Abstract
The sarcomeric myosin heavy chain (MyHC) proteins are a family of molecular motors responsible for the transduction of chemical energy into mechanical work in striated muscle. The vertebrate genome contains multiple copies of the MyHC gene, and expression of different isoforms correlates with differences in the physiological properties of muscle fibers. Most MyHC isoforms are found in two arrays, one containing the “fast-twitch” skeletal muscle isoforms and the other the “slow-twitch” or cardiac isoforms. To extend our understanding of MyHC evolution, we have examined the genome of the anuran Xenopus tropicalis. The X. tropicalis genome includes15 full-length MyHC genes organized in seven genomic locations. One unique array of MyHC genes is similar to the mammalian fast-skeletal array, but is not found in amniotes. The isoforms in this array are expressed during larval stages and in muscles of the adult larynx. Duplication of the fast-skeletal MyHC array appears to have led to expression divergence of muscle proteins in the larval and adult stages of the anuran life cycle. A striking similarity of gene order between regions flanking X. tropicalis MyHC arrays and human arrays was evident; genomic organization of MyHC isoforms may thus be highly conserved across tetrapods.
Keywords: Gene duplication, Comparative genomics, Evolution, Xenopus, Myosin heavy chain
Introduction
Vertebrate striated and cardiac muscle is capable of producing contractions that vary in a number of parameters including maximum force generation (Lutz and Lieber 2000), contraction velocity (Pette and Staron 2000), and temperature optima (Watabe 2002). These characteristics of muscle can change in response to changes in load, temperature (Watabe 2002), or as part of a developmental program (Sassoon et al. 1987). Studies examining the source of this variation have concentrated on the multiple isoforms of sarcoplasmic type II myosin heavy chain (MyHC), the molecular motor responsible for the transduction of chemical energy into mechanical work in muscle. Vertebrate genomes contain several copies of the MyHC gene, and many studies have correlated changes in expression of MyHC isoforms with changes in contractile properties (Pette and Staron 2000).
Preparations of single muscle fibers are particularly robust in anurans, and this characteristic has contributed to our understanding of MyHC isoform functions in a variety of frog species, particularly Rana pipiens and Xenopus laevis (Lutz and Lieber 2000; Andruchova et al. 2006). However, while many mammalian and avian MyHC sequences are available, to date, only fragments of a few anuran MyHC genes have been cloned and sequenced (Radice and Malacinski 1989; Lutz et al. 1998), leaving a significant gap between function (well studied in anurans) and sequence. To understand how MyHC sequence contributes to muscle properties, it would be useful to have a wider range of MyHC genes to examine in organisms, such as frogs, that are subject to physiological constraints (aquatic habitat, poikilothermy) beyond those that operate in mammals.
Studies in anurans may also shed light on the evolution of MyHC isoforms. In mammals, there are examples of duplication resulting in both expansion of a locus within an array and also in the formation of new arrays. In humans, for example, the fast skeletal isoforms are found in a six-member tandem array on chromosome 17 (Weiss et al. 1999) and encode genes whose expression is regulated both developmentally (MyHC-EO, perinatal MyHC, and embryonic MyHC) and by physiological demands (MyHC IIb, IId/x, IIa). The cardiac isoforms are found on chromosome 14 and are also expressed in slow-twitch skeletal muscle. In addition, three unarrayed MyHC genes of unknown function have been found (Desjardins et al. 2002). This distribution of MyHC genes led Desjardins et al. (2002) as well as others (Schachat and Briggs 2002) to conclude that the ancestral vertebrate had four MyHC genes, three of which led directly to the three unclustered genes, the fourth giving rise, by duplication and translocation, to both the cardiac and fast skeletal isoforms. Examination of MyHC genes in the mouse (Weiss et al. 1999) and the chicken (Moore et al. 1993) shows a distribution of isoforms similar to that of humans, with the majority of fast skeletal isoforms found in one tandem array and cardiac isoforms found in another. In fish, MyHC genes are also found in arrays (McGuigan et al. 2004); gene order between fishes and amniotes is not, however, sufficiently conserved to be useful in establishing the homology of genomic locations. Thus, knowledge of anuran MyHC sequence and genomic organization could be important in understanding the degree of conservation of the amniote MyHC genomic organization as well as the mechanisms by which this important gene family expands and diversifies.
