Abstract
Breast tumors expressing estrogen receptor-α (ER) respond well to therapeutic strategies using selective ER modulators, such as tamoxifen. However, ~ 30% of invasive breast cancers are hormone independent because they lack ER expression due to hypermethylation of ER promoter. Treatment of ER-negative breast cancer cells with demethylating agents [5-aza-2′-deoxycytidine (5-aza-dC)] and histone deacetylase (HDAC) inhibitors (trichostatin A) leads to expression of ER mRNA and functional protein. Here, we examined whether epigenetically reactivated ER is a target for tamoxifen therapy. Following treatment with trichostatin A and 5-aza-dC, the formerly unresponsive ER-negative MDA-MB-231 breast cancer cells became responsive to tamoxifen. Tamoxifen-mediated inhibition of cell growth in these cells is mediated at least in part by the tamoxifen-bound ER. Tamoxifen-bound reactivated ER induces transcriptional repression at estrogen-responsive genes by ordered recruitment of multiple distinct chromatin-modifying complexes. Using chromatin immunoprecipitation, we show recruitment of two different corepressor complexes to ER-responsive promoters in a mutually exclusive and sequential manner: the nuclear receptor corepressor-HDAC3 complex followed by nucleosome remodeling and histone deacetylation complex. The mechanistic insight provided by this study might help in designing therapeutic strategies directed toward epigenetic mechanisms in the prevention or treatment of breast cancer.
Introduction
Estrogen is a key regulator for normal growth and differentiation of mammary glands as well as the malignant progression of breast cancer (1). Estrogen exerts its effects by binding to estrogen receptor (ER) that functions as transcription factor controlling cell proliferation and differentiation (2). Breast tumors that express ER are generally more amenable to endocrine therapy compared with ER-negative tumors that exhibit de novo resistance (3). This loss of ER is transcriptional in nature, without a high frequency of deletion, mutation, or other structural changes in ER gene (4). It is suggested that epigenetic modifications of cytosine residues in DNA and the NH2 termini of histone proteins are responsible for the silencing of ER expression in ER-negative breast tumors (5). Because all endocrine therapies are designed to block ER function in some way, the identification of new therapies or strategies for sensitization of ER-negative breast cancer cells to selective ER modulators (SERM) treatment has become very important.
Recent studies revealed that distinct coregulatory complexes modulate transcriptional activity of nuclear receptors. Ligand-bound nuclear receptors recruit multiple coactivator/corepressor complexes to modulate gene expression (6). Some of these coactivator complexes possess histone acetyltransferase activity and/or histone methyltransferase activity (7). In addition, nuclear receptors are involved in transrepression and active repression in the absence and presence of ligand through interactions with various coregulatory complexes (8). Whereas the role of coactivators for ER is very well established, the importance of corepressors is still under investigation. An increasing number of ER corepressors have been reported in last few years, which interact with ER in the AF-1, DNA-binding domain/hinge, and ligand-binding domain (LBD)/AF-2 regions (9). SERMs, such as tamoxifen, are thought to inhibit ER function by passive processes, such as repositioning of helix 12, thereby blocking the coactivator binding, and active repression via recruitment of corepressor complexes (10). Some studies have indicated that tamoxifen-bound ER might interact with corepressor complexes containing histone deacetylase (HDAC) activity leading to chromatin condensation and gene silencing (9).
Three class I HDAC-containing multiprotein complexes have been purified and characterized: the HDAC1/HDAC2-containing Sin3 and Mi2/nucleosome remodeling and histone deacetylation (NuRD) complexes and the HDAC3-containing nuclear receptor corepressor (NCoR) complex (11, 12). Some of the components of NuRD complex, such as HDAC1 and HDAC2 and two histone-binding proteins (RbAp46 and RbAp48), are also found in the Sin3 complex. In addition to HDAC activity, the NuRD complex has ATP-dependent nucleosome remodeling activity. A distinguishing feature of NuRD is the presence of metastasis-associated proteins MTA1 and/or MTA2. The unique components of the Sin3 complex include Sin3, Sin-associated protein (SAP) 18, and SAP30. Specific components of these complexes may serve to couple them to specific repression systems (11). The NCoR complex was initially found to be involved in repression associated with unliganded retinoic acid receptor and thyroid hormone receptor (13). Subsequently, it was shown that ER can also interact with this corepressor in the presence of antagonist (14). NCoR is associated with HDAC3, TBL1 (transducin β-like protein), and TBLR1 in large protein complexes (15). Both TBL and TBLR1 might function as histone-binding proteins preferentially binding to histones H2B and H4 through their NH2-terminal region, correlating with their transcriptional repression function (16).
Earlier studies have shown that one mechanism leading to loss of ER expression in ER-negative breast cancer cells involves epigenetic silencing associated with hypermethylation of the ER promoter. Treatment of such cells with DNA methyltransferase (DNMT) and/or HDAC inhibitors leads to demethylation of the ER promoter and reactivation of ER expression (17, 18). The findings that reactivated ER in ER-negative cells can function as a transcription factor for ER-responsive genes prompted us to ask whether this can also make hormone-unresponsive MDA-MB-231 cells receptive to the antagonistic actions of tamoxifen. Here, we explored this combinatory approach using both HDAC and DNMT inhibitors and found that it sensitizes ER-negative cells to tamoxifen, leading to inhibition of cell proliferation. We further examined the modulation of expression of ER target genes in response to both agonist and antagonist treatment. Using chromatin immunoprecipitation and reimmunoprecipitation analysis to examine the specific components of the corepressor complexes involved in active repression mediated by antagonist-bound reactivated ER, we show recruitment of the NuRD and NCoR complexes to ER-responsive promoters in response to treatment with tamoxifen. Interestingly, these multiprotein complexes bind in a distinctive manner, and time-course analysis indicates ordered recruitment, with the binding of the NCoR complex preceding that of NuRD complex.
Materials and Methods
Antibodies
Antibodies against poly(ADP-ribose) polymerase (PARP), caspase-3, cleaved caspase-3 (Asp175), HDAC1, HDAC2, HDAC3, HDAC5, HDAC6, and HDAC7 were purchased from Cell Signaling Technology (Danvers, MA). Anti-actin antibody was procured from Sigma (St. Louis, MO). The anti-ERα, Bax, Bcl-2, NCoR, TBL1, mSin3A, SAP18, SAP30, MTA2, and Mi2 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-rabbit IgG and anti-mouse IgG were obtained from Upstate Biotechnology (Charlottesville, VA).
Cell culture, reagents, and treatments
The human breast cancer cell lines MDA-MB-231 and MCF7 were grown in DMEM supplemented with 5% fetal bovine serum (FBS; Hyclone, Logan, UT) and 2 µmol/L l-glutamine (Invitrogen, Carlsbad, CA). For treatment, cells were seeded at a density of 5 × 105 per 100-mm tissue culture dish in phenol red–free DMEM supplemented with 5% charcoal-treated FBS (estrogen-free medium). After 24 hours, the estrogen-free medium was changed to estrogen-free medium containing 2.5 µmol/L 5-aza-2′-deoxycytidine (5-aza-dC; Sigma) for 72 hours or 100 ng/mL trichostatin A (Wako Pure Chemical Industries Ltd., Osaka, Japan) for 12 hours. For the combination study, 5-aza-dC was present in culture for 72 hours and trichostatin A was added for the last 12 hours. For treatments with 17β-estradiol (E2) and 4-hydroxytamoxifen, cells untreated or pretreated with 5-aza-dC/trichostatin A were treated with E2 (100 nmol/L for 2 hours), 4-hydroxytamoxifen (1 µmol/L for 2 hours), or vehicle in fresh estrogen-free medium for indicated time periods.
RNA isolation and reverse transcription-PCR
Total cellular RNA was extracted using the Trizol reagent kit (Life Technologies, Inc., Rockville, MD) and quantified by UV absorption. Reverse transcription-PCR (RT-PCR) was carried out according to our previously described method (18) using specific sense and antisense PCR primers for amplification. PCR products were resolved by 2% agarose gel electrophoresis and visualized by ethidium bromide staining. The primers used were ER-binding fragment-associated antigen 9 (EBAG9) forward primer and 5′-GCTACACAAGATTCTGCCT-3′ and reverse primer 5′-CTTCTTCATTAGCCGTTGTG-3′, cathepsin D forward primer 5′-TACATGATCCCCTGTGAGAAGGT-3′ and reverse primer 5′-GGGACAGCTTGTAGCCTTTGC-3′, c-Myc forward primer 5′GCCACGTCTCCACACATCAG-3′ and reverse primer 5′-TCTTGGCAGCAGGATAGTCCTT-3′, insulin-like growth factor-I (IGF-I) forward primer 5′-TGCTCTTCAGTTCGTGTGTG-3′ and reverse primer 5′-TGGCATGTCACTCTTCACTC-3′, and progesterone receptor (PR) forward primer 5′-TCATTACCTCAGAAGATTTGTTTAATC-3′ and reverse primer 5′-TGATCTATGCAGGACTAGACAA-3′.
