Abstract
The Krüppel-like transcription factors are zinc finger proteins that activate and suppress target gene transcription. Although KLF factors have been implicated in regulating many developmental processes, a comprehensive gene expression analysis has not been reported. Here we present the chicken KLF gene family and expression during the first five days of embryonic development. Fourteen chicken KLF genes or expressed sequences have been previously identified. Through synteny analysis and cDNA mapping we have identified the KLF9 gene and determined that the gene presently named KLF1 is the true ortholog of KLF17 in other species. In situ hybridization expression analyses show that in general KLFs are broadly expressed in multiple cell and tissue types. Expression of KLFs 3, 7, 8, and 9, is widespread at all stages examined. KLFs 2, 4, 5, 6, 10, 11, 15 and 17 show more restricted patterns that suggest multiple functions during early stages of embryonic development.
Keywords: Chicken Embryo, In Situ Hybridization, Krüppel-like factor, KLF
Introduction
The Krüppel-Like Factor (KLF) family of genes codes for a subset of the zinc finger transcription factors. Seventeen mammalian homologs (KLF1-17) of Drosophila Krüppel have been identified (Suske et al., 2005; Pearson et al., 2008). KLFs are identified by the presence of a triplet of Cys2His2 zinc-finger DNA binding domains coded for near the C-terminus of each protein, and a conserved amino acid sequence (TGEKPY/FX) between the zinc fingers (Dang et al., 2002; Haldar et al., 2007). The KLFs show relatively little homology outside of these domains. In Drosophila, the eponymous Krüppel is a gap class segmentation gene that codes for a protein with five zinc fingers with the conserved sequence HTGEKP between the last His of one finger and the first Cys of the next (Nusslein-Volhard and Wieschaus, 1980; Jackle et al., 1985). Vertebrate homologs of Krüppel include the KLFs which typically contain three zinc fingers, the related SP1 and SP6 genes containing six and three zinc fingers, respectively, and the Gli/Glis-family which contain five zinc fingers (Kaczynski et al., 2003).
KLFs function as both transcriptional activators and repressors (Dang et al., 2002; Kaczynski et al., 2003; Ghaleb et al., 2005; Suzuki et al., 2005; Fisch et al., 2007; Haldar et al., 2007; Nemer and Horb, 2007; Pearson et al., 2008). KLFs have been implicated in numerous developmental processes, including maintenance of pluripotency, cell proliferation, erythrogenesis, skeletal muscle development, cardiovascular development, neurogenesis, skin and bone development, and adipogenesis (Groenendijk et al., 2004; Suzuki et al., 2005; Takahashi and Yamanaka, 2006; Haldar et al., 2007). Several recent reviews have highlighted the general functions of individual KLFs (Kaczynski et al., 2003; Suske et al., 2005; Pearson et al., 2008), however a comprehensive embryonic expression analysis has not been reported.
Here we re-examine the chicken KLF gene family with regard to genomic location and identity. We identify the chicken KLF9 gene, and show that the chicken gene presently named KLF1 is the true ortholog of KLF17. We also present a comprehensive in situ hybridization expression analysis of the fifteen known chicken KLF genes during the first five days of embryo development.
RESULTS AND DISCUSSION
The KLF gene family in chicken
The KLF gene family encodes transcriptional regulatory proteins that are defined by the number and organization of the zinc finger motifs and a conserved amino acid sequence between them. The closely related Gli/Glis gene family and the SP1 and SP6 genes also code for proteins with these motifs, though with lower sequence conservation. While these proteins may function similarly to the KLF family members, for this study we limit our focus to chicken orthologs of the canonical human KLF gene family (Pearson et al., 2008).
Of the seventeen KLF family members that have been identified in vertebrate genomes, fourteen have thus far been identified in chicken. An earlier study conducted prior to sequencing of the chicken genome identified nine chicken KLF family members through analysis of EST sequences (Basu et al., 2004). Annotation of the assembled chicken genome identified five additional genes. Chicken orthologs for KLF 14 and 16 are missing from the sequence assembly and remain unidentified. Genes flanking their orthologs in other species are also missing from the chicken genome, raising the possibility that chicken orthologs to KLFs 14 and 16 exist but have not yet been identified due to gaps and errors in the present chicken genome assembly. Expressed sequences corresponding to KLF9 have been identified (Basu et al., 2004), however a corresponding gene model is not present on the genome assembly.
