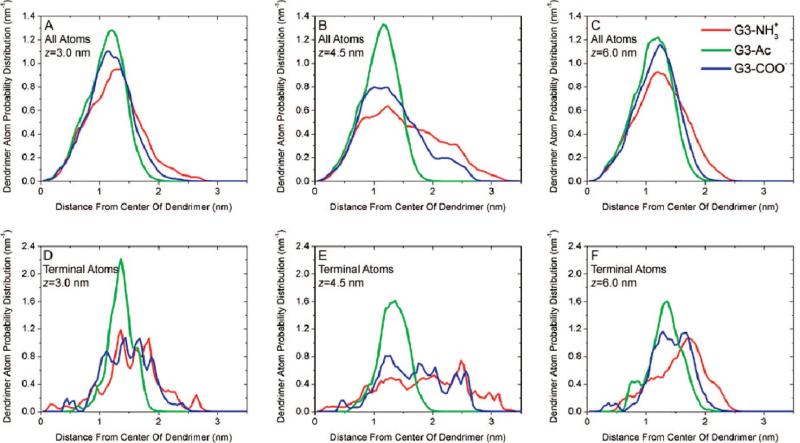
Turning from these average quantities, it is useful to examine the distribution of the dendrimer atoms in more detail. This can provide an understanding of the dendrimer structural change upon binding. Normalized histograms of radial atomic positions from the dendrimers’ centers of mass demonstrate the distribution of dendrimer atoms (Figure 6A–C) and the terminations (Figure 6D–F). At large bilayer–dendrimer separation, the atoms within the dendrimer are distributed with nearly uniform density. Specifically, the dendrimer terminal groups are distributed throughout the dendrimer with both large and small separation distances. This may be surprising, as the terminal groups might be expected to form an outer shell due to electrostatics. At z = 4.5 nm the charged terminal groups are pulled away from the dendrimer center as they become attracted to the DMPC bilayer.
Figure 6.
Atomic distributions for G3–NH3+, G3–Ac, and G3–COO– at varying interaction coordinates. The vertical axis is a normalized probability distribution of (A–C) all dendrimer atoms or (D–F) the 32 most terminal carbon or nitrogen atoms in each dendrimer. The horizontal axis is the radial distance from dendrimer center of mass. Results have been averaged over a 4 ns simulation within each sampling window: (A and D) z = 3.0 nm, (B and E) z = 4.5 nm, and (C and F) z = 6.0 nm.
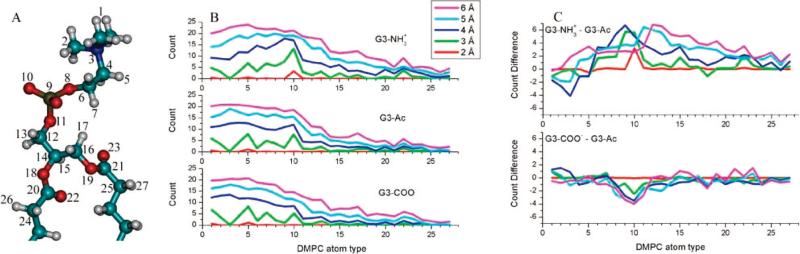
The atomic details of the dendrimer-lipid interaction have been examined at the equilibrium separation distance for all three dendrimers on the DMPC bilayer. Each atom in the DMPC molecule has been compared by its proximity to the dendrimer, and differences in the binding morphology of the three dendrimers are identified. The individual atoms on the lipid molecules, as labeled in Figure 7A, have been examined via counting how many of each DMPC atom type are within 2, 3, 4, 5, or 6 Å of the dendrimer (Figure 7B). This analysis considers all DMPC atoms of identical structure as a single type, and the resulting count is normalized by the degeneracy of each atom type. The data of Figure 7B has been further compared in Figure 7C by subtracting the data of G3–Ac from the data of G3–NH3+ and G3–COO–. In this way the binding morphology of the charged terminal groups can be more clearly observed. Conclusions from this data include (1) fewer positively charged choline atoms are close to G3–NH3+, (2) more negatively charged phosphate atoms are close to G3–NH3+, (3) fewer negatively charged phosphate atoms are close to G3–COO–, and (4) G3–NH3+ are able to penetrate deeper into the DMPC bilayer and closer to more of the glycerol atoms.