Western blot
Whole-cell lysates were prepared as described previously (19). Proteins were quantified using the BCA protein assay kit (Pierce, Rockford, IL). Proteins were resolved on SDS-polyacrylamide gels and transferred to nitrocellulose membranes, and Western blot analyses were done using previously described antibodies. Immunodetection was done using Enhanced Chemiluminescence System (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) according to the manufacturer’s instructions.
Cell viability assay
Cell viability assay was done by estimating reduction of 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide inner salt (XTT) using a commercially available kit (Roche, Basel, Switzerland) following the manufacturer’s instructions. MCF7 and MDA-MB-231 cells were plated in 96-well plates at an initial density of 4 × 103 per well for 24 hours. Cells were treated as described above, except that 4-hydroxytamoxifen treatment (1 µmol/L) was given for 24 hours either alone or following 72-hour 5-aza-dC/trichostatin A treatment. XTT labeling reagent was added to each culture well to attain a final concentration of 0.3 mg/mL. After 4-hour exposure at 37° C, absorbance was measured at 450 and 690 nm using a 96-well plate reader (SPECTRAmax PLUS, Molecular Devices, Sunnyvale, CA). Pilot experiments verified that the cell densities were within the linear range of the XTT assay. A standard curve was prepared using cell densities from 1 × 103 to 1 × 106, and the results were calculated with respect to the number of cells.
Chromatin immunoprecipitation
Chromatin immunoprecipitation analyses were done using a published procedure (20) with following modifications. Chromatin samples were sonicated on ice thrice for 10 seconds each (i.e., until the average length of sheared genomic DNA was 1–1.5 kb) followed by centrifugation for 10 minutes. The immunoprecipitated DNA was ethanol precipitated and resuspended in 25 µL H2O. Total input samples were resuspended in 100 µL H2O and diluted 1:100 before PCR analysis. The primers for chromatin immunoprecipitation were EBAG9 forward primer 5′-ATTGTCTGCCCTTCGCCGT-3′ and reverse primer 5′-TTTGGAGGCTGCGTGCTTT-3′, cathepsin D forward primer 5′-GGTTTCTCTGGAAGCCCTGTAG-3′ and reverse primer 5′-TCCTGCACCTGCTCCTCC-3′, c-Myc forward primer 5′-AGGCGCGCGTAGTTAATTCAT-3′ and reverse primer 5′-CGCCCTCTGCTTTGGGA-3′, and IGF-I forward primer 5′-TTGTCACCATGCCCAAAAAA-3′ and reverse primer 5′-TTGCGCAGGCTCTATCTGC-3′. Initially, PCR was done with different numbers of cycles and/or dilutions of input DNA to determine the linear range of amplification; all results shown fall within this range. Following 28 to 30 cycles of amplification, PCR products were run on 1% agarose gel and analyzed by ethidium bromide staining. All chromatin immunoprecipitation assays were done at least thrice with similar results.
Chromatin immunoprecipitation/reimmunoprecipitation
Chromatin immunoprecipitation/reimmunoprecipitations on supernatants were done following the same procedure as the primary immunoprecipitations. Bead eluates from the first cycle of immunoprecipitation were incubated with 10 mmol/L DTT at 37° C for 30 minutes and diluted 1:50 in dilution buffer containing 1% Triton X-100, 2 mmol/L EDTA, 20 mmol/L Tris-HCl (pH 8.1), and 150 mmol/L NaCl followed by reimmunoprecipitation with specific second antibodies.
Results
ER reexpression is induced by the HDAC and DNMT inhibitors
Although antiestrogen therapy targeting ER is the most successful therapy for breast cancer, a major problem is that its use is limited to ER-positive breast cancers that generally have a better prognosis (1, 3). Because ER-negative breast cancers are more aggressive (21), alternative combinatorial therapies targeting ER-negative breast cancers are urgently needed. Earlier studies have shown that the ER promoter is hypermethylated and ER mRNA is absent in ER-negative breast cancer cells (5). One efficient way for targeted therapy for ER-negative hormone-independent breast cancers could be to transform ER-negative into ER-positive breast cancer cells by gene therapy or ER gene reexpression. We used the MDA-MB-231 breast cancer cells as a model of ER-negative breast cancers. This human cell line is particularly suitable for preclinical studies because it is highly aggressive both in vitro and in vivo (22).
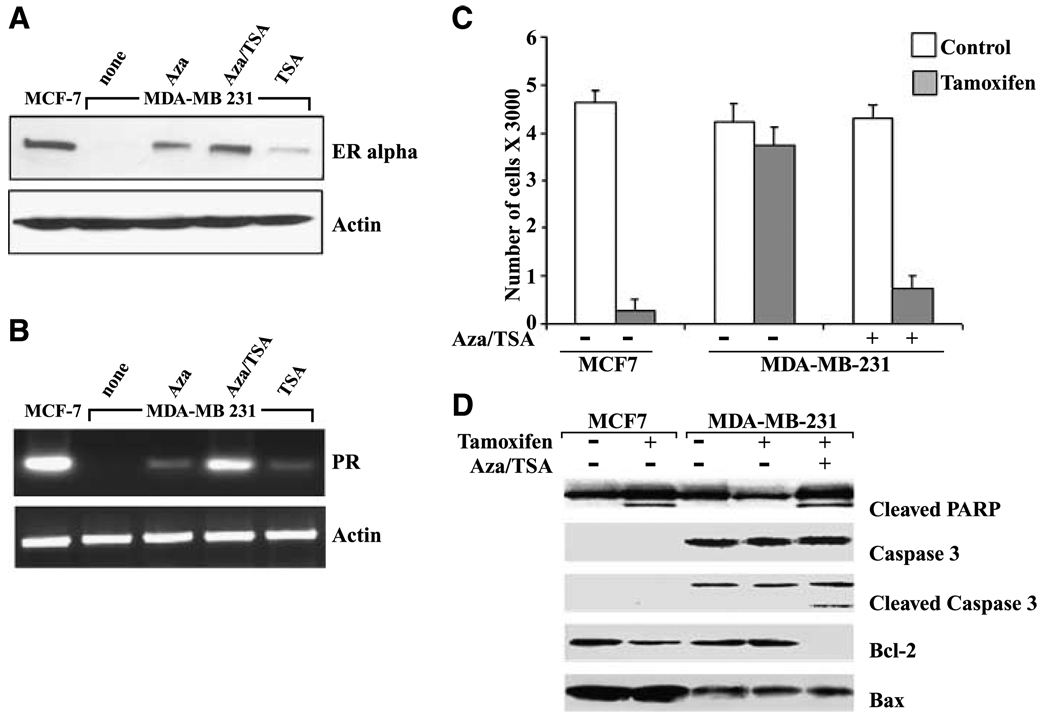
First, we analyzed the reversible nature of ER expression in MDA-MB-231 cells using methylation-specific PCR and RT-PCR (data not shown), confirming the previous findings (18) that the ER CpG island is methylated in MDA-MB-231 cells and treatment with 5-aza-dC/trichostatin A results in reexpression of ER mRNA. Importantly, we observed the reexpression of ER protein in MDA-MB-231 cells treated with 5-aza-dC/trichostatin A (Fig. 1A). We also observed ER protein reexpression in MDA-MB-231 cells treated with trichostatin A alone (Fig. 1A). Trichostatin A induces changes in histone acetylation resulting in chromatin decondensation (18); hence, trichostatin A treatment results in ER protein reexpression albeit at low levels. Next, we examined if the reexpressed ER in MDA-MB-231 cells can act as a functionally active transcription factor and control the expression of ER-responsive genes. As evident in Fig. 1B, 5-aza-dC/trichostatin A–treated ER-negative cells induced greater PR expression compared with either treatment alone, whereas untreated cells showed no PR expression.
Figure 1.