Through analysis of the chicken genome assembly, KLF cDNAs, ESTs, and BAC sequences, we have identified the location and genomic organization of chicken KLF9 and have predicted a full-length protein sequence. Comparison of the genomic locations of KLF9 in human and mouse with the syntenic region of the chicken genome identified the chicken gene LOC770238 (similar to GC box binding protein) on Chromosome Z in the NCBI genome annotation at the predicted location of KLF9 (Fig. 1A). The gene model for LOC770238 was predicted by automated computational analysis and contains two exons (Fig. 1B). Mapping the cDNA of the human KLF9 coding region to the chicken genome showed that the second exon of LOC770238 is virtually identical to the 5' end of the second exon of human KLF9. The first exon of LOC770238 is unrelated to any expressed sequence in chicken or other species.
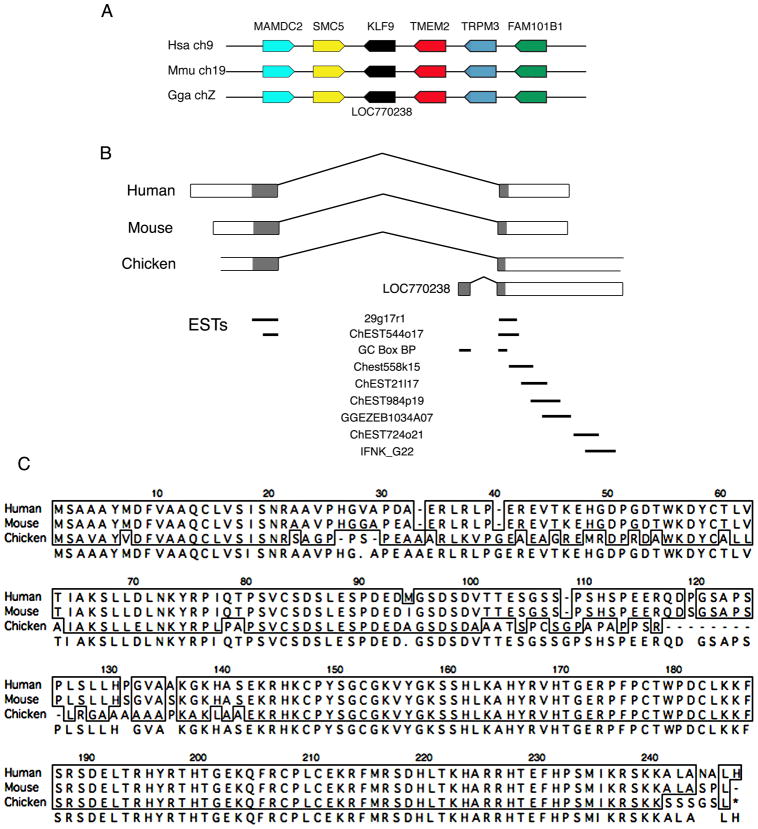
Figure 1.
Identification of the chicken KLF9 gene and predicted amino acid sequence. A) Syntenic regions of the human and mouse genomes containing KLF9, and the corresponding region of the chicken genome. An NCBI gene model is present at the proper location for chicken KLF9 (LOC770238). B) Exon organization of human, mouse and chicken KLF genes. LOC770238 comprises the second exon of KLF9 plus an additional 5’ exon. Mapping of ESTs to the predicted KLF9 exons is shown; 29g17r1 and ChEST544o17 cross the exon-intron boundaries of the KLF9 gene model. C) Predicted amino acid sequence of chicken KLF9 protein compared to the known human and mouse KLF proteins. The chicken protein is 83% identical to human and mouse KLF9.
Exon 1 of human KLF9 showed no similarity to any sequence in the chicken genome, however gaps are present in this region of the assembly. Mapping the cDNA of the human KLF9 coding region to a chicken BAC spanning this genomic region (BAC CH261-48J8) identified a sequence several kilobases 5’ of the two exons of LOC7770238 that was highly similar to exon 1 of the human KLF9 cDNA. Two chicken EST sequences (29g17r1, ChEST544o17) show 100% identity with this upstream exon, and also span the intron separating this upstream exon and the 3' exon of LOC7770238 that shows high percentage identity to the second exon of KLF9 in mouse and human. The EST ChEST544o17 was previously identified as coding for a portion of chicken KLF9 (Basu et al. 2004). Combining the two exons produced a cDNA with a predicted open reading frame showing 83% amino acid identity to human and mouse KLF9 (Figure 1C). Synteny, high nucleotide and amino acid homology, and the existence of multiple cDNA sequences (ESTs) that cross the exon-intron boundary indicate that we have identified the chicken KLF9 gene.
Based upon cDNA and protein homology comparisons, the chicken KLF1 gene name has been assigned to a gene model on Chromosome 8 (Chervenak et al., 2006). However, alignment of this chicken genomic region to the syntenic regions of the human, mouse and Xenopus genomes shows that this chicken KLF gene is the true ortholog of KLF17 in the other species (Fig. 2A). Comparing the KLF1-containing regions of the human and mouse genomes with the chicken genome failed to identify a syntenic region in chicken. Chicken orthologs to genes surrounding human KLF1 are found in unassembled genomic sequence, and so it is possible that a chicken KLF1 ortholog exists but has not yet been identified.