Materials and methods
Sequence identification
The X. tropicalis genome was searched for putative MyHC sequences using the Basic Local Alignment Search Tool (BLAST) implemented at the Joint Genome Initiative (JGI), X. tropicalis website (http://www.jgi.doe.gov/xenopus/). The human MyHC-beta and MyHC-EO (translations of DNA with Genbank accession numbers X52889 and AF111782, respectively) were used for the initial alignment, producing an identical set of hits containing >70% sequence identity covering >90% of the length of hMyHC-beta and hMyHC-EO. Each scaffold thus identified was downloaded, and fragments containing the BLAST hits were submitted to Twinscan using the human models for comparison to identify putative transcripts and exon/intron boundaries. At this point, a nomenclature was established to identify each putative transcript. Each transcript is identified by its scaffold (as present in the X. tropicalis genome v. 2, released Oct. 2004) and the order in which it appears using the scaffold orientation deposited in the JGI database.
Wise2 (Birney and Durbin 2000) was used to refine the exon/intron boundaries using the hMyHC-beta and hMyHC-EO sequences as templates, yielding identical results from the two sequences. Sequences were further refined, and in some cases, 5′ and 3′ untranslated regions (UTRs) were identified using the Gurdon full-length complementary DNA (cDNA) project (Gilchrist et al. 2004) and expressed sequence tag sequences deposited in Genbank. Sequence was identified from these databases having >98% identity to a single MyHC isoform identified from the X tropicalis genome.
Dotplots of X. tropicalis scaffolds were generated using Dotmatcher, comparing scaffolds identified by BLAST as containing MyHC sequences (y-axis) and human MyHC-EO sequence (x-axis). For each region, trials were run using a 50-bp window and a threshold of 55% or 90%. Except for higher “background,” when a threshold of 55% was used, no substantial differences were observed for any of the scaffolds. Results using a threshold of 90% are shown.
Synteny
For each scaffold, the human proteins track table was downloaded from the UCSC genome browser (www.genome.ucsc.edu). Only those sequences with >50% identity more than 80% of the query sequence were used. A table showing all of the hits for each sequence against the tropicalis genome was then downloaded, and any sequence that had a hit elsewhere in the genome that was greater in either extent or percent identity was removed from the list. Human genomic coordinates for each of the remaining genes thus identified were obtained, and the full table of human genes was obtained for each identified human genomic region. The tables of human and tropicalis genes were then mapped onto diagrams and lines drawn between orthologous human and tropicalis genes.
Animals
Animals were housed in polycarbonate tanks containing 8 l of filtered tap water at 22°C. Embryos were generated by injecting a male and female adult with 200 IU of human chorionic gonadotropin (Sigma) and allowing the animals to mate overnight. Eggs were collected, and feeding was begun when the tadpoles began swimming. Tadpoles were fed Larval Z every other day, and frogs were fed ‘Frog Chow’ (Nasco) two times per week. Stages are as described in Nieuwkoop and Faber (1956).
Primers
Primers (S2) were generated to the 3′ UTR of each gene. For xtMyHC-270a, sequence within the coding region was used, as no 3′ UTR could be identified. These primers shared only limited identity to the most similar MyHC gene and did not produce any apparent cross-hybridization. xtMyHC-270d and xtMyHC-270e were nearly identical at the 3′ end, and so primers were designed to detect both isoforms (270d/e). An additional primer set to EF1α was used as a positive control.
Tissue and collection
Adults
Adults were anesthetized by immersion in 0.1% MS-222 (Sigma). Tissue was dissected from adult X. tropicalis male and female animals and frozen in OCT medium. For each tissue, about twenty 30-μm sections were collected and used for RNA extraction.
Tadpoles and embryos
Tadpoles were anesthetized by immersion in 0.1% MS-222. Whole tadpoles or grossly dissected tadpoles were frozen on dry ice and processed whole for RNA extraction.
RNA extraction, cDNA synthesis, and PCR
RNA was extracted using Tri-reagent (Sigma) following the manufacturer’s instructions. Remaining DNA was digested using DNAseI (Roche). cDNA synthesis was carried out using the MMTV reverse transcriptase (Invitrogen) and poly dT primers (Invitrogen). For each RNA preparation, an additional tube was processed without MMTV and used as a control for DNA contamination (NoRT).
For each cDNA and its NoRT control, the reaction was added to jumpstart readymix DNA polymerase (Sigma), which was then mixed with each primer set. Polymerase chain reaction (PCR) was performed for 27 cycles using a 95°C dissociation step for 45 s, a 58°C annealing step for 45 s, and a 72°C extension step for 1 min. Samples were then run on a 2% agarose gel. A sample was considered positive if there was a clear band of the correct size from the cDNA reaction but no detectable band from the NoRT control. Band intensity relative to EF-1 was subjectively rated as +, ++, or +++. For each primer set, the identity of the band was verified by blunt cloning the PCR product into the pCR2.1 vector (Invitrogen) and sequencing the insert.