Treatment with HDAC and DNMT inhibitors reexpress ER protein and enhanced the responsiveness to 4-hydroxytamoxifen in ER-negative MDA-MB-231 cells. A, ER protein reexpression was analyzed after treatment of MDA-MB-231 cells with 5-aza-dC (Aza; 2.5 µmol/L for 72 hours), trichostatin A (TSA; 100 ng/mL for 12 hours), or 5-aza-dC and trichostatin A (Aza/TSA) as described in Materials and Methods. ER-positive MCF7 cells were used as positive control, whereas untreated ER-negative MDA-MB-231 cells were used as negative control. Western blotting using specific antibodies for ER shows ER protein reexpression after 5-aza-dC and trichostatin A treatments in MDA-MB-231 cells. B, expression of the ER-responsive gene PR was used as an indicator of functionally active ER. RT-PCR analysis showed PR mRNA reexpression after 5-aza-dC and trichostatin A treatments in MDA-MB-231 cells. The ER-positive prototype, MCF7 cells, was used as a positive control. The RT-PCR product of β-actin was included as a control. C, growth inhibition by combined 4-hydroxytamoxifen (1 µmol/L) and 5-aza-dC/trichostatin A treatment was assayed using XTT assay. Both MCF7 and MDA-MB-231 cells were treated with 4-hydroxytamoxifen for 24 hours before assay. In addition, MDA-MB-231 cells were treated with a combination of 5-aza-dC/trichostatin A as described along with 4-hydroxytamoxifen treatment before XTT assay. D, MDA-MB-231 cells were treated as indicated. Whole-cell lysates were prepared, fractionated by SDS-PAGE, and transferred to membranes. Changes in apoptotic proteins were detected by Western blotting with specific antibodies against cleaved PARP, caspase-3, cleaved caspase-3, Bcl-2, and Bax as indicated.
Inhibition of HDAC and DNMT activity sensitizes ER-negative breast cancer cells to estrogen antagonist tamoxifen
Based on our observation that simultaneous HDAC and DNMT inhibition by trichostatin A and 5-aza-dC leads to reexpression of ER mRNA and protein in ER-negative breast cancer cells, we next investigated whether this reexpression could be physiologically linked to tamoxifen responsiveness. Treatment with trichostatin A and 5-aza-dC rendered ER-negative MDA-MB-231 cells responsive to tamoxifen (Fig. 1C). Combined treatment of MDA-MB-231 cells with trichostatin A and 5-aza-dC along with 1 µmol/L 4-hydroxytamoxifen resulted in a significant increase in cell growth inhibition compared with cells treated with either 5-aza-dC/trichostatin A combination only or 4-hydroxytamoxifen alone. As expected, cell proliferation was inhibited considerably in response to 4-hydroxytamoxifen treatment in ER-positive MCF7 cells. It has been shown that tamoxifen inhibits cell proliferation and induces apoptosis in breast cancer cells (23). Several apoptosis-associated proteins have been shown to play critical roles in regulating cell death, including caspases, Bcl-2 family members, and PARP (24). To determine whether these proteins are involved in tamoxifen-induced cell death in MDA-MB-231 cells pretreated with 5-aza-dC and trichostatin A, we examined their expression by Western blotting. Our results (Fig. 1D) are in concurrence with the previous finding (25) that caspase-3 is not expressed in MCF7 cells. Treatment of MCF7 cells with 4-hydroxytamoxifen resulted in PARP cleavage and reduced expression of Bcl-2, whereas Bax protein expression remained unaltered. MDA-MB-231 cells treated with 5-aza-dC/trichostatin A in combination with tamoxifen showed cleaved, active caspase-3 along with cleaved PARP, its downstream target. Furthermore, Bcl-2 protein was down-regulated, and Bax protein expression remained unaltered. Our findings suggest that a combinatory treatment of ER-negative breast cancer cells with 5-aza-dC/trichostatin A along with 4-hydroxytamoxifen induces cell death as indicated by the activation of caspase-3 and cleavage of PARP.
Reexpression of ER in ER-negative cells renders ER-responsive genes responsive to the antagonistic effects of 4-hydroxytamoxifen
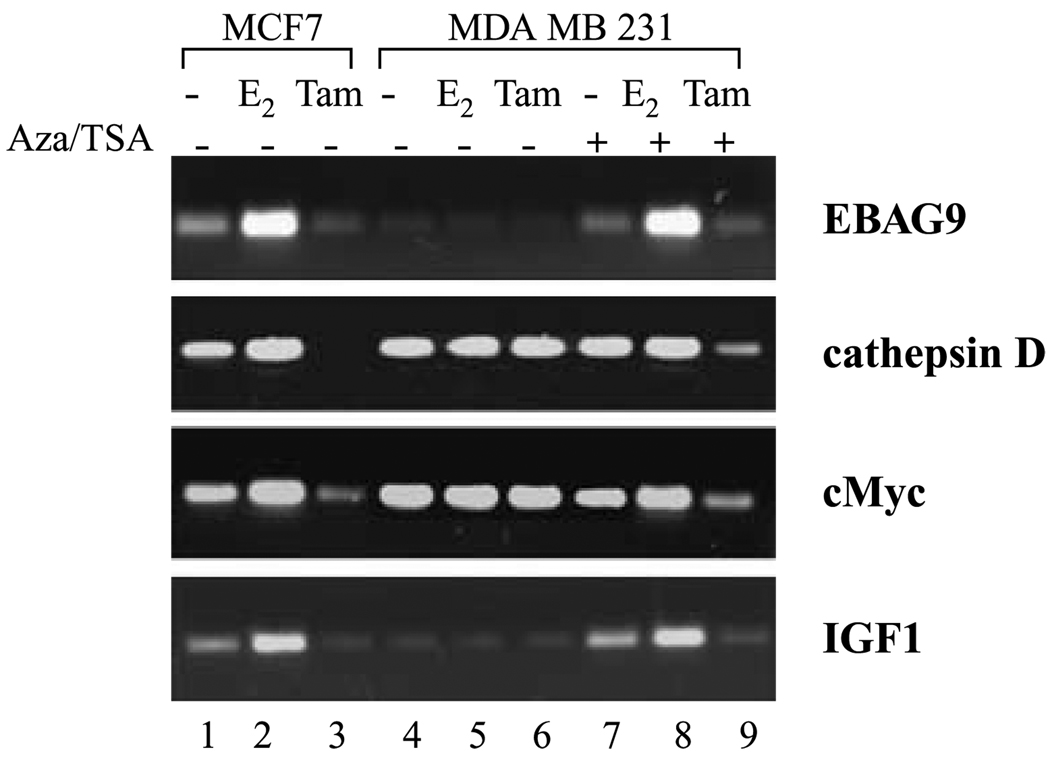
Steroid receptors, such as ER, regulate gene transcription either by binding directly to the promoter of target genes or by binding indirectly through other transcription factors (1, 2). Genes regulated through direct ER binding, such as EBAG9 and cathepsin D, typically harbor a classic hormone-responsive element (HRE) and are activated slowly (26, 27). In contrast, genes regulated by indirect binding of ER to nonclassic HREs are quickly activated, such as c-Myc and IGF-I (28, 29). We next examined the difference in agonist-modulated versus antagonist-modulated expression of these genes in our experimental system. The ER-responsive genes showed a strong increase in expression in the presence of E2 in ER-positive MCF7 cells, whereas tamoxifen treatment repressed the expression of these same genes (Fig. 2). ER-negative MDA-MB-231 cells exhibited no detectable expression of EBAG9, whereas IGF-I showed very low levels of expression. ER-negative cells show robust levels of both cathepsin D and c-Myc similar to ER-positive cells. Importantly, the basal levels of expression were unaffected by both E2 and tamoxifen treatment in ER-negative cells. Reexpression of ER in ER-negative cells with 5-aza-dC/trichostatin A treatment restored basal levels of expression of ER-responsive genes EBAG9 and IGF-I and the expression was inducible by E2 treatment. Treatment with tamoxifen inhibited the expression of ER-responsive genes (Fig. 2). More importantly, ER-responsive genes that are constitutively expressed in MDA-MB-231 cells and previously remained unaltered with either tamoxifen or E2 treatment became responsive to antagonist treatment in ER-negative cells reexpressing ER. Expression of both cathepsin D and c-Myc was inhibited strongly by tamoxifen treatment in MDA-MB-231 cells pretreated with 5-aza-dC/trichostatin A. These data indicate that 5-aza-dC/trichostatin A pretreatment of ER-negative cells restores not only E2-dependent activation but also tamoxifen-dependent repression of ER target genes via reexpression of ER.
Figure 2.