Figure 7.
Plot of which DMPC atoms are in close proximity to each dendrimer at the equilibrium dendrimer–lipid separation distance. The atoms in each DMPC molecule have been assigned a number, according to (A), and plotted on the horizontal axis of (B) and (C). Identical DMPC atoms have been assigned the same number, and the results have been normalized appropriately. The vertical axis of (B) represents the average number of DMPC atoms of each type that are within 2, 3, 4, 5, or 6 Å of the dendrimer at the equilibrium interaction coordinate. (C) shows the difference between the charged and uncharged dendrimers.
The morphology of the equilibrated structure is examined in terms of the area that the dendrimers occupy over the lipid bilayer. The dendrimers cover an area of 9.6, 8.2, or 7.9 nm2 for G3–NH3+, G3–Ac, or G3–COO–, respectively, with G3–NH3+ flattening out the most in covering the greatest area of lipids. This analysis measures the footprint, or the area of their shadow on the lipids. The structures have also been examined by the area of the smallest circle inscribing the projection of the dendrimer onto the bilayer plane. The smallest circles containing the dendrimer have area of 14.9, 9.4, or 10.4 nm2 for G3–NH3+, G3–Ac, or G3–COO–, respectively. By comparing the size of the inscribing circle to the size of the dendrimer footprint, it is revealed how spherically symmetrical and dense the dendrimers are over the bilayer. G3–NH3+ stretches out the farthest on the bilayer, as represented by its largest inscribing circle. These effects are observed qualitatively in the top view of the bound dendrimers in Figure 8. Interestingly, G3–COO– is the largest mass dendrimer but displays the smallest footprint upon binding to the lipids. This is one of the many ways by which the longer termination of G3–COO– affects atomic structure and yields considerable differences from the other terminations.
Figure 8.
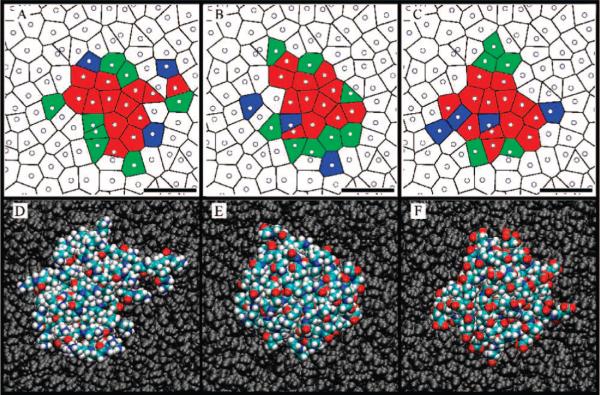
(A–C) Voronoi diagram of the top leaflet of the DMPC bilayer with coloration according to portion of time each DMPC molecule is within 3 Å of the dendrimer.(D–F) Top view of dendrimers bound to the bilayer. (A and D) G3–NH3+, (B and E) G3–Ac, and (C and F) G3–COO–. (A–C) Circles correspond to each lipid molecule's center of mass projected onto the bilayer plane, and the thin black lines represent the boundaries between lipid molecules. Lipid molecules colored red represent those that are within 3 Å of the dendrimer in 100% of the simulation frames; green indicates close proximity 50–100% of the time, and blue indicates 1–50%. The scale bar in each image is 2 nm. (D–F) Lipids are colored gray and dendrimers are colored cyan, blue, white, and red for atom types C, N, H, and O, respectively.
A final analysis of the morphology of the dendrimers equilibrated on the DMPC bilayers was performed with a Voronoi diagram (Figure 8A–C).58 Each circle represents the center of mass of a DMPC molecule on the top bilayer leaflet projected onto the bilayer plane. The thin black lines represent the division between DMPC molecules and those points that are halfway between the two closest lipids. This representation provides a mapping of the top DMPC leaflet with which we can map which DMPC molecules are strongly interacting with the bound dendrimers. This has been represented by a coloration of the DMPC molecules on the Voronoi diagram. DMPC molecules that are within 3 Å of the dendrimer for 100% of the acquired simulation frames of the equilibrated structure are colored red; those that are within 3 Å of the dendrimer in 50–100% of frames are colored green, and 1–50% are colored blue.