Phylogenetic analysis
Sequences
Human MyHC sequences were identified in Genbank (S2). Clustal was then used to align the amino acid sequences, and the sequences were translated back into nucleic acids. Several regions were removed for phylogenetic analysis due to their lack of good alignment: the 5′ ends [ending at amino acid (aa) 117], the two loop regions aa 205–214 and aa 618–643, the region of non-MyHC sequence from xtMyHC-270d (aa 327–351), and the 3′ end (beginning at aa 1920). All sequence numbers are given as amino acid position relative to the hMyHC-emb start codon. This sequence was then used for the analysis of the whole molecule.
Bayesian analysis
Bayesian analysis was performed using MrBayes v3.1. A 4 × 4 nucleotide model with an F81 substitution model was used with no rate variation across sites. Nucleotide state frequencies were given equal priors and followed a Dirichlet distribution. Topology was equal a priori and branch lengths were unconstrained, following an exponential distribution. Two independent MCMC chains were run for 1,000,000 generations with sampling every 50 generations, resulting in a standard deviation of <0.01 between the runs. Trees were calculated after excluding the first 1,000 generations as burn in because the likelihoods of the subsequent generations were similar. The probability scale reduction factor was near 1 for all parameters. The final tree was rooted with cf MyHC16, as this has been shown previously to be the earliest branch of the sarcoplasmic MyHC sequences (Desjardins et al. 2002).
Results and specific discussion
Identification of 15 sarcoplasmic MyHC genes and one MyHC pseudo-gene
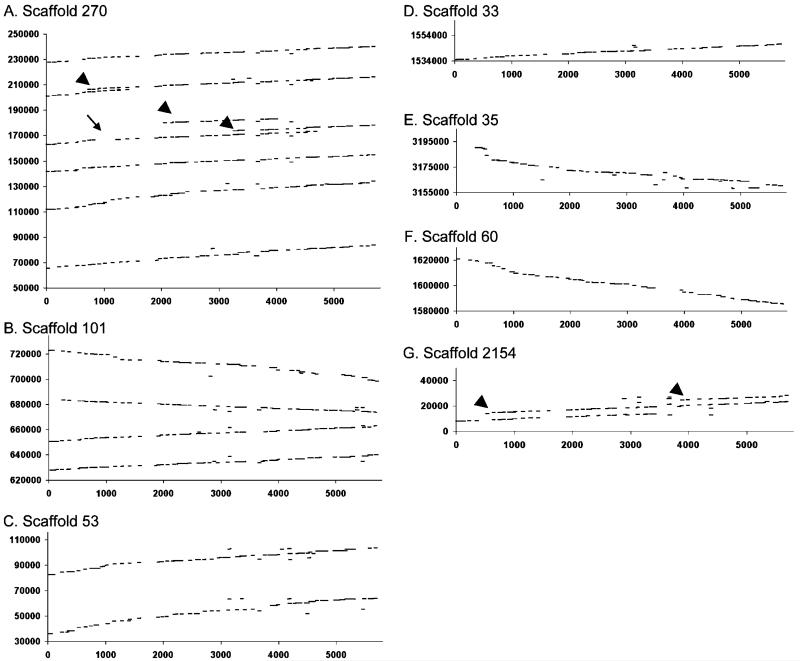
Using BLAST and the published sequence of the X. tropicalis genome (http://www.jgi.doe.gov/xenopus/), we identified seven scaffolds with similarity to human MyHC-EO and MyHC-alpha across the entire length of the human MyHC coding sequences. To compare the hMyHC-EO nucleic acid sequence with the loci identified by BLAST as containing one or more X. tropicalis MyHC genes, dotplots were constructed using Dotmatcher executed from EMBOSS (http://emboss.bioinformatics.nl/; Fig. 1). As is the case for other identified MyHC arrays (Weiss et al. 1999), those on scaffold 270 and scaffold 53 (Fig. 1a and c) are arranged in head to tail orientation, while the array of scaffold 101(Fig. 1b) has two isoforms on the “+” strand adjacent to two additional isoforms on the “−” strand. Scaffold 270 (Fig. 1a, arrowheads between 160,000 and 180,000 bp and again between 200,000 and 210,000 bp) and 2154 (Fig. 1g, arrowheads) also contain partial duplications of the MyHC sequence. In addition, one pair of tandemly arrayed MyHC isoforms is found on scaffold 53 (Fig. 2c), and single isoforms are found on scaffolds 33, 35, and 60 (Fig. 2d–f).
Fig. 1.