Expression levels of ER-responsive genes in ER-negative cells reexpressing ER in response to E2 and 4-hydroxytamoxifen. MDA-MB-231 cells were grown in estrogen-free medium and treated with 5-aza-dC and trichostatin A for 72 and 12 hours, respectively, to reexpress ER followed by fresh medium containing no treatment or treatment with E2 or 4-hydroxytamoxifen (Tam) for 2 hours as described in Materials and Methods. Total RNA was isolated, quantified, and subjected to cDNA synthesis followed by RT-PCR analysis using specific primer pairs. Differential modulation of expression of ER-responsive genes was observed in agonist- and antagonist-treated MDA-MB-231 cells reexpressing ER.
Both HDAC2 and HDAC3 bind to the promoters of ER-responsive genes in response to antagonist treatment
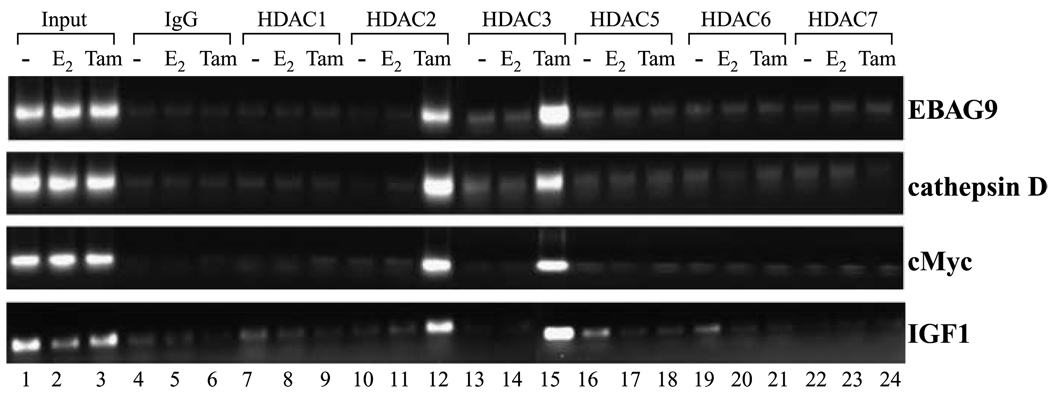
Recruitment of corepressors to the ER results in the formation of multisubunit corepressor complexes, including various HDACs, facilitating chromatin condensation (6, 9). To investigate the specific corepressor complexes involved in antagonist-induced repression of ER-responsive genes in MDA-MB-231 cells reexpressing ER (MDA-MB-231/ER), we first sought to investigate the pattern of recruitment of various HDAC molecules on these ER-responsive gene promoters using chromatin immunoprecipitation. Treatment of 5-aza-dC/trichostatin A–pretreated MDA-MB-231 cells with 4-hydroxytamoxifen induced a dramatic increase in the occupancy of HDAC2 and HDAC3 at the EBAG9, cathepsin D, c-Myc, and IGF-I gene promoters (Fig. 3). In contrast, HDAC2 and HDAC3 were not recruited to the same genes in response to E2. Importantly, another member of class I HDACs, HDAC1, and certain members of class II HDACs, including HDAC5, HDAC6, and HDAC7, were not recruited to ER-responsive promoters (Fig. 3).
Figure 3.
Profile of various HDAC molecules on the promoters of ER-responsive genes. Soluble chromatin was prepared from MDA-MB-231 cells pretreated with 5-aza-dC (2.5 µmol/L for 72 hours) and trichostatin A (100 ng/mL for 12 hours) followed by treatment with E2 (100 nmol/L for 2 hours), 4-hydroxytamoxifen (1 µmol/L for 2 hours), vehicle (−) as described in Materials and Methods and immunoprecipitated with 5 µg specific antibodies against various class 1 and II HDAC proteins overnight at 4° C. The immune complexes were pulled down with protein A agarose/salmon sperm DNA beads and washed extensively as described in Materials and Methods, and cross-linking was reversed. The purified DNA was analyzed by PCR using primers spanning the EREs of EBAG9, cathepsin D, c-Myc, and IGF-I gene promoters. Recruitment of HDAC2 and HDAC3 was observed on all four promoters in response to tamoxifen treatment, whereas HDAC1, HDAC5, HDAC6, and HDAC7 were not recruited.
Tamoxifen-bound reactivated ER recruits Mi2/NuRD corepressor complex but not Sin3A corepressor complex to ER-responsive genes
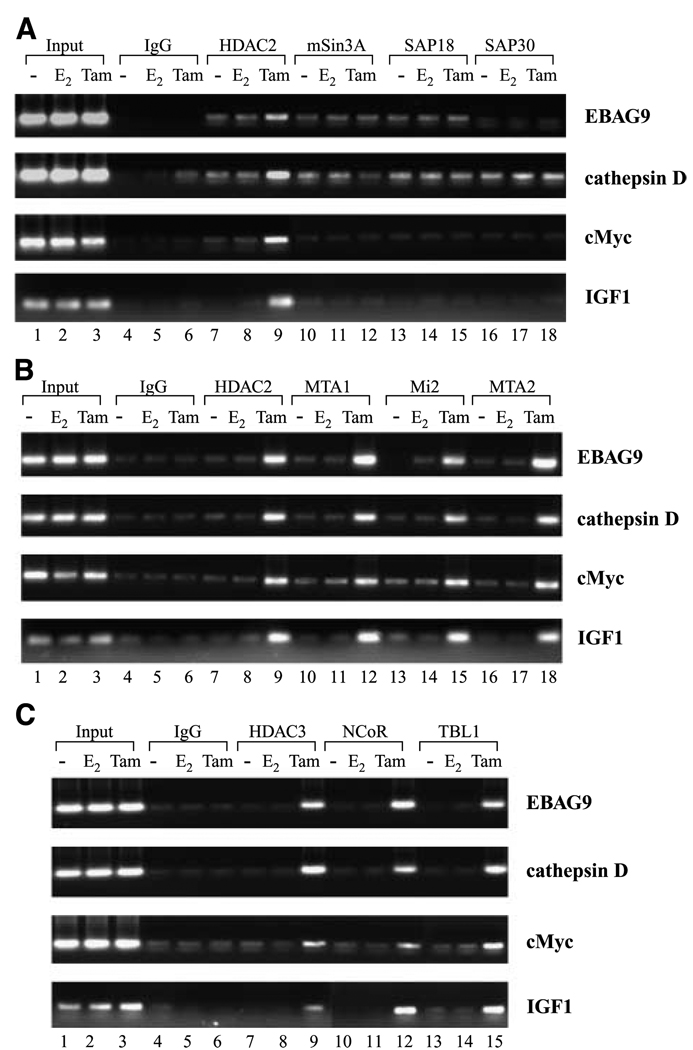
Biochemical studies from various laboratories have characterized three class I HDAC-containing corepressor complexes [i.e., the HDAC1/HDAC2-containing Sin3 (11) and Mi2/NuRD (12) complex and the HDAC3-containing NCoR complexes (13)]. Having shown that tamoxifen-bound reactivated ER is able to induce occupancy of ER-responsive promoters by HDAC2 and HDAC3, we sought to determine which corepressor complexes are also getting recruited to achieve active repression of these target genes. Chromatin immunoprecipitation analysis with specific antibodies to HDAC2 showed that tamoxifen-bound reactivated ER specifically recruits HDAC2 to the promoter region of EBAG9, cathepsin D, c-Myc, and IGF-I in an antagonist-dependent manner (Fig. 4A). Importantly, chromatin immunoprecipitation analysis using specific antibodies to the components of Sin3 complex, mSin3A, SAP18, and SAP30 revealed that neither agonist nor antagonist treatment of MDA-MB-231 cells reexpressing ER resulted in the recruitment of members of Sin3 complex at the promoter region of ER-responsive genes (Fig. 4A). In contrast, reactivated ER recruited distinct components of NuRD complex in MDA-MB-231 cells treated with tamoxifen (Fig. 4B). 4-Hydroxytamoxifen stimulated the ER-dependent recruitment of MTA1 and Mi2 to ER-responsive promoters, whereas E2-bound reactivated ER does not induce recruitment of these proteins. Our results clearly show that antagonist-bound reactivated ER induces active repression of target genes via engaging distinctive corepressor complexes. The recruitment of NuRD components occurred irrespective of the type of estrogen-responsive element (ERE) in the various gene promoters.
Figure 4.