The surprising result from these Voronoi diagrams is the similarity between the number and distribution of lipids in close proximity to the dendrimers, regardless of the dendrimer termination. All three dendrimers are within 3 Å of 12–13 particular lipid molecules in all of the simulation frames, 5–8 lipids are within 3 Å for 50–100% of the frames, and 3–4 lipids are within 3 Å for 1–50% of the frames. Variations between dendrimer binding morphologies (number of lipids, likelihood of switching between lipids, etc.) as a function of dendrimer termination are possible, although not observed over this time scale. Combined analysis of Figure 8 and Table 3 provides a quantitative understanding of bound configuration of a G3 PAMAM dendrimer on a DMPC bilayer.
TABLE 3.
Area Occupied by the Dendrimers on the Lipid Bilayer, Analyzed as the Average Area They Shadow or as the Average Area of the Smallest Circle Which Inscribes the Dendrimer Projected onto the Bilayer Planea
| area of dendrimer footprint (nm2) | area of inscribing circle (nm2) | |
|---|---|---|
| G3–NH3+ | 9.6 | 14.9 |
| G3–Ac | 8.2 | 9.4 |
| G3–COO– | 7.9 | 10.4 |
The standard deviation of observed areas was 2% for the footprints and 6% for the inscribing circles.
Conclusions
Simulations using an implicit solvent model have been performed to quantitatively analyze the interaction between G3 PAMAM dendrimers, of varying terminal chemistry, to DMPC bilayers. These simulations have the advantage of making quantitative PMF calculations computationally feasible but are unable to include solvent or counterion entropic effects. The qualitative conclusions from these simulations are identical to those observed experimentally: charged dendrimers more favorably interact with zwitterionic membranes than do neutral dendrimers. Additionally, these simulations provide a quantitative description of this interaction. An average force of 35 pN attracts the charged dendrimers to the lipid before the dendrimer and lipid make contact with each other. In contrast, neutral dendrimers have negligible interactions. After the dendrimer has made contact with the lipids, average attractive forces of 170, 200, and 240 pN are observed for G3–NH3+, G3–Ac, and G3–COO–, respectively. A net free energy release for dendrimer binding to the DMPC bilayers is measured to be 36, 26, 47 kcal/mol and 5.2, 3.2, 4.7 × 10–3 kcal/g for G3–NH3+, G3–Ac, and G3–COO–, respectively.
These results are directly applicable to nanodevice design, or other nanoparticles, for medical purposes. This research contributes to the growing understanding of the critical importance of nanoparticle size and surface chemistry to biological activity. Nanodevices utilized for gene delivery applications commonly exploit cationic nanoparticles for strong binding to nucleic acids.9,10,60–62 However, targeting these gene delivery nanodevices is challenging due to the influence of the remaining charged terminal groups. By further comparing the forces and energies of nonspecific nanoparticle binding to relevant system components, such as targeting moieties and cellular receptors, optimized nanoparticles can be designed and implemented.
Supplementary Material
Acknowledgment
C.V.K. received fellowship support from the NIH Michigan Molecular Biophysics Training Program (T32 GM008270-20), the Applied Physics Program, and the Graham Environmental Sustainability Institute. Computational time was provided by the Center for Advanced Computing at the University of Michigan and Lawrence Livermore National Laboratory. The authors thank Christine Orme and Timothy Sullivan. This research was supported by a Grant from the National Institute of Biomedical Imaging and BioEngineering (R01-EB005028). I.A. gratefully acknowledges support from the NSF CAREER award program (CHE-0548047).