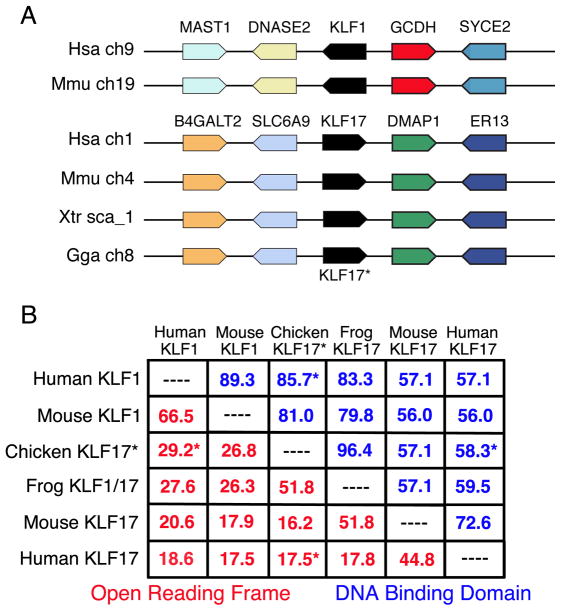
Figure 2.
A) Syntenic regions of the human and mouse genomes containing KLF1, compared with the region of the chicken genome containing the gene presently called KLF1 (shown as KLF17*) and regions of the human, mouse and frog genomes containing the KLF17 gene. Chicken KLF17 is syntenic with KLF17 in other species but not with mammalian KLF1. B) Relatedness between the chicken KLF17 protein versus KLF1 and KLF17 proteins from other species. Chicken KLF17 is 29.2% identical to human KLF1 across the entire protein, but only 17.5% identical to human KLF17 (asterisks, red numbers). The chicken KLF17 DNA binding domain shows 85.7% identity with human KLF1 but only 58.3% identity with human KLF17 (asterisks, blue numbers).
Confusion regarding the assignment of the KLF1 gene name likely arose because the predicted chicken KLF17 protein is more homologous to mammalian KLF1 proteins than to KLF17 proteins. Chicken and human KLF17 proteins are 17.5% identical (58.3% identical within the DNA binding domain), while chicken KLF17 and human KLF1 proteins show 29.2% identity (85.7% identity within DNA binding domain; Figure 2B). Nevertheless, the syntenic analysis demonstrates that the KLF gene on chromosome 8 is the chicken ortholog of KLF17. The cDNA and protein sequences presented in Chervenak et al (2006) match KLF17, and so their situ hybridization (ISH) analyses show expression of KLF17 rather than KLF1.
KLF gene expression analysis
Table I presents official and alternate gene names, NCBI and Ensembl IDs, IDs for the cDNA templates used to prepare antisense RNA probes for the fifteen known chicken KLF genes, and a summary of gene expression patterns. To obtain information about potential regulatory functions of the KLFs, ISH expression analyses were performed in chicken embryos between 0.5 and 5 days of development for all known chicken KLFs. KLFs 3, 7, 8, and 9 were broadly expressed at all stages examined, while probes for KLFs 12 and 13 produced very weak or no detectable hybridization signal at stages 3–24 (not shown). Expression patterns of KLFs showing temporal and spatial restricted expression are described below.
Table I.
| Gene Name | Alternative Names | Entrez GeneID | Ensembl gene ID | cDNA Template* | Expression Pattern |
|---|---|---|---|---|---|
| KLF2 | LKLF | 420148 | ENSGALG00000003939 | pgp1n.pk010.g3 | Endothelial cells, endocardium, limb bud mesenchyme and cartilage |
| KLF3 | BKLF | 429811 | ENSGALG00000000177 | ChEST36p14 | Widespread |
| KLF4 | GKLF | 770254 | ChEST913e4 | Area opaca, posterior streak, neural plate, neural tube, allantois, tailbud, craniofacial, myotome, limb bud mesenchyme and cartilage | |
| KLF5 | BTEB2 | 118084763 | ENSGALG00000016927 | ChEST429a18 | Ectoderm, myotome, branchial arches, pharynx, allantois, amnion. |
| KLF6 | 71896661 | ENSGALG00000007056 | ChEST837d22 | Ectoderm, endoderm, myotome | |
| KLF7 | UKLF | 429011 | ENSGALG00000008501 | pgr1n.UA001.3466 | Widespread |
| KLF8 | BKLF3 | 430697 | ENSGALG00000000373 | pgp1n.pk003.o23 | Widespread |
| KLF9 | BTEB1 | 770238 | ChEST0724o21 | Widespread | |
| KLF10 | TIEG1, EGR-alpha | 420255 | ENSGALG00000016063 | ChEST296k22 | Primitive streak, epiblast, presomitic mesoderm, somites, ectoderm, endoderm |
| 429593 | |||||
| KLF11 | TIEG2, FKLF | 421934 | ENSGALG00000016440 | ChEST927g14 | Area opaca, posterior primitive streak, neural plate, neural tube. Ectoderm, endoderm |
| KLF12 | AP-2rep | 418817 | ENSGALG00000016926 | ChEST388a8 | Not detected |
| KLF13 | BTEB3 | 427493 | ENSGALG00000003809 | ChEST210h10 | Not detected |
| KLF15 | KKLF | 427588 | ENSGALG00000006245 | pgl1n.pk015.j24 | Liver, limb buds |
| KLF17 | 424577 | ENSGALG00000010101 | pgp1n.pk003.f7 | Area opaca, primitive streak, blood islands, anterior neural tube/anterior neural ridge |
EST clone ID used as template for antisense RNA probe synthesis
KLF1
Based upon the synteny analysis discussed above (Fig. 2), we propose that the gene presently known as KLF1 be re named KLF17. KLF17 expression patterns are described below.