Recruitment of HDAC2-Sin3, HDAC2-NuRD, and HDAC3-NCoR corepressor complex to ER-responsive gene promoters. A, formaldehyde cross-linked chromatin was immunoprecipitated with antibodies specific for HDAC2, mSin3A, SAP18, and SAP30 from MDA-MB-231 cells treated as in Fig. 3. The immune complexes were pulled down with protein A agarose/salmon sperm DNA beads and washed extensively as described in Materials and Methods, and cross-linking was reversed. The purified DNA was analyzed by PCR using specific primers spanning the EREs of EBAG9, cathepsin D, c-Myc, and IGF-I gene promoters. Recruitment of signature subunits of the HDAC2-Sin3 complex was not observed. B, cross-linked chromatin-protein complexes were immunoprecipitated with antibodies specific for HDAC2, MTA2, Mi2, and MTA1 from MDA-MB-231 cells treated as indicated. The purified DNA was analyzed by PCR using primers spanning the EREs of EBAG9, cathepsin D, c-Myc, and IGF-I gene promoters. Recruitment of all four signature subunits of the HDAC2-NuRD complex was observed. C, formaldehyde cross-linked chromatin was immunoprecipitated with antibodies specific for HDAC3, NCoR, and TBL1 from MDA-MB-231 cells treated as in Fig. 3. The immune complexes were pulled down with protein A agarose/salmon sperm DNA beads and washed extensively as described in Materials and Methods, and cross-linking was reversed. The purified DNA was analyzed by PCR using primers spanning the EREs of EBAG9, cathepsin D, c-Myc, and IGF-I gene promoters. Recruitment of all three signature subunits of the NCoR-HDAC3 complex was observed.
HDAC3 recruited by tamoxifen-bound reactivated ER is a part of NCoR complex
Previous in vitro studies indicate that ER interacts with NCoR and SMRT corepressors in the presence of antagonist (14) and biochemical purification and characterization of these complexes showed that both are associated with HDAC3 (15). We thus sought to understand the participation of NCoR corepressor complex in the mediation of active repression of ER-responsive genes via antagonist-bound reactivated ER. Interestingly, as observed for components of NuRD complex, the members of NCoR complex, HDAC3, NCoR, and TBL1, associated with the promoters of all ER-responsive genes included in this study in an antagonist-dependent manner (Fig. 4C). In contrast, E2 treatment did not induce recruitment of the NCoR complex components over IgG and vehicle controls. These data indicate a role of NCoR complex in active repression achieved by antagonist-bound reactivated ER in MDA-MB-231 cells.
Tamoxifen-bound reactivated ER uses multiple corepressor complexes
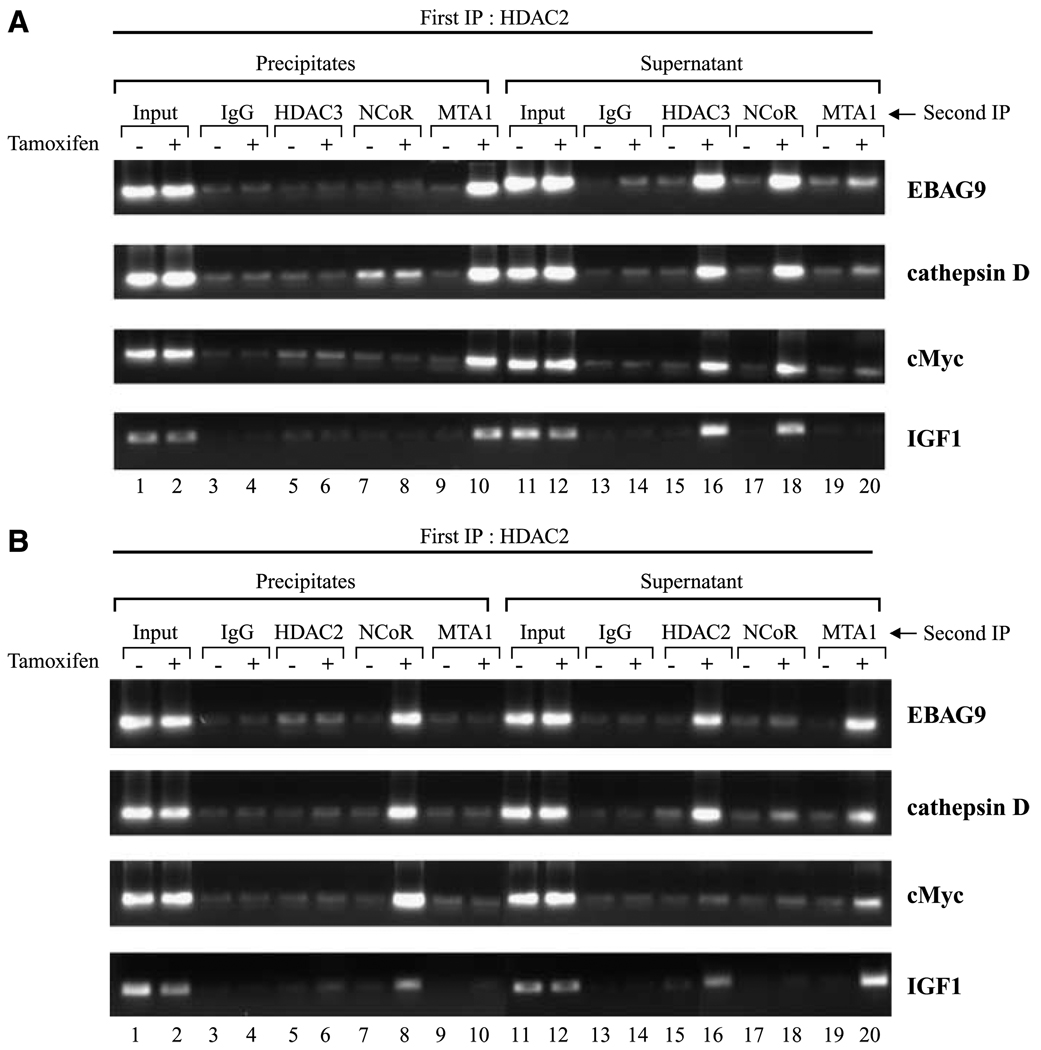
Having established the antagonist-specific recruitment of the NuRD and NCoR corepressor complexes at the ER-responsive promoters by reactivated ER, we next investigated the pattern of recruitment of these complexes. The presence of two different multiprotein corepressor complexes could be explained by one of the following scenarios: both complexes get recruited to the promoter region simultaneously either through different anchor subunits or by a member of one multiprotein complex acting as an anchor for the other multiprotein complex. NCoR has been found to interact with components of NuRD complex (30). Alternatively, the complexes may be recruited in an ordered manner with one complex promoting or excluding the binding of the other. To examine this, we used chromatin immunoprecipitation/reimmunoprecipitation to determine which subunits were present on the promoter at the same time. In these experiments, DNA bound by HDAC2 was immunoprecipitated using HDAC2 antibodies. Then, both the precipitates and the supernatants were subjected to reimmunoprecipitation with antibodies against HDAC3, NCoR, or MTA1 representing the NCoR (HDAC3/NCoR) and NuRD (MTA1) complex, respectively. The ER-responsive promoters were then analyzed by PCR. As shown in Fig. 4B and C, the presence of both NuRD and NCoR multiprotein complex could be detected on the promoter of EBAG9, cathepsin D, c-Myc, and IGF-I in MDA-MB-231 cells reexpressing ER in an antagonist-dependent manner. Our chromatin immunoprecipitation/reimmunoprecipitation experiments show that ER-responsive gene promoters immunoprecipitated with antibodies against HDAC2 could be reimmunoprecipitated with antibodies against MTA1, whereas no detection was observed with HDAC3 and NCoR antibodies (Fig. 5A). In contrast, the supernatants from the primary chromatin immunoprecipitation with HDAC2 antibodies showed weak detection of the promoter regions when reimmunoprecipitation was done with antibodies against MTA1, but reimmunoprecipitation with antibodies against HDAC3 and NCoR resulted in robust enrichment of ER-responsive gene promoters (Fig. 5A). In a reciprocal experiment, precipitates from the first chromatin immunoprecipitation with HDAC3 antibodies were subjected to chromatin immunoprecipitation/reimmunoprecipitation analysis with antibodies against HDAC2, NCoR, and MTA1. As shown in Fig. 5B, NCoR protein is recruited to the ER-responsive promoters along with HDAC3 in an antagonist-dependent manner, whereas both HDAC2 and MTA1 showed no binding. In contrast, supernatants from initial chromatin immunoprecipitation with HDAC3 antibodies showed occupancy of ER-responsive promoters by both HDAC2 and MTA1, whereas NCoR showed no recruitment. Antibody specificities were checked by immunoprecipitation experiments followed by Western blotting. These data suggest that both HDAC2/NuRD complex and NCoR/HDAC3 complex might get recruited to the ER-responsive gene promoters in a mutually exclusive manner. Importantly, the pattern of association of these corepressor complexes with the promoters of genes that harbor classic EREs was the same as genes harboring nonclassic EREs.