Dotplots comparing X. tropicalis scaffolds ( y-axis) and human MyHC-EO (x-axis) by plotting 50-bp windows having >90% identity between the two genes. The presence of a complete MyHC gene can be inferred by a region of largely uninterrupted homology (few gaps) across the x-axis. Regions of scaffolds 270 (a), 101 (b), and 53 (c) contain multiple such regions of homology to hMyHC-EO. Scaffolds 33 (d), 35 (e), and 60 (f) each contain a single region of homology, while scaffold 2154 (g) appears to contain two overlapping sequences having homology to hMyHC-EO. Some regions contain incomplete repetitions of MyHC sequence (arrowheads), and one repeat has a region lacking homology to hMyHC-EO (arrow), which is present in other alignments
Fig. 2.

Except for scaffold 2154, all of the scaffolds identified as containing MyHC isoforms were found to contain multiple homologous genes in an order similar to one or more human chromosomal loci. For each of these six scaffolds, the area around the MyHC gene or MyHC array is represented as the lower horizontal line of the figure, with a black box representing the position and size of homologous human genes. Above this line is a representation of the human chromosomal locus on which these homologous genes are found. Human genes are represented by a white box, and a vertical line connects homologous genes between the human and Xenopus loci. Some human genes not having homologs on the Xenopus scaffold are labeled on the human chromosome, but many of these names were removed for clarity. MyHC genes or MyHC arrays are depicted as a gray boxes and are labeled on both the Xenopus and human locus maps. The specific region represented by each map is labeled at the beginning and end of each map. Note that scales may be different between human and Xenopus maps. The loci presented are as follows: a human Chromosome 17 and Xenopus scaffold 270, b human Chromosome 14 and Xenopus scaffold 53, c human Chromosome 20 and Xenopus scaffold 35, d human Chromosome 3 and Xenopus scaffold 60, e human Chromsome 14 and Xenopus scaffold 33, and f human Chromosome X and Xenopus scaffold 101
Putative messenger RNA (mRNA) sequences were compiled using a combination of exon/intron boundary identification tools as well as publicly available cDNA databases (S1). In total, 15 copies of the MYH gene in X. tropicalis produced good alignments without significant gaps with hMyHC-EO. In addition to these full-length MyHC genes, one isoform (xtMyHC-270d) encodes a nearly complete MyHC sequence, but is missing the region of amino acids 300–409. This deletion is apparent in the dot plot comparison (Fig. 1a, arrow) as a region that aligns well in other isoforms, but is missing in xtMyHC-270d. Alternative sequence was identified as part of the gene using Twinscan (Korf et al. 2001), but searches of public databases revealed no cDNA to support this region or the surrounding sequence. One cDNA corresponding to a more 3′ region of this gene contains a stop codon and does not align with other MyHC genes, suggesting that xtMyHC-270d is a MyHC pseudo-gene. Although the isoform is unlikely to be functional, we have included it in our analysis because it is nearly complete.
The MyHC isoform of scaffold 2154 contains at least two overlapping partial repeats (Fig. 1g). This organization may allow for alternative spice variants, and indeed, cDNAs were found which correspond to both of the possible 3′ ends of the MyHC gene on scaffold 2154 and share internal sequence.
The X. tropicalis genome contains both conserved and novel MyHC loci
The sarcoplasmic MyHC isoforms of chicken, humans, and mouse have a well-conserved genomic organization (Moore et al. 1993; Weiss et al. 1999). Both mammalian genomes contain 11 MyHC isoforms in five genomic locations, two of which contain arrays of MyHC isoforms. We used conservation of gene order between X. tropicalis scaffolds and the human genome to determine whether this feature was maintained in more phylogenetically distant species.
With one exception (2154), all of the X. tropicalis scaffolds that included MyHC isoforms also contained several other genes with high homology to human genes. For example, scaffold 270 contains a tandem array of six MyHC isoforms, and the order of genes surrounding the MyHC array is similar to that of the region of human chromosome 17 that contains the array of fast skeletal isoforms. Both human and tropicalis arrays have the same gene order (GAS-7, RCV-1, GPL2R) flanking the MyHC array (Fig. 2a). Scaffold 53 includes a tandem array of two MyHC isoforms and is homologous with the region of human chromosome 14 that contains the two cardiac MyHC isoforms (Fig. 2b). Scaffolds 60 and 35 each contain only one copy of a MyHC isoform; these are homologous to the regions of chromosomes 20 and 3, respectively, containing hMyHC7b and hMyHC15 (Fig. 2c and d).