KLF2
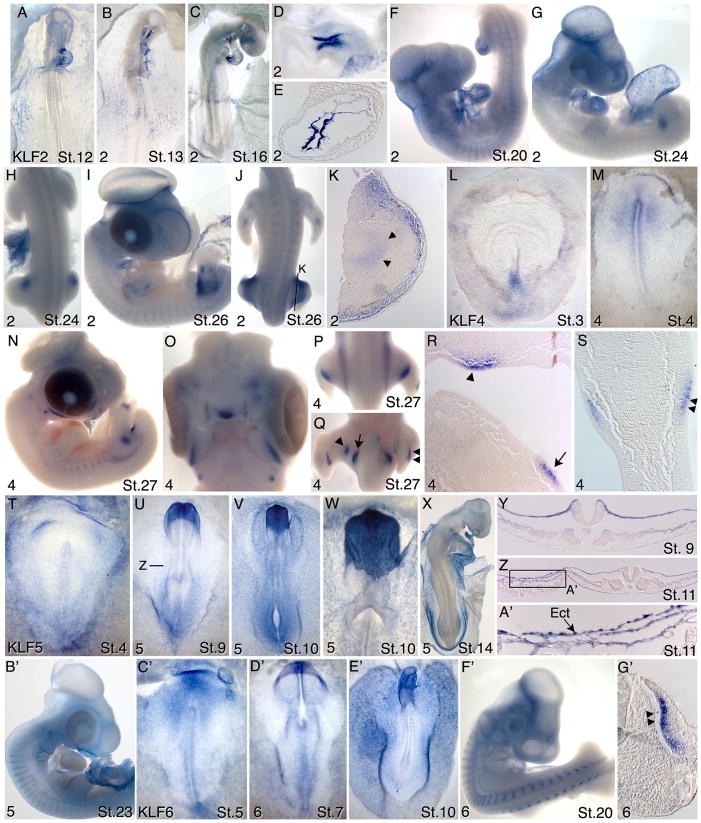
KLF2 mRNAs were not detected prior to Hamburger Hamilton stages 8–9 (Hamburger and Hamilton, 1951; Hamburger and Hamilton, 1992). Beginning at stages 9–10, expression was detected in some endothelial cells within the forming vasculature (not shown). At stages 12–13, expression was detected in endothelial cells of the forming blood vessels and in the endocardium (Figs. 3A,B). Hybridization signal was variable between embryos and in different regions of the vasculature within the same embryo. For example, some embryos showed expression within the endocardium with highest expression in the inflow and outflow tract (Fig. 3A), while another embryo failed to show expression in the endocardium but showed high level expression in the dorsal aortas (Fig 3B). At stage 16, high-level expression was observed in the endocardium of the atrio-ventricular canal, in the outflow tract and variably in endothelial cells throughout the vasculature (Fig. 3C–E). At stage 20, KLF2 transcripts were detected generally throughout the vascular endothelium and in the endocardium (Fig. 3F). Beginning at stage 21 and continuing at least through stage 26, KLF2 transcripts were also detected in the anterior proximal mesoderm of the leg bud (Figs. 3G,H). At stage 26, expression in wing and leg buds was also detected in endothelial cells of the forming vasculature and in the chondrogenic condensations at the sites of bone formation (Figs 3I–K).
Figure 3.