Figure 5.
Pattern of recruitment of tamoxifen-bound reactivated ER-NCoR-HDAC3 complex and tamoxifen-bound reactivated ER-NuRD-HDAC2 complex. A, cross-linked chromatin was immunoprecipitated with antibodies specific for HDAC2 from MDA-MB-231 cells treated as detailed in Fig. 3 and the immune complexes were pulled down with protein A/G agarose/salmon sperm DNA beads, supernatants were saved, and beads were washed extensively as described in Materials and Methods. Bead eluates from the first immunoprecipitation were incubated with 10 mmol/L DTT at 37° C for 30 minutes and diluted 1:50 in dilution buffer followed by reimmunoprecipitation with the second antibodies as indicated. Reimmunoprecipitation on supernatants was done essentially the same as primary immunoprecipitations. After second immunoprecipitations, cross-linking was reversed and the purified DNA was analyzed by PCR using primers spanning the EREs of EBAG9, cathepsin D, c-Myc, and IGF-I gene promoters. B, chromatin was immunoprecipitated first with antibodies specific for HDAC3 and the experiment was done exactly the same way as in (A) using second antibodies as indicated and the purified DNA was analyzed by PCR using primers spanning the EREs of EBAG9, cathepsin D, c-Myc, and IGF-I gene promoters.
Multiprotein corepressor complexes assembled by tamoxifen-bound reactivated ER show ordered recruitment
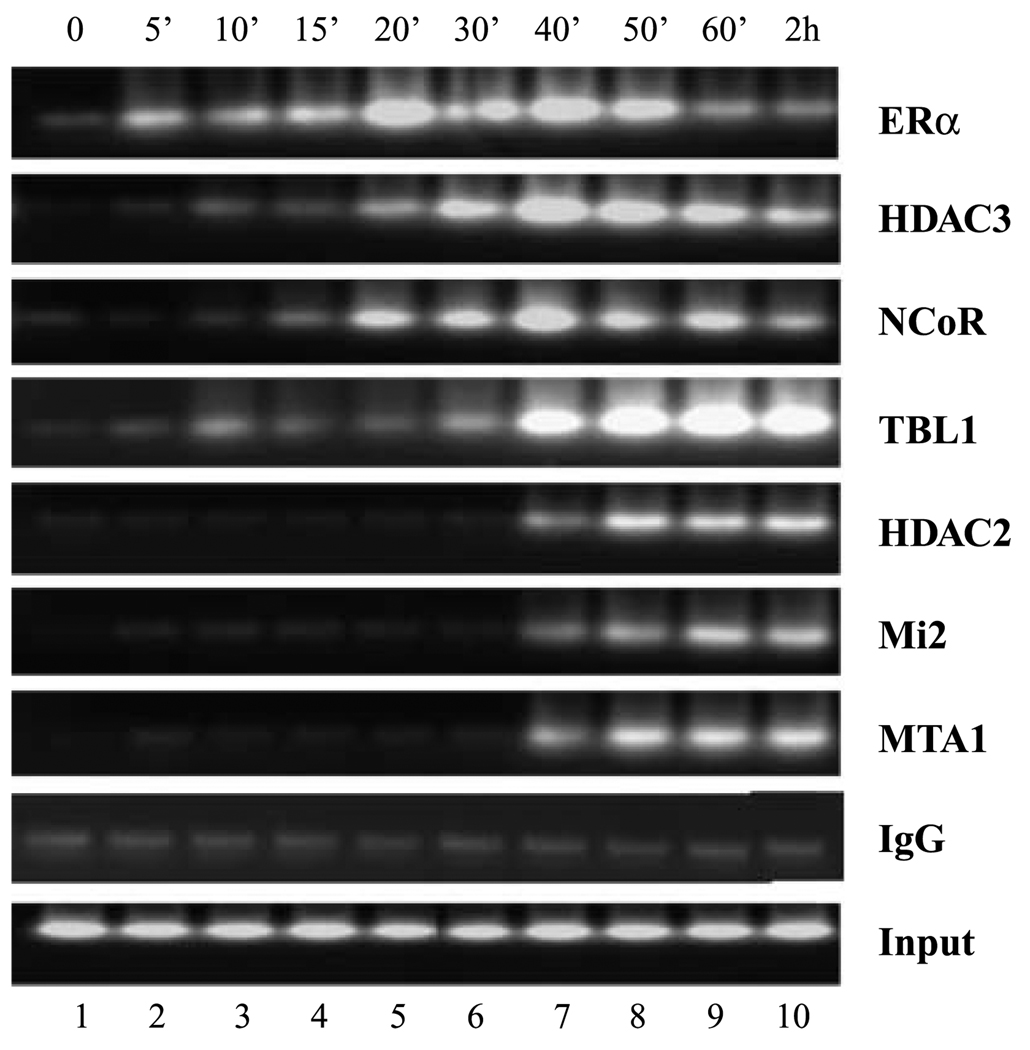
Our findings that the 4-hydroxytamoxifen-dependent occupancy of distinct ER corepressor complexes might occur in a mutually exclusive manner could be explained in two ways. The recruitment of the corepressor complexes could be stochastic, such that at any one time individual promoter DNA molecules are bound by one complex or the other. Alternatively, all promoter DNA molecules may be bound by one complex, which is then temporally replaced by the other. To examine this, we next sought to determine the kinetics of specific corepressor recruitment in time-course experiments. Chromatin immunoprecipitation analysis with antibodies to ER indicated that ER was associated with the EBAG9 promoter in the presence of antagonist (Fig. 6). Examination of the recruitment kinetics of specific repressor proteins revealed association of NCoR/HDAC3 complex components within 20 to 30 minutes of tamoxifen treatment. Maximal levels of HDAC3 and NCoR binding were achieved within 40 minutes followed by a slight decline in promoter occupancy 2 hours post-treatment (Fig. 6). In contrast, recruitment of the NuRD complex components, HDAC2, Mi2, and MTA1, displayed considerably slower kinetics. Detectable occupancy of the EBAG9 promoter by NuRD components did not occur until 40 minutes after tamoxifen treatment, with maximal levels occurring 50 to 60 minutes post-treatment (Fig. 6). In summary, data in Fig. 6 suggest that tamoxifen-mediated repression of ER-responsive genes involves the sequential recruitment of multi-protein corepressor complexes with components of the NCoR/HDAC3 complex preceding that of NuRD/HDAC2 complex. Collectively, these result indicate that epigenetic reactivation the ER gene in MDA-MB-231 cells sensitizes ER-negative cells the antagonistic effects of tamoxifen involving modulation expression of ER-responsive genes via ordered recruitment distinct corepressor complexes.
Figure 6.
Dynamics of assembly of tamoxifen-bound reactivated ER-NCoR-HDAC3 complex and tamoxifen-bound reactivated ER-NuRD-HDAC2 complex. Chromatin immunoprecipitation assays were done as described earlier. MDA-MB-231 cells were pretreated with 5-aza-dC (2.5 µmol/L for 72 hours) and trichostatin A (100 ng/mL for 12 hours) as described in Materials and Methods, and cells were treated with 1 µmol/L 4-hydroxytamoxifen for varying lengths of time as indicated above the lanes. Right, antibodies used for chromatin immunoprecipitation. Recruitment of tamoxifen-bound reactivated ER to EBAG9 promoter was observed within 5 minutes of tamoxifen treatment, whereas NCoR-TBL1-HDAC3 complex gets recruited in 20 to 30 minutes followed by the binding of the components of NuRD complex by 40 minutes of tamoxifen treatment.
Discussion
The effects of endocrine therapy are primarily mediated through the ER; therefore, ER expression is a strong predictor of response SERM treatment. Indeed, lack of ER expression is the dominant mechanism of de novo resistance to SERMs, such as tamoxifen (1–3). In addition, during breast cancer progression, many initially ER-positive tumors lose ER expression and attain hormone unresponsiveness (4, 5). ER-negative tumors are more aggressive, and considering the ability of these tumors to metastasize and their heterogeneity, new therapies or strategies for sensitization ER-negative tumors to endocrine treatment are required.