Footnotes
Supporting Information Available: Animations of G3–NH3+, G3–Ac, and G3–COO– binding to gel phase DMPC bilayer. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Hong SP, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi XY, Balogh L, Orr BG, Baker JR, Holl MMB. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: Hole formation and the relation to transport. Bioconjugate Chem. 2004;15(4):774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 2.Hong SP, Leroueil PR, Janus EK, Peters JL, Kober MM, Islam MT, Orr BG, Baker JR, Holl MMB. Interaction of polycationic polymers with supported lipid bilayers and cells: Nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chem. 2006;17(3):728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 3.Leroueil PR, Hong SY, Mecke A, Baker JR, Orr BG, Holl MMB. Nanoparticle interaction with biological membranes: Does nanotechnology present a janus face. Acc. Chem. Res. 2007;40(5):335–342. doi: 10.1021/ar600012y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiao R, Roberts AP, Mount AS, Klaine SJ, Ke PC. Translocation of C-60 and its derivatives across a lipid bilayer. Nano Lett. 2007;7(3):614–619. doi: 10.1021/nl062515f. [DOI] [PubMed] [Google Scholar]
- 5.Xing XL, He XX, Peng JF, Wang KM, Tan WH. Uptake of silica-coated nanoparticles by HeLa cells. J. Nanosci. Nanotechnol. 2005;5(10):1688–1693. doi: 10.1166/jnn.2005.199. [DOI] [PubMed] [Google Scholar]
- 6.Manunta M, Nichols BJ, Tan PH, Sagoo P, Harper J, George AJT. Gene delivery by dendrimers operates via different pathways in different cells, but is enhanced by the presence of caveolin. J. Immunol. Methods. 2006;314(1–2):134–146. doi: 10.1016/j.jim.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Chithrani BD, Ghazani AA, Chan WCW. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006;6(4):662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 8.Manunta M, Tan PH, Sagoo P, Kashefi K, George AJT. Gene delivery by dendrimers operates via a cholesterol dependent pathway. Nucleic Acids Res. 2004;32(9):2730–2739. doi: 10.1093/nar/gkh595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat. Rev. Drug Discovery. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 10.Quintana A, Raczka E, Piehler L, Lee I, Myc A, Majoros I, Patri AK, Thomas T, Mule J, Baker JR. Design and function of a dendrimer-based therapeutic nanodevice targeted to tumor cells through the folate receptor. Pharm. Res. 2002;19(9):1310–1316. doi: 10.1023/a:1020398624602. [DOI] [PubMed] [Google Scholar]
- 11.Duncan R, Izzo L. Dendrimer biocompatibility and toxicity. Adv. Drug Delivery Rev. 2005;57(15):2215–2237. doi: 10.1016/j.addr.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Tomalia DA. Birth of a new macromolecular architecture: dendrimers as quantized building blocks for nanoscale synthetic polymer chemistry. Prog. Polym. Sci. 2005;30(3–4):294–324. [Google Scholar]
- 13.Landmark KJ, DiMaggio S, Ward J, Kelly CV, Vogt S, Hong S, Kotlyar A, Penner-Hahn JE, James R, Baker J, Holl MMB, Orr BG. Synthesis, characterization, and in vitro testing of superparamagnetic iron oxide nanoparticles targeted using folic acid-conjugated dendrimers. ACS Nano. 2008;2(4):773–783. doi: 10.1021/nn800034w. [DOI] [PubMed] [Google Scholar]
- 14.Majoros IJ, Thomas TP, Mehta CB, Baker JR. Poly(amidoamine) dendrimer-based multifunctional engineered nanodevice for cancer therapy. J. Med. Chem. 2005;48(19):5892–5899. doi: 10.1021/jm0401863. [DOI] [PubMed] [Google Scholar]
- 15.Patri AK, Majoros IJ, Baker JR. Dendritic polymer macromolecular carriers for drug delivery. Curr. Opin. Chem. Biol. 2002;6(4):466–471. doi: 10.1016/s1367-5931(02)00347-2. [DOI] [PubMed] [Google Scholar]
- 16.Choi Y, Baker JR. Targeting cancer cells with DNA-assembled dendrimerss—A mix and match strategy for cancer. Cell Cycle. 2005;4(5):669–671. doi: 10.4161/cc.4.5.1684. [DOI] [PubMed] [Google Scholar]