Expression patterns of KLFs 2, 4 and 6 in chicken embryos between stages 1–27. A–K) KLF2; D) higher magnification view of the atrioventricular canal region of the embryo in C); E) section through the atrioventricular canal region of the heart of the embryo in C). H) dorsal view of the embryo in G; J) dorsal view of the embryo in I; K) section through the leg of the embryo shown in J, arrowheads indicate faint labeling of the chondrogenic condensations. L–S) KLF4; O) ventral view of the facial structures of embryo in N); P,Q) dorsal and ventral views of the posterior region of embryo in N; R) section through leg region of embryo in Q) showing the location of label corresponding to the arrowhead and arrow in Q); S) transverse section through the left leg bud in Q), showing the dorsal (double arrowheads) and ventral labeling of mesoderm beneath the ectoderm. T-B’) KLF5; Y) transverse section of embryo in U) showing labeling of non neural ectoderm and lateral mesoderm; Z) transverse section through the posterior region of a stage 11 embryo, showing labeling of non neural ectoderm A’) Magnification of boxed region in Z). C’-G’) KLF6; G’) Transverse section through embryo in F’, showing myotome labeling (double arrowheads). See text for detailed descriptions of these expression patterns.
KLF4
KLF4 expression was detected at stages 2–3 in the caudal primitive streak (Fig. 3L). At stage 4, expression was detected in epiblast anterior to and surrounding the anterior primitive streak (Fig 3M). By stages 8–9, expression was detected in the neural folds and weakly along the neural tube (not shown). At stage 27, KLF4 expression was detected in the face and neck region, in a punctate pattern along the body wall that likely represents the rib primordia, and in the developing limbs (Fig. 3N–S). Labeling in the leg was restricted to two pairs of spots one more distal than the other (Fig. 3Q,R, single and double arrowheads). Each pair was located at the same proximo-distal level in the dorsal and ventral mesoderm immediately beneath the ectoderm (Fig. 3S). A spot also localized to the body wall near the limb (Fig. 3Q,R, arrowheads). These patterns were not observed in the wing bud. KLF4 expression was also detected in the chondrogenic condensations in the wing and leg buds associated with bone formation (not shown).
KLF5
At stage 4, KLF5 transcripts were detected primarily in the epiblast, with highest expression in the lateral and extraembryonic regions (Fig 3T). At stages 8–10, KLF5 was expressed broadly in the non-neural ectoderm, with particularly high expression in the most anterior ectoderm surrounding the head (Figs. 3U–W, Y,). Expression levels between non-neural ectoderm cells varied, resulting in a punctate staining pattern (Fig. 3W). Punctate staining was particularly prominent at the boundary between the embryonic and extraembryonic ectoderm and extending into the extraembryonic region (Fig. 3Z,A’). Ectoderm cells in this region are exceedingly thin and closely associated with a correspondingly thin layer of somatic mesoderm. Moderate expression was also detected in the dorsal aspects of the more rostral somites in the region corresponding to the future dermamyotome, and in the lateral and extraembryonic mesoderm excluding the blood islands and endothelial cells (Fig. 3V,X). At stage 23, KLF5 transcripts were detected in the myotome, the surface ectoderm around the branchial arches and pharynx, and in the amnion (Fig. 3B’). KLF5 expression was also evident in the allantois (not shown).
KLF6
Moderate expression of KLF6 was broadly detected at stages 4–6, with higher expression in the epiblast rostral to the streak (Fig. 3C’). At stage 7, KLF6 transcripts were detected in the epiblast and endoderm, with higher levels rostrally (Fig. 3D’). At stage 10, KLF6 was expressed in the non-neural ectoderm and in a punctate pattern within the endoderm (Fig. 3E’). In rostral regions, expression was detected broadly throughout the non-neural ectoderm, while more caudally expression was confined to the more lateral ectoderm. KLF6 transcripts were also detected in the lateral and extraembryonic mesoderm surrounding blood vessel endothelial cells (not shown). By stage 20, KLF6 transcript levels were much reduced in the ectoderm and endoderm (Fig. 3F’). Beginning at stage 14 and extending at least through stage 25, expression was prominent in the myotome (Fig. 3F’, G’). From stage 20 onward, KLF6 transcripts were also detected in the motor horns of the neural tube (not shown).