Several enzymatic inhibitors targeting HDACs have been developed with good in vivo bioavailability and intracellular capability to inhibit HDAC. Preclinical studies and initial clinical trials indicate that HDAC inhibitors from different structural classes are very well tolerated and exhibit clinical activity against a variety of human cancers (31, 32). The hydroxamate trichostatin A was shown previously to have an in vivo antitumor activity with daily parenteral dosing associated with little systemic toxicity (33). The greatest potential of HDAC inhibitors lies in their ability to modulate the activity of other therapeutic agents. Demethylating agents, such as 5-aza-dC, are particularly interesting candidates, owing to the interaction of DNA methylation with histone deacetylation in gene silencing of tumor suppressor genes. Combined treatment of trichostatin A or depsipeptide with 5-aza-dC has been shown to synergistically reactivate silenced tumor suppressor genes in human cancer cells, including MLH1, TIMP3, CDKN2B, CDKN2A, gelsolin, and maspin (34, 35).
We have shown previously that treatment of ER-negative breast cancer cells with DNMT and HDAC inhibitors leads to reactivation of expression of functional ER (18). We explored this strategy for sensitizing ER-negative breast cancer cells to tamoxifen. We found that ER-negative breast cancer cells can be sensitized to antitumor effects of tamoxifen by combined treatment with 5-aza-dC/trichostatin A, underscoring the importance of drugs having the potential to derepress the expression of epigenetically silenced key genes in cancer therapeutics. Previously, other laboratories have transfected ERα recombinant cDNA into the MDA-MB-231 cells in an attempt to recover normal estrogen and antiestrogen responsiveness. The unliganded ER was found to inhibit the invasiveness and growth of MDA-MB-231 cells transfected with ER (36). However, whereas E2 inhibited cell growth and invasiveness to some extent, tamoxifen had no effect (37, 38), indicating that transfecting ER into ER-negative breast cancer cells is not sufficient to sensitize these unresponsive cells to endocrine therapy. In addition, this approach is not directly translatable to clinical studies.
Tamoxifen is a potent ER antagonist and its pharmacology has been reviewed extensively (39). Many studies focusing on the mechanism of action of tamoxifen have indicated that this compound acts in both a cytostatic (causing G0-G1 arrest) and a cytotoxic (inducing apoptosis) manner (40, 41). We show here that tamoxifen-induced growth inhibition of 5-aza-dC/trichostatin A–pretreated cells involves apoptosis as indicated by cleavage of caspase-3 and its downstream target PARP. Tamoxifen induced down-regulation of Bcl-2 in MDA-MB-231 cells reexpressing ER without any alteration at Bax expression as reported earlier for MCF7 cells (42), indicating the involvement of Bcl-2 family in tamoxifen-mediated cell death.
The findings that reactivation of ER can direct tamoxifen-dependent repression of endogenous ER target genes indicates that 5-aza-dC/trichostatin A reactivated ER is able to interact with both agonists and antagonists to modulate transcription. Tamoxifen mainly exerts its antiproliferative action by binding competitively to ER, modulating the ER-mediated transcriptional cascade. Agonist-bound ER adopts a conformation in which α-helices (3, 5, and 12) in the LBD form a hydrophobic cleft (AF-2), providing a binding surface for NR boxes (LXXLL motifs) in coactivators. ER antagonists, such as tamoxifen, have a bulky side chain that sterically modulates the conformation of the hydrophobic cleft (AF-2) with helix 12 binding to the AF-2 cleft with its own intrinsic NR box, occluding the binding of coactivators. Antagonist-mediated inhibition of ER not only is a passive process resulting from repositioning of helix 12, thereby blocking the coactivator binding (10), but also involves the active recruitment of corepressors to form repressive ER complex at ER target genes.
To comprehend the molecular basis of repression of ER-responsive genes by tamoxifen-bound reactivated ER in ER-negative breast cancer cells, it is important to decipher the nature of corepressor complex involved in these antagonistic actions. Our studies with tamoxifen-bound reactivated ER show the formation of a distinct complex containing HDAC3, NCoR, and TBL1 on promoter regions of ER-responsive genes. Other studies have also shown that HDAC3 is the major HDAC associated with NCoR/SMRT complexes and NCoR interacts directly with HDAC3 through a deacetylase-activating domain activating HDAC3 activity (15, 16). TBL1 then recognizes and binds the resultant deacetylated histone tails, further stabilizing the binding of this multiprotein complex leading to repression. TBL1 and TBLR1 are not required for HDAC3 activity or initial binding of the NCoR/SMRT complex to nuclear receptors, but they can interact with core histones to stabilize the binding. This is similar to the role of RbAp46 and RbAp48 in NuRD complex. Whereas RbAp48 binds to H2A, H3, and H4, TBL1 bind preferentially to H2B and H4. We observed binding of NuRD complex to the ER-responsive promoters in ER-negative breast cancer cells reexpressing functional ER, and this recruitment was specifically dependent on tamoxifen treatment as no binding was observed in the presence of estrogen.
Combinatorial utilization of multiple corepressor complexes may be required to achieve physiologic levels of repression on some promoters, whereas on other promoters different complexes might get recruited independent of each other. NCoR directly interacts with nuclear receptors via its NR box-related conserved bipartite NR interaction domain containing L/IXXI/VI sequence (16), anchoring NCoR/HDAC3 multiprotein complex. NCoR can also interact with components of both the SAP and the NuRD complexes (30), suggesting that NCoR and NuRD complexes could be corecruited to ER or other nuclear receptor gene targets. Our chromatin immunoprecipitation/reimmunoprecipitation data show that both NCoR/HDAC3 and NuRD complex bind to ER-responsive promoters containing either classic or nonclassic ERE promoters in a mutually exclusive manner. Mutually exclusive binding of both NCoR and NuRD corepressor complexes rules out the possibility of NCoR-mediated recruitment of NuRD complex at least in tamoxifen-bound reactivated ER. Because human NuRD complex is a multisubunit protein complex, it is possible that it gets recruited using one of its own subunits as the anchoring protein. Biochemical and immunofluorescence studies have shown that MTA1 interacts directly with the ER (43). However, whether MTA1 targets the NuRD complex to ER-responsive promoter has not been elucidated. Other candidate subunits of the NuRD complex are methyl-binding proteins, such as MBD2 and MBD3. Whereas human MBD3 does not recognize methylated DNA (44), MBD2 might direct the recruitment of NuRD complex to methylated loci at target gene promoters (45). Our time-course experiments, however, suggest that a DNA methylation-mediated mechanism is unlikely, as NuRD complex components were observed at the EBAG9 promoter within 40 minutes of tamoxifen treatment. In addition, NuRD complex purified with HDAC1 contains MBD2 (46), whereas a similar immunoaffinity purification of HDAC2 generated a NuRD complex with no detectable MBD2 (46). Our data show the recruitment of HDAC2 but not HDAC1 containing NuRD complex at the ER-responsive promoters, suggesting that MBD2 is not involved in tamoxifen-mediated repression by reactivated ER. Further studies are needed to clearly show how NuRD complex binds to tamoxifen-bound reactivated ER and mediate repression of ER-responsive genes.
Our findings also provide evidence for an ordered recruitment of NCoR complex followed by NuRD complex at distinct ER target promoters in ER-negative breast cancer cells via tamoxifen-bound reexpressed ER. Sequential recruitment of various cofactors has been reported for regulation of various mammalian genes (19, 20). Given the ordered recruitment of corepressor complexes shown here, our findings support a multistep model of tamoxifen-mediated repression by reactivated ER. NCoR complex could directly interact with tamoxifen-bound reactivated ER resulting in deacetylation of local histones through recruitment of HDAC activity (15, 16). One possibility is that removal of the acetyl groups from K9 and K14 of histone H3 (47) creates an environment that promotes the binding of Suv39H1/Clr4. The methylation of H3-K9 by Suv39H1/Clr4 after HDACs remove the acetyl groups from K9 and K14 of histone H3 (47) then serves as a binding site for the chromodomain of HP1/Swi6 (48, 49). NuRD complex contains Mi2/CHD family proteins that have a chromodomain (12) and biochemical analysis have shown that the NuRD complex associates with histone H3 when Lys9 is methylated (50). This model is in accordance with the histone code hypothesis as the pattern of histone tail modifications serves as a recognition code for the recruitment of cofactors resulting in modulation of chromatin structure and function.
In conclusion, our findings are clinically important, as endocrine therapy targeting ER has proven its efficacy with the development of antiestrogens and aromatase inhibitors. Sensitizing hormone-resistant ER-negative breast cancer cells to endocrine therapy by combined treatment with DNMT inhibitors and HDAC inhibitors provides new treatment options for patients with de novo resistance. In addition, the elucidation of the specific corepressor complexes involved in the tamoxifen-bound reactivated ER-mediated repression of endogenous ER-responsive genes might help in designing more combined therapies using other therapeutic agents and innovative drug delivery strategies.