- 17.Mecke A, Lee I, Baker JR, Holl MMB, Orr BG. Deformability of poly(amidoamine) dendrimers. Eur. Phys. J. E. 2004;14(1):7–16. doi: 10.1140/epje/i2003-10087-5. [DOI] [PubMed] [Google Scholar]
- 18.Mecke A, Majoros IJ, Patri AK, Baker JR, Holl MMB, Orr BG. Lipid bilayer disruption by polycationic polymers: The roles of size and chemical functional group. Langmuir. 2005;21(23):10348–10354. doi: 10.1021/la050629l. [DOI] [PubMed] [Google Scholar]
- 19.Mecke A, Uppuluri S, Sassanella TM, Lee DK, Ramamoorthy A, Baker JR, Orr BG, Holl MMB. Direct observation of lipid bilayer disruption by poly(amidoamine) dendrimers. Chem. Phys. Lipids. 2004;132(1):3–14. doi: 10.1016/j.chemphyslip.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Ginzburg VV, Balijepalli S. Modeling the thermodynamics of the interaction of nanoparticles with cell membranes. Nano Lett. 2007;7(12):3716–3722. doi: 10.1021/nl072053l. [DOI] [PubMed] [Google Scholar]
- 21.Lee H, Larson RG. Molecular dynamics simulations of PAMAM dendrimer-induced pore formation in DPPC bilayers with a coarse-grained model. J. Phys. Chem. B. 2006;110:18204–18211. doi: 10.1021/jp0630830. [DOI] [PubMed] [Google Scholar]
- 22.Torrie GM, Valleau JP. Nonphysical sampling distributions in Monte-Carlo free-energy estimationsumbrella sampling. J. Comput. Phys. 1977;23(2):187–199. [Google Scholar]
- 23.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for free-energy calculations on biomolecules 0.1. the method. J. Comput. Chem. 1992;13(8):1011–1021. [Google Scholar]
- 24.Kirkwood J. Statistical mechanics of fluid mixtures. J. Chem. Phys. 1935;3:300–313. [Google Scholar]
- 25.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmms—a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4(2):187–217. [Google Scholar]
- 26.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Roux B. The calculation of the potential of mean force using computer-simulations. Comput. Phys. Commun. 1995;91(1–3):275–282. [Google Scholar]
- 28.Mitchell JBO, Laskowski RA, Alex A, Thornton JM. BLEEP—Potential of mean force describing protein–ligand interactions: I. Generating potential. J. Comput. Chem. 1999;20:1165–1176. [Google Scholar]
- 29.Roux B, Karplus M. Ion-transport in a model gramicidin channels—structure and thermodynamics. Biophys. J. 1991;59:961–981. doi: 10.1016/S0006-3495(91)82311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprik M, Ciccotti G. Free energy from constrained molecular dynamics. J. Chem. Phys. 1998;109:7737–7744. [Google Scholar]
- 31.Johnson SJ, Bayerl TM, McDermott DC, Adam GW, Rennie AR, Thomas RK, Sackmann E. Structure of an adsorbed dimyristoylphosphatidylcholine bilayer measured with specular reflection of neutrons. Biophys. J. 1991;59(2):289–294. doi: 10.1016/S0006-3495(91)82222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kucerka N, Liu YF, Chu NJ, Petrache HI, Tristram-Nagle ST, Nagle JF. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 2005;88(4):2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang ZY, Smith BD. High-generation polycationic dendrimers are unusually effective at disrupting anionic vesicles: Membrane bending model. Bioconjugate Chem. 2000;11(6):805–814. doi: 10.1021/bc000018z. [DOI] [PubMed] [Google Scholar]
- 34.Karoonuthaisiri N, Titiyevskiy K, Thomas JL. Destabilization of fatty acid-containing liposomes by polyamidoamine dendrimers. Colloids Surf., B. 2003;27(4):365–375. [Google Scholar]
- 35.Fischer D, Li YX, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 36.Kelly CV, Leroueil PR, Orr BG, Holl MMB, Andricioaei I. Poly(amidoamine) dendrimers on lipid bilayers II: effects of bilayer phase and dendrimer termination. J. Phys. Chem. B. 2008;112:xxxx. doi: 10.1021/jp8013783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feller SE, MacKerell AD. An improved empirical potential energy function for molecular simulations of phospholipids. J. Phys. Chem. B. 2000;104(31):7510–7515. [Google Scholar]
- 38.Mercier GA. Dendrimer Builder. 1996 http://server.ccl.net/chemistry/ resources/messages/1996/05/20.009-dir/index.html.