Gene order on scaffold 101 shows a more complex relationship with the human genome. While the genes immediately surrounding the MyHC cluster (TKTL, ARD1, and ARHGAP4) are found—in that order—in close proximity on the human X chromosome (Fig. 2f), other flanking genes correspond to regions of chromosome 3 (nudix and AK023995, Fig. 2f). No MyHC pseudo-gene or remnant of sequence was found in either the corresponding region of the X chromosome or chromosome 3. The X. tropicalis genome also contains MyHC genes at loci that do not appear to contain MyHC genes in humans (Fig. 2). Scaffold 33 has apparent homology with a region of human chromosome 14 which is ~33 Mbp away from the cardiac MyHC isoforms (Fig. 2e); this region does not contain a known MyHC gene in humans. There were no signs of a MyHC pseudo-gene, and pairwise tblastn alignment between the homologous region and human MyHC-IIa protein resulted in no apparent similarity, as did a similar alignment between human MyHC-IIa and the homologous region of chicken chromosome 5 (not shown).
The MyHC containing loci of scaffold 33 and scaffold 101 may be novel to the amphibian lineage.
Novel MyHC loci are restricted to embryonic and larval expression
In X. tropicalis, larval and embryonic muscles express distinct sets of mRNA corresponding to different MyHC isoforms (Table 1). In adults, xtMyHC-35, xtMyHC-270b, xtMyHC-270c, xtMyHC-270d/e, and xtMyHC-270f are expressed in a variety of limb and body wall muscles. At the tailbud stage (the earliest stage examined), embryos express xtMyHC-101c, xtMyHC-2154a, xtMyHC-33, xtMyHC-35, xtMyHC-53b, and xtMyHC-60 (Table 1). By stage 42, tadpoles express xtMyHC-101a, xtMyHC-101b, xtMyHC-2154, and xtMyHC-53a. Older tadpoles (stage 49) also express xtMyHC-270b and 270c. At stage 54, as metamorphosis begins and limb buds become well formed, xtMyHC-270d/e, xtMyHC-270c, xtMyHC-270b, and xtMyHC-270a are expressed together with those isoforms seen in earlier stages.
Table 1.
Expression of MyHC isoforms across stages and tissue in X. tropicalis
| Tissue | 101a | 101b | 101c | 101d | 270a | 270b | 270c | 270d/e | 270f | 2154 | 2154a | 33 | 35 | 53a | 53b | 60 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gastrocnemius | − | − | − | − | − | +++ | − | +++ | − | − | − | − | + | − | − | − |
| Gracialis major | − | − | − | − | − | ++ | − | +++ | − | − | − | − | − | − | − | − |
| Heart | − | − | − | − | − | − | − | − | − | − | − | − | ++ | − | ++ | ++ |
| Upper arm | − | − | − | − | − | ++ | +++ | +++ | +++ | − | − | − | ++ | − | − | − |
| Laryngeal muscle (female) |
− | − | + | + | N/Aa | +++ | +++ | ++ | ++ | − | − | − | − | − | − | − |
| Laryngeal muscle (male) |
− | − | − | ++ | N/Aa | − | N/Aa | − | − | − | − | − | − | − | − | − |
| St. 39 embryo | − | − | +++ | − | − | − | − | − | − | − | ++ | + | + | − | + | − |
| St. 42 tadpole | ++ | ++ | +++ | − | − | +++ | + | − | − | + | + | + | N/Aa | ++ | ++ | +/− |
| St. 49 Heart | + | + | +++ | − | − | +/− | − | − | − | − | − | + | − | +++ | ++ | |
| St. 49 body wall | ++ | + | +++ | − | − | +/− | + | − | − | − | − | + | + | ++ | ++ | − |
| St. 49 tail | ++ | ++ | +++ | − | N/Aa | ++ | N/Aa | − | − | ++ | + | − | ++ | + | + | − |
| St. 49 jaw | +++ | ++ | +++ | − | N/Aa | +++ | N/Aa | − | − | − | − | − | − | ++ | ++ | + |
| St. 49 head | ++ | ++ | +++ | − | N/Aa | ++ | N/Aa | − | − | + | + | − | + | + | + | − |
| St. 54 body wall | ++ | ++ | ++ | − | ++ | ++ | +++ | − | − | − | − | − | − | − | − | − |
| St 54. limb bud | ++ | + | ++ | − | − | +++ | + | + | − | − | − | − | − | − | − | − |
| St. 54 tail | +++ | +++ | +++ | − | ++ | ++ | ++ | ++ | − | ++ | + | + | + | N/Aa | N/Aa | − |
| St. 54 jaw | ++ | ++ | + | − | N/Aa | +++ | N/Aa | ++ | − | + | − | − | − | ++ | ++ | − |
| St. 54. head | ++ | ++ | ++ | − | − | +++ | +++ | ++ | + | + | − | − | + | + | ++ | − |
Primer pairs for each of the identified MyHC genes were used to amplify target genes from cDNA pools made from muscles of different staged animals. The absence of a band (−) or band intensity relative to elongation factor-1 (+, ++, +++) for each primer set is given. Gene names are as listed in Table S1, except xtMyHC 270d/e, the primers for which amplify both xtMyHC-270d and xtMyHC-270e.