KLF10
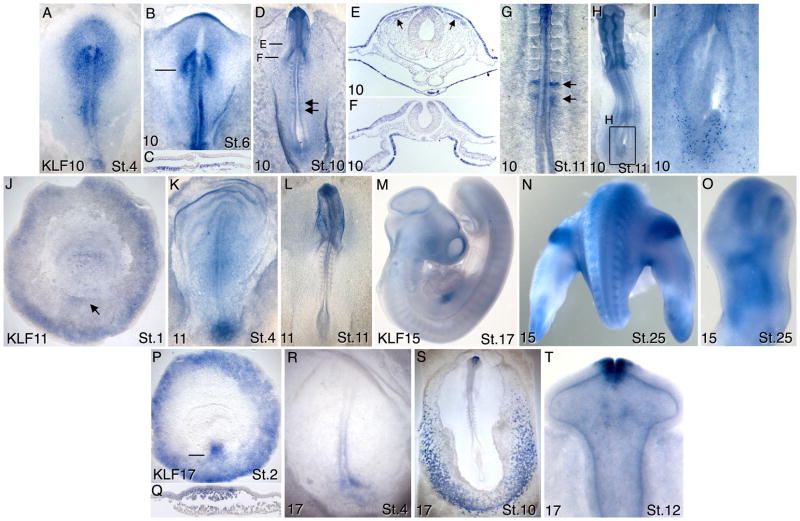
At stage 4, KLF10 expression was evident in the epiblast around the primitive streak, and also in the ectoderm and mesoderm surrounding Hensen’s node (Fig. 4A). By stage 6, KLF10 transcripts were localized to epiblast adjacent to the primitive streak and in the presumptive neural plate except in the neural ectoderm overlying the notochord (Fig. 4B). At this stage KLF10 expression was also prominent in chevrons marking the newly forming somite (Fig. 4B,C). At stage 10 expression was observed in endoderm, ectoderm, dorsal regions of the neural tube (Fig. 4D–G) and in migrating cranial neural crest cells (Fig 4E, arrows). KLF10 expression was also observed in the presomitic mesoderm and first forming somite in a dynamic pattern indicative of genes regulated by the somite segmentation clock (arrows in Fig. 4D,G). KLF10 has been previously identified as a somite segmentation gene (Dequeant et al., 2006). KLF10 transcripts were also observed within individual cells scattered throughout the embryo and concentrated in the caudal endodermal layer (Fig. 4H,I). Moderate general KLF10 expression was observed in stage 14–25 embryos along with continued expression in the presomitic mesoderm associated with the forming somites (not shown).
Figure 4.
Expression patterns of KLFs 10, 11, 15 and 17 in chicken embryos between stages 1–26. A–I) KLF10; E–F) transverse sections through embryo in D); arrows in D) and G) show expression in the forming somites; arrows in E) show migrating cranial neural crest. I) Magnification of boxed region in H), showing staining of individual endoderm cells. J–L) KLF11; arrow in J) points to Koller’s sickle. M–O) KLF15; P–T) KLF17; Q) transverse section of embryo in P) at the indicated level. See text for detailed descriptions of these expression patterns.
KLF11
At pregastrula stages, KLF11 expression was observed around the periphery of the area opaca and in the vicinity of Koller’s sickle (Fig. 4J). Expression caudal to the primitive streak persisted through stage 4 (Fig. 4K). As neurulation progressed, KLF11 expression became higher in the neural plate and neural tube, with highest expression rostrally (Fig. 4L).
KLF15
KLF15 transcripts were detected at stage 17 in the anterior myotomes and in the liver (Fig. 4M). At stage 25, KLF15 expression was also evident in the dorsal proximal mesoderm of the leg buds (Fig. 4N), and in the chondrogenic condensations of the limb bones (Fig. 4O).
KLF17
The embryonic expression patterns of KLF17 have been reported, although the gene was identified as KLF1 (Chervenak et al., 2006). Early expression of KLF17 was observed around the periphery of the area opaca from pregastrula stages through at least stage 6 (Fig. 4P). At the onset of gastrulation, KLF17 transcripts were detected within epiblast and middle layer cells of the forming primitive streak (Fig. 4P, Q). Posterior streak expression persisted through stage 4 (Fig. 4R). KLF17 expression within hematopoietic cells of the blood islands was detected as early as stage 5, was robust at stage 10 (Fig. 4S), and was evident at least through stage 14. Beginning at stage 8 and persisting at least through stage 14, KLF17 transcripts were also evident in fusing edges of the anterior neural tube and anterior neural ridge (Figs. 4S,T).
Summary
In contrast to numerous other developmentally regulated transcription factor families that show highly restricted temporospatial patterns of expression, in general KLFs are broadly expressed in multiple cell and tissue types. KLFs 3, 7, 8, and 9 are widely expressed at all stages examined, and even KLFs that show more restricted patterns tend to be expressed at varying levels in multiple cell layers and embryo regions. This observation is consistent with expression data for individual KLFs from other organisms (Pearson et al., 2008). Of KLFs showing more restricted expression patterns, KLFs 4, 11 and 17 are co-expressed prior to and during gastrulation in the epiblast around the periphery of the area opaca. While the functional significance of expression in this location is not known, components of the BMP and WNT signaling pathways are also expressed in the peripheral area opaca epiblast. These include BMPs 4 and 7 (Streit et al., 1998), WNTs 5A and 8A (Skromne and Stern, 2001) and CTNNB1 (Schmidt et al., 2004). KLFs 4, 11 and 17 are also expressed at the posterior of the pregastrula embryo in a region that includes Koller’s sickle, which contains cells that will give rise to Hensen’s node and have streak inducing properties (Izpisua-Belmonte et al., 1993; Callebaut et al., 2003). KLF4 is one of four genes first used to generate induced pluripotent stem cells (Takahashi and Yamanaka, 2006), and so it is worth considering that KLF expression within Koller’s sickle and the posterior streak is related to pluripotency.