- 39.Maiti PK, Cagin T, Lin ST, Goddard WA. Effect of solvent and pH on the structure of PAMAM dendrimers. Macromolecules. 2005;38(3):979–991. [Google Scholar]
- 40.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics. 1996;14(1):33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 41.Pickersgill RW. A rapid method of calculating charge charge interaction energies in proteins. Protein Eng. 1988;2:247–248. doi: 10.1093/protein/2.3.247. [DOI] [PubMed] [Google Scholar]
- 42.Lee I, Athey BD, Wetzel AW, Meixner W, Baker JR. Structural molecular dynamics studies on polyamidoamine dendrimers for a therapeutic application: Effects of pH and generation. Macromolecules. 2002;35(11):4510–4520. [Google Scholar]
- 43.Paulo PMR, Lopes JNC, Costa SMB. Molecular dynamics simulations of charged dendrimers: Low-to-intermediate half-generation PAMAMs. J. Phys. Chem. B. 2007;111(36):10651–10664. doi: 10.1021/jp072211x. [DOI] [PubMed] [Google Scholar]
- 44.Brooks CL, Karplus M, Pettitt BM. Proteins: A Theoretical PerspectiVe of Dynamics, Structure, and Thermodynamics. John Wiley & Sons; 1990. [Google Scholar]
- 45.Chang R, Violi A. Insights into the effect of combustion-generated carbon nanoparticles on biological membranes: A computer simulation study. J. Phys. Chem. B. 2006;110(10):5073–5083. doi: 10.1021/jp0565148. [DOI] [PubMed] [Google Scholar]
- 46.Feig M, Brooks CL. Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr. Opin. Struct. Biol. 2004;14(2):217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 47.Bashford D, Case DA. Generalized born models of macromolecular solvation effects. Annu. Rev. Phys. Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 48.Qiu D, Shenkin PS, Hollinger FP, Still WC. The GB/SA continuum model for solvation. A fast analytical method for the calculation of approximate Born radii. J. Phys. Chem. A. 1997;101(16):3005–3014. [Google Scholar]
- 49.Dominy BN, Brooks CL. Development of a generalized born model parametrization for proteins and nucleic acids. J. Phys. Chem. B. 1999;103(18):3765–3773. [Google Scholar]
- 50.Schaefer M, Karplus M. A comprehensive analytical treatment of continuum electrostatics. J. Phys. Chem. 1996;100:1578–1599. [Google Scholar]
- 51.Schaefer M, Bartels CM. Solution conformations and thermodynamics of structured peptides: Molecular dynamics simulation with an implicit solvation model. J. Mol. Biol. 1998;284:835–848. doi: 10.1006/jmbi.1998.2172. [DOI] [PubMed] [Google Scholar]
- 52.Calimet N, Schaefer M, Simonson T. Protein molecular dynamics with the generalized Born/ACE solvent model. Proteins. 2001;45:144–158. doi: 10.1002/prot.1134. [DOI] [PubMed] [Google Scholar]
- 53.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins: Struct. Funct. Genet. 1999;35(2):133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 54.Rhee YM, Sorin EJ, Jayachandran G, Lindahl E, Pande VS. Simulations of the role of water in the protein-folding mechanism. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6456–6461. doi: 10.1073/pnas.0307898101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.ten Wolde PR, Chandler D. Drying-induced hydrophobic polymer collapse. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6539–6543. doi: 10.1073/pnas.052153299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou RH, Huang XH, Margulis CJ, Berne BJ. Hydrophobic collapse in multidomain protein folding. Science. 2004;305:1605–1609. doi: 10.1126/science.1101176. [DOI] [PubMed] [Google Scholar]
- 57.Rudnick J, Gaspari G. The asphericity of random walks. J. Phys. A: Math. Gen. 1986;19:L191–L193. [Google Scholar]
- 58.Voronoi GF. Nouvelles applications des paramètres continus à la théorie des formes quadratiques. J. Reine Angew. Math. 1907;133:97178. [Google Scholar]
- 59.Moy VT, Florin EL, Gaub HE. Intermolecular forces and energies between ligands and receptors. Science. 1994;266(5183):257–259. doi: 10.1126/science.7939660. [DOI] [PubMed] [Google Scholar]
- 60.Han S, Mahato RI, Sung YK, Kim SW. Development of biomaterials for gene therapy. Mol. Ther. 2000;2(4):302–317. doi: 10.1006/mthe.2000.0142. [DOI] [PubMed] [Google Scholar]
- 61.Tang MX, Redemann CT, Szoka FC. In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjugate Chem. 1996;7(6):703–714. doi: 10.1021/bc9600630. [DOI] [PubMed] [Google Scholar]
- 62.Haensler J, Szoka FC. Polyamidoamin cascade polymers mediate efficient transfection of cells in culture. Bioconjugate Chem. 1993;4(5):372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.