Data not available.
This pattern of expression confirms earlier studies that show a difference in MyHC isoform expression between larval and adult muscle (Radice and Malacinski 1989). Furthermore, the novel MyHC-containing loci in X. tropicalis identified via comparison with mammalian genomes—those found on scaffolds 101, 33, and 2154—are primarily restricted in expression to larval muscles. Expression of isoforms on scaffold 270, a homologous location to the fast-twitch MyHC isoforms of human chromosome 17 (Fig. 2a), occurs at both adult and larval stages. More isoforms of this array are expressed in adults and metamorphic tadpoles (Table 1).
A notable exception to the larval expression of isoforms from the array on scaffold 101 is xtMyHC-101d, whose expression is restricted to laryngeal muscle of adults. In the related species, X. laevis (Catz et al. 1992), a laryngeal-specific isoform, is thought to contribute to generating the rapid muscle contraction required for the fast vocal trills produced by adult males (Catz et al. 1992).
Cardiac muscle expresses xtMyHC-53b, which is found in a genomic region homologous to the human cardiac array (Table 1, Fig. 2b). In addition, we found expression of xtMyHC-60 and xtMyHC-35 in adult heart muscle. These three mRNA isoforms are also found in the tadpole heart. Tissue containing the tadpole heart expresses xtMyHC-101a, b, and c; the latter two isoforms are also expressed in the body wall, and their presence in the heart sample may reflect difficulty in isolating this very small organ at tadpole stages. In adults, xtMyHC-53b is restricted to the heart; in tadpoles, however, this isoform is more broadly expressed. In mammals, the type-I MyHC isoforms are found in both slow-twitch skeletal muscles as well as cardiac muscles. These genes occur within the two-isoform array on chromosome 14, a region homologous to the tropicalis scaffold 53. Thus, expression of these isoforms in both cardiac and skeletal muscle may be feature retained by both the amniote and amphibian lineages.
Phylogenetic analysis of tropicalis myosin heavy chain
Bayesian analysis of an alignment of the 16 X. tropicalis, ten human, and one Canis familiaris sarcoplasmic MyHC coding regions yielded a well-supported tree (Fig. 3), with the 95% credible set containing two trees; all branches were well supported, with the lowest posterior probability being 0.94. The C. familiaris MyHC16 sequence was used because the human sequence had been shown to be a pseudogene (Schachat and Briggs 1999), although the human MyHC16 sequence segregated exactly as the cf MyHC16 (not shown).
Fig. 3.

Phylogeny generated using full-length MyHC sequences from humans (using cf MyHC16 to replace hMyHC16) and X. tropicalis. The major clades include the MyHC7b/MyHC15 (node A) group, the cardiac/fast skeletal group (node B), and MyHC16. The cardiac/fast skeletal group splits into the cardiac (node C) and fast-skeletal groups (Node D). The tropicalis fast skeletal isoforms form a single clade (node E). This clade subdivides by both array and temporal expression into adult and larval subgroups. Posterior probabilities are given as internal nodes, and all non-Xenopus sequences are listed in Supplementary material
The subfamily groupings are consistent with those previously shown for human MyHC sequences (Desjardins et al. 2002) where the three initial clades include one containing cf MyHC16, a second containing the hMyHC15/hMyHC7b (Fig. 3, node A), and a third containing all of the skeletal and cardiac isoforms (Fig. 3, node B). No tropicalis MyHC sequence was found in the MyHC16 clade, while xtMyHC-60 and hMyHC-15 formed a clade within the MyHC15/7b grouping, as would be expected from the fact that the genes are syntenous. Although the two genes are found in homologous genomic locations, xtMyHC-35 does not form a clade with hMyHC7b. This result may reflect the apparently divergent function of hMyHC7b, as evidenced by its expression in only very specialized human muscles (Desjardins et al. 2002).
The remaining MyHC isoforms fall into the cardiac/fast-skeletal clade, which is then divided into the cardiac (Fig. 3, node C) and fast-skeletal groups (Fig. 3, node D). The cardiac group is made up of the two arrayed MyHC isoforms from human chromosome 14 and the pair of arrayed MyHC isoforms on tropicalis scaffold 53. As is often the case with arrayed genes, the two tropicalis isoforms form a group distinct from the arrayed genes of the human cardiac array. This pattern could be due either to duplications in the tropicalis and human lineages or to ongoing gene conversion, which has been found to occur at this locus in mammals (Epp et al. 1995).