KLF2 is the only KLF family member found to be expressed in vascular endothelial cells. Comparing expression in embryos processed in parallel, we find that the pattern of KLF2 expression in subsets of endothelial cells varies between embryos (Figs. 3A–G). KLF2 is also expressed in vascular endothelial cells in mouse, zebrafish and Xenopus (Kuo et al., 1997; Oates et al., 2001; Meadows et al., 2009), and its expression is upregulated in response to sheer stress (Dekker et al., 2002; Lee et al., 2006). The variable vascular expression patterns that we observed likely reflect variations in blood flow within the developing vasculature.
The expression analyses conducted in this study identified novel leg-specific expression patterns for KLFs 2, 4 and 15 that have not been previously described. KLF2 expression is observed in the anterior proximal mesoderm of the leg bud, and KLF4 transcripts are detected in several localized spots along the leg bud axis and in the adjacent ventral body wall. KLF15 transcripts are localized to the dorsal and ventral mesoderm of the proximal leg bud. We are not aware of similar expression patterns in the developing leg buds, thus this suggests previously unrecognized functions for KLFs during limb development. KLF2 transcripts are also observed in the developing limb vasculature, and all three KLFs are expressed in the chondrogenic cells of bone forming regions of both the wing and leg buds.
Although availability of the assembled chicken genome has greatly aided in gene identification and nomenclature, many genes remain unidentified or misnamed. Many gaps are present in the genome assembly, and approximately 5% of sequence remains unassembled. The KLF9 gene is located within an incomplete portion of the chicken genome. An NCBI gene model was generated based upon a cDNA corresponding to the 3’ portion KLF9 (LOC770238), however this was not recognized as KLF9 because the genomic region corresponding to exon 1 of this gene was not present in the assembly. Through analysis of a chicken BAC sequence, exon 1 was identified and a gene model was generated that closely resembles the KLF9 gene in other species. The predicted protein sequence also is highly similar to mammalian KLF9.
The chicken KLF17 gene was identified through cDNA mapping and synteny analysis while we were confirming the location of all KLF genes. The chicken gene presently called KLF1 is syntenic to KLF17 in other species, confirming its identity as KLF17. Interestingly however, chicken KLF17 is expressed in a pattern similar to KLF1 in mice (Miller and Bieker, 1993; Chervenak et al., 2006), and the chicken KLF17 protein is more homologous to human KLF1 than to human KLF17. These similarities likely led to confusion regarding gene identity. A gene syntenic to mammalian KLF1 is not present in the chicken genome, although genes neighboring the mammalian KLF1 genes are also missing from chicken. Improvement in the chicken genome assembly may ultimately lead to identification of genes coding for KLF1 and KLF14, the two remaining chicken KLF family members that have not yet been identified.
METHODS
Embryo collection and preparation
Fertile chicken eggs (Hy-Line International; not a commercially available source) were incubated in a forced-draft, humidified incubator at 38°5 C for 6–120 hours, depending on the stages desired. Embryos were collected into chilled chick saline (123mM NaCl), removed from the vitelline membrane and cleaned of yolk. Extra-embryonic membranes and large body cavities (brain vesicles, atria, allantois, eye) were opened to minimize trapping of the in situ reagents. Embryos were fixed overnight at 4 C in freshly prepared 4% paraformaldehyde.
Embryos were rinsed in PBS, then in PBS plus 0.1% Tween-20 (PBT), and dehydrated by steps (25, 50, 75, 100, 100%) into methanol before being cooled to −20°C overnight (or up to 10 days). Rehydration reversed this series. Embryos were rinsed 2 × in PBS and older embryos were treated with proteinase K: stages 8–13 and 14–18 at 10 μg/ml of proteinase K for 10 and 20 min., respectively; stages 19 and older at 20 μg/mL of proteinase K for 20 min. Embryos were rinsed repeatedly in PBT to stop the digestion, and were then transferred to prehybridization solution. Embryos were stored until use either at the methanol step or in prehybridization solution at −20°C for fewer than 10 days.