Acknowledgments
Grant support: U.S. Army Medical Research and Material Command W81WXH-04-BC-030963, Susan G. Komen Breast Cancer Research Foundation grant BCTR 0503526 (D. Sharma), and NIH grants CA88843 (N.E. Davidson) and CA077337 (P.M. Vertino).
References
- 1.Platet N, Cathiard AM, Gleizes M, Garcia M. Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit Rev Oncol Hematol. 2004;51:55–67. doi: 10.1016/j.critrevonc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston SR, Dowsett M, Smith IE. Towards a molecular basis for tamoxifen resistance in breast cancer. Ann Oncol. 1992;3:503–511. doi: 10.1093/oxfordjournals.annonc.a058251. [DOI] [PubMed] [Google Scholar]
- 4.Roodi N, Bailey LR, Kao WY, et al. Estrogen receptor gene analysis in estrogen receptor-positive and estrogen receptor negative primary breast cancer. J Natl Cancer Inst. 1995;87:446–451. doi: 10.1093/jnci/87.6.446. [DOI] [PubMed] [Google Scholar]
- 5.Ottaviano YL, Issa J-P, Parl FF, Smith HS, Baylin SB, Davidson NE. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 1994;54:2552–2555. [PubMed] [Google Scholar]
- 6.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 7.Spencer TE, Jenster G, Burcin MM, et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 8.Zamir I, Harding HP, Atkins GB, et al. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrzycka KM, Townson SM, Jiang S, Oesterreich S. Estrogen receptor corepressors—a role in human breast cancer. Endocr Relat Cancer. 2003;10:517–536. doi: 10.1677/erc.0.0100517. [DOI] [PubMed] [Google Scholar]
- 10.Shiau AK, Barstad D, Loria PM, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 12.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 13.Horlein AJ, Naar AM, Heinzel T, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 14.Smith CL, Nawaz Z, O’Malley BW. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogens, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–666. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and NCoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon HG, Chan DW, Huang ZQ, et al. Purification and functional characterization of the human NCoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X, Phillips D, Ferguson AT, Nelson WG, Herman JG, Davidson NE. Synergistic activation of functional estrogen receptor (ER)-α by DNA methyl-transferase and histone deacetylase inhibition in human ER-α-negative breast cancer cells. Cancer Res. 2001;61:7025–7029. [PubMed] [Google Scholar]
- 18.Sharma D, Blum J, Yang X, Beaulieu N, Macleod AR, Davidson NE. Release of methyl CpG binding proteins and histone deacetylase 1 from the estrogen receptor α (ER) promoter upon reactivation in ER-negative human breast cancer cells. Mol Endocrinol. 2005;19:1740–1751. doi: 10.1210/me.2004-0011. [DOI] [PubMed] [Google Scholar]
- 19.Sharma D, Fondell JD. Temporal formation of distinct thyroid hormone receptor coactivator complexes in HeLa cells. Mol Endocrinol. 2000;14:2001–2009. doi: 10.1210/mend.14.12.0567. [DOI] [PubMed] [Google Scholar]
- 20.Sharma D, Fondell JD. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci U S A. 2002;99:7934–7939. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheikh MS, Garcia M, Pujol P, Fontana JA, Rochefort H. Why are estrogen-receptor-negative breast cancers more aggressive than the estrogen-receptor-positive breast cancers? Invasion Metastasis. 1995;14:329–336. [PubMed] [Google Scholar]
- 22.Price JE, Polyzos A, Zhang RD, Daniels MD. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res. 1990;50:717–721. [PubMed] [Google Scholar]
- 23.Mandlekar S, Kong ANT. Mechanisms of tamoxifen-induced apoptosis. Apoptosis. 2001;6:469–477. doi: 10.1023/a:1012437607881. [DOI] [PubMed] [Google Scholar]
- 24.Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 25.Li F, Srinivasan A, Wang Y, Armstrong RC, Tomaselli KJ, Fritz LC. Cell-specific induction of apoptosis by microinjection of cytochrome c. Bcl-xL has activity independent of cytochrome c release. J Biol Chem. 1997;272:30299–30305. doi: 10.1074/jbc.272.48.30299. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchiya F, Ikeda K, Tsutsumi O, et al. Molecular cloning and characterization of mouse EBAG9, homolog of a human cancer associated surface antigen: expression and regulation by estrogen. Biochem Biophys Res Commun. 2001;284:2–10. doi: 10.1006/bbrc.2001.4892. [DOI] [PubMed] [Google Scholar]
- 27.Augereau P, Miralles F, Cavailles V, Gaudelet C, Parker M, Rochefort H. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Mol Endocrinol. 1994;51:693–703. doi: 10.1210/mend.8.6.7935485. [DOI] [PubMed] [Google Scholar]
- 28.Dubik D, Shiu RP. Mechanism of estrogen activation of c-myc oncogene expression. Oncogene. 1992;7:1587–1594. [PubMed] [Google Scholar]
- 29.Umayahara Y, Kawamori R, Watada H, et al. Estrogen regulation of the insulin-like growth factor I gene transcription involves an AP-1 enhancer. J Biol Chem. 1994;269:16433–16442. [PubMed] [Google Scholar]
- 30.Li J, Lin Q, Wang W, Wade P, Wong J. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 2002;16:687–692. doi: 10.1101/gad.962502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 32.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylase (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vigushin DM, Ali S, Pace PE, et al. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin Cancer Res. 2001;7:971–976. [PubMed] [Google Scholar]
- 34.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 35.Primeau M, Gagnon J, Momparler RL. Synergistic antineoplastic action of DNA methylation inhibitor 5-aza-2′-deoxycytidine and histone deacetylase inhibitor depsipeptide on human breast carcinoma cells. Int J Cancer. 2003;103:177–184. doi: 10.1002/ijc.10789. [DOI] [PubMed] [Google Scholar]
- 36.Platet N, Cunat S, Chalbos D, Rochefort H, Garcia M. Unliganded and liganded estrogen receptors protect against cancer invasion via different mechanisms. Mol Endocrinol. 2000;14:999–1009. doi: 10.1210/mend.14.7.0492. [DOI] [PubMed] [Google Scholar]
- 37.Garcia M, Derocq D, Freiss G, Rochefort H. Activation of estrogen receptor transfected into a receptor-negative breast cancer cell line decreases the metastatic and invasive potential of the cells. Proc Natl Acad Sci U S A. 1992;89:11538–11542. doi: 10.1073/pnas.89.23.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang SY, Jordon VC. Growth regulation of estrogen receptor-negative breast cancer cells transfected with complementary DNAs for estrogen receptor. J Natl Cancer Inst. 1992;84:580–591. doi: 10.1093/jnci/84.8.580. [DOI] [PubMed] [Google Scholar]
- 39.Furr BJA, Jordan VC. The pharmacology and clinical uses of tamoxifen. Pharmacol Ther. 1984;25:127–205. doi: 10.1016/0163-7258(84)90043-3. [DOI] [PubMed] [Google Scholar]
- 40.Chen HM, Tritton TR, Kenny N, Absher M, Chiu JF. Tamoxifen induces TGF β1 activity and apoptosis of human MCF-7 breast cancer cells in vitro. J Cell Biochem. 1996;61:9–17. doi: 10.1002/(SICI)1097-4644(19960401)61:1%3C9::AID-JCB2%3E3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 41.Osbourne CK, Boldt DH, Clark GM, Trent JM. Effects of tamoxifen on human breast cancer cell cycle kinetics: accumulation of cells in early G1 phase. Cancer Res. 1983;43:3583–3585. [PubMed] [Google Scholar]
- 42.Zhang GJ, Kimijima I, Onda M. Tamoxifen-induced apoptosis in breast cancer cells relates to down-regulation of bcl-2, but not bax and bcl-X(L), without alteration of p53 protein levels. Clin Cancer Res. 1999;5:2971–2977. [PubMed] [Google Scholar]
- 43.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey GW, Wang Y, Russanova VR, et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- 46.Mazumdar A, Wang RA, Mishra SK, et al. Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat Cell Biol. 2001;3:30–37. doi: 10.1038/35050532. [DOI] [PubMed] [Google Scholar]
- 47.Rea S, Eisenhaber F, O’Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyl-transferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 48.Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromodomain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 49.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 50.Zegerman P, Canas B, Pappin D, Kouzarides T. Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex. J Biol Chem. 2002;277:11621–11624. doi: 10.1074/jbc.C200045200. [DOI] [PubMed] [Google Scholar]