The family of fast-skeletal MyHC isoforms (Fig. 3, node D) contains the isoforms found in the human chromosome 17 array as well as those found on the tropicalis scaffolds 2154, 33, and the arrays on scaffolds 270 and 101. Our analysis confirms previous results (Schachat and Briggs 2002) that hMyHC-EO is the oldest isoform in this group (a duplication that predated the split between the amniotes and anamniotes). However, despite its apparent ancient origin, we could not find a direct tropicalis homolog to hMyHC-EO, suggesting that the isoform was either lost or was subjected to gene conversion that obscured its true evolutionary history.
The remaining members of the fast-skeletal group cluster by species, with all tropicalis MyHC sequences falling into one clade (Fig. 3, node E) and with the next major division separating the isoforms of scaffold 270 from the remaining isoforms, all expressed only in larval muscle (Table 1). The two unarrayed fast skeletal MyHCs, xtMyHC-2154, and xtMyHC-33 are most closely related to the isoforms of scaffold 101. With the exception of the laryngeal xtMyHC-101d, these isoforms are all expressed in pre-metamorphic tadpoles. The tropicalis fast-skeletal MyHC isoforms thus consist of an adult cluster, the isoforms of scaffold 270, and a larval cluster, consisting of both the isoforms of scaffold 101 and the two unarrayed fast-skeletal isoforms. Both clusters are more related to each other than to the human fast skeletal cluster, suggesting that these two groups formed after the split between amniotes and amphibians.
Thus, at some point after divergence of the tetrapod lineages, additional duplications and translocations of MyHC isoforms established distinct ‘larval’ and ‘adult’ sets of MyHC isoforms at distinct genomic locations. While all other described species of Xenopus are polyploid, X. tropicalis is diploid, and thus, MyHC duplications are not a consequence of the genetic introgression otherwise prevalent in the genus (Evans et al. 2004, 2005). The expression of different MyHC isoforms underlies the contractile properties of muscle (Lutz et al. 2002) and may also be related to optimizing performance at specific temperatures in poikilotherms (Watabe 2002). Additional MyHC isoforms may have provided the substrate for variation permitting functional and developmental specialization of isoform expression.
In summary then, we have screened the assembly of the complete genome of the pipid frog, Xenopus tropicalis and found 15 full-length copies of the myosin heavy chain gene as well as one probable pseudo-gene and several partial repeats. Using phylogenetic analysis and examination of conserved gene order with the human genome, we find that in addition to the previously known vertebrate MyHC containing loci, X. tropicalis has three additional genomic locations containing MyHC genes. These three clusters represent copies of the fast-skeletal isoforms that are suspected to be copies of the six isoforms that are homologous to the human fast-skeletal isoforms of chromosome 17. By assaying for the expression of each of the X. tropicalis MyHC genes in a variety of muscles, we have shown that while the isoforms syntenic with the human fast-skeletal isoforms are expressed at both larval and adult stages of development, MyHC isoforms from the novel loci appear to only be expressed during larval stages, except for one, which is expressed in the adult male vocal organ. These additional MyHC isoforms may have facilitated adaptation to the amphibian life cycle with a distinct aquatic larval stage and a tetrapod adult stage.
Supplementary Material
Acknowledgments
The authors would like to thank Elizabeth Wilbanks, Tanya Gonzales, and Gianna Cricco Lizza for help with tissue collection and RT-PCR. We would also like to thank Joe Thornton, Ben Evans, and Eun-Jin Yang for critical reading of the manuscript and the Cold Spring Harbor Genome Access Course for technical assistance. This work was supported by a Ruth L. Kirschstein pre-doctoral NRSA fellowship DC007567-03 to BTN and NS23684 to DBK.
Footnotes
Communicated by T. Hollemann
Electronic supplementary material The online version of this article (doi: 10.1007/s00427-008-0225-0) contains supplementary material, which is available to authorized users.
Contributor Information
Brian T. Nasipak, Department of Biological Sciences, Columbia University, New York, NY 10027, USA; Department of Cell Biology, University of Massachusetts Medical School, 55 Lake Ave. North, Worcester, MA 01655, USA
Darcy B. Kelley, Department of Biological Sciences, Columbia University, New York, NY 10027, USA
References
- Andruchova O, Stephenson GM, Andruchov O, Stephenson DG, Galler S. Myosin heavy chain isoform composition and stretch activation kinetics in single fibres of Xenopus laevis iliofibularis muscle. J Physiol. 2006;574:307–317. doi: 10.1113/jphysiol.2006.109926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Durbin R. Using GeneWise in the Drosophila annotation experiment. Genome Res. 2000;10:547–500. doi: 10.1101/gr.10.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catz DS, Fischer LM, Moschella MC, Tobias ML, Kelley DB. Sexually dimorphic expression of a laryngeal-specific, androgen-regulated myosin heavy chain gene during Xenopus laevis development. Dev Biol. 1992;154:366–376. doi: 10.1016/0012-1606(92)90075-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins PR, Burkman JM, Shrager JB, Allmond LA, Stedman HH. Evolutionary implications of three novel members of the human sarcomeric myosin heavy chain gene family. Mol Biol Evol. 2002;19:375–393. doi: 10.1093/oxfordjournals.molbev.a004093. [DOI] [PubMed] [Google Scholar]
- Epp TA, Wang R, Sole MJ, Liew CC. Concerted evolution of mammalian cardiac myosin heavy chain genes. J Mol Evol. 1995;41:284–292. doi: 10.1007/BF00186540. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Melnick DJ, Cannatella DC. Evolution of RAG-1 in polyploid clawed frogs. Mol Biol Evol. 2005;22(5):1193–1207. doi: 10.1093/molbev/msi104. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implications for polyploid evolution. Mol Phylogenet Evol. 2004;33(1):197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Gilchrist M, Zorn AM, Voigt J, Smith JC, Papalopulu N, Amaya E. Defining a large set of full length clones from a Xenopus tropicalis EST project. Dev Biol. 2004;271:498–516. doi: 10.1016/j.ydbio.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Korf I, Flicek P, Duan D, Brent MR. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17(Suppl 1):S140–S148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- Lutz GJ, Lieber RL. Myosin isoforms in anuran skeletal muscle: their influence on contractile properties and in vivo muscle function. Microsc Res Tech. 2000;50:443–457. doi: 10.1002/1097-0029(20000915)50:6<443::AID-JEMT3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Lutz GJ, Cuizon DB, Ryan AF, Lieber RL. Four novel myosin heavy chain transcripts define a molecular basis for muscle fibre types in Rana pipiens. J Physiol. 1998;508(Pt 3):667–680. doi: 10.1111/j.1469-7793.1998.667bp.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz GJ, Sirsi RL, Shapard-Palmer RL, Bremner SN, Lieber RL. Influence of myosin isoforms on contractile properties of intact muscle fibers from Rana pipiens. Am J Physiol Cell Physiol. 2002;282:C835–C844. doi: 10.1152/ajpcell.00482.2001. [DOI] [PubMed] [Google Scholar]
- McGuigan K, Phillips P, Postlethwait J. Evolution of sarcomeric myosin heavy chain genes: evidence from fish. Mol Biol Evol. 2004;21:1042–1056. doi: 10.1093/molbev/msh103. [DOI] [PubMed] [Google Scholar]
- Moore LA, Tidyman WE, Arrizubieta MJ, Bandman E. The evolutionary relationship of avian and mammalian myosin heavy-chain genes. J Mol Evol. 1993;36:21–30. doi: 10.1007/BF02407303. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (Daudin) North Holland; Amsterdam: 1956. [Google Scholar]
- Pette D, Staron RS. Myosin isoforms, muscle fiber types, and transitions. Microsc Res Tech. 2000;50:500–509. doi: 10.1002/1097-0029(20000915)50:6<500::AID-JEMT7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Radice GP, Malacinski GM. Expression of myosin heavy chain transcripts during Xenopus laevis development. Dev Biol. 1989;133:562–568. doi: 10.1016/0012-1606(89)90058-4. [DOI] [PubMed] [Google Scholar]
- Sassoon DA, Gray GE, Kelley DB. Androgen regulation of muscle fiber type in the sexually dimorphic larynx of Xenopus laevis. J Neurosci. 1987;7:3198–3206. doi: 10.1523/JNEUROSCI.07-10-03198.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachat F, Briggs MM. Identification of two patterns of exon organization in the human striated muscle myosin heavy chain genes. Mol Biol Cell. 1999;10s:34a. [Google Scholar]
- Schachat F, Briggs MM. Phylogenetic implications of the superfast myosin in extraocular muscles. J Exp Biol. 2002;205:2189–2201. doi: 10.1242/jeb.205.15.2189. [DOI] [PubMed] [Google Scholar]
- Watabe S. Temperature plasticity of contractile proteins in fish muscle. J Exp Biol. 2002;205:2231–2236. doi: 10.1242/jeb.205.15.2231. [DOI] [PubMed] [Google Scholar]
- Weiss A, McDonough D, Wertman B, Acakpo-Satchivi L, Montgomery K, Kucherlapati R, Leinwand L, Krauter K. Organization of human and mouse skeletal myosin heavy chain gene clusters is highly conserved. Proc Natl Acad Sci U S A. 1999;96:2958–2963. doi: 10.1073/pnas.96.6.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.