In situ hybridization
EST clones corresponding to each expressed KLF sequence were identified through the NCBI Unigene database (http://www.ncbi.nlm.nih.gov/unigene). ESTs from the BBSRC Chick EST Database (clonesIDs beginning with “ChEST” in Table I; http://www.chick.manchester.ac.uk/) were obtained through the MRC Geneservice. EST clones from the University of Delaware Chick EST Database (most clone IDs beginning with “pg” in Table I; http://www.chickest.udel.edu/) were purchased from the Delaware Biotechnology Institute. The clone for KLF7 was the generous gift of Douglas Rhoads (Univ. of Arkansas). Antisense probe preparation and in situ hybridizations were carried out as described in Nieto et al (Nieto et al., 1996) with minor modifications (Baker and Antin, 2004). Probes were prepared using the entire cDNA insert in each EST clone. Embryos were photographed on a Leica PlanApo stereomicroscope using a digital acquisition system and transmitted, lateral and/or direct illumination. Some embryos were embedded in paraffin, sectioned at 12–14 μm and viewed on a Leica DMRE microscope using DIC and brightfield optics. Additional images of KLF gene expression patterns are accessible on the GEISHA gene expression database (http://geisha.arizona.edu).
Acknowledgments
This work was supported by NIH grant HD044767 to PBA.
Grant Sponsor: NIH; Grant Number HD044767
Abbreviations
- KLF
Krüppel-like factor
- ISH
In situ hybridization
References
- Baker RK, Antin PB. Ephs and Ephrins during early stages of chick embryo development. Devel Dyn. 2004;299:677–687. doi: 10.1002/dvdy.10354. [DOI] [PubMed] [Google Scholar]
- Basu P, Sargent TG, Redmond LC, Aisenberg JC, Kransdorf EP, Wang SZ, Ginder GD, Lloyd JA. Evolutionary conservation of KLF transcription factors and functional conservation of human gamma-globin gene regulation in chicken. Genomics. 2004;84:311–319. doi: 10.1016/j.ygeno.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Callebaut M, Van Nueten E, Bortier H, Harrisson F. Positional information by Rauber's sickle and a new look at the mechanisms of primitive streak initiation in avian blastoderms. J Morphol. 2003;255:315–327. doi: 10.1002/jmor.10065. [DOI] [PubMed] [Google Scholar]
- Chervenak AP, Basu P, Shin M, Redmond LC, Sheng GJ, Lloyd JA. Identification, characterization, and expression pattern of the chicken EKLF gene. Developmental Dynamics. 2006;235:1933–1940. doi: 10.1002/dvdy.20829. [DOI] [PubMed] [Google Scholar]
- Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 2002;30:2736–2741. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJ. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- Dequeant ML, Glynn E, Gaudenz K, Wahl M, Chen J, Mushegian A, Pourquie O. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
- Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, Das H, Petersen CA, Mende U, Burleigh BA, Zhu Y, Pinto YM, Liao R, Jain MK. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A. 2007;104:7074–7079. doi: 10.1073/pnas.0701981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenendijk BC, Hierck BP, Gittenberger-De Groot AC, Poelmann RE. Development-related changes in the expression of shear stress responsive genes KLF-2, ET-1, and NOS-3 in the developing cardiovascular system of chicken embryos. Dev Dyn. 2004;230:57–68. doi: 10.1002/dvdy.20029. [DOI] [PubMed] [Google Scholar]
- Haldar SM, Ibrahim OA, Jain MK. Kruppel-like Factors (KLFs) in muscle biology. J Mol Cell Cardiol. 2007;43:1–10. doi: 10.1016/j.yjmcc.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A Series Of Normal Stages In The Development Of The Chick-Embryo, (Reprinted From Journal Of Morphology, Vol 88, 1951) Devel Dyn. 1992;195:231. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Izpisua-Belmonte JC, De Robertis EM, Storey KG, Stern CD. The homeobox gene goosecoid and the origin of organizer cells in the early chick blastoderm. Cell. 1993;74:645–659. doi: 10.1016/0092-8674(93)90512-o. [DOI] [PubMed] [Google Scholar]
- Jackle H, Rosenberg UB, Preiss A, Seifert E, Knipple DC, Kienlin A, Lehmann R. Molecular analysis of Kruppel, a segmentation gene of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1985;50:465–473. doi: 10.1101/sqb.1985.050.01.058. [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Yu Q, Shin JT, Sebzda E, Bertozzi C, Chen M, Mericko P, Stadtfeld M, Zhou D, Cheng L, Graf T, MacRae CA, Lepore JJ, Lo CW, Kahn ML. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11:845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Meadows SM, Salanga MC, Krieg PA. Kruppel-like factor 2 cooperates with the ETS family protein ERG to activate Flk1 expression during vascular development. Development. 2009;136:1115–1125. doi: 10.1242/dev.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemer M, Horb ME. The KLF family of transcriptional regulators in cardiomyocyte proliferation and differentiation. Cell Cycle. 2007;6:117–121. doi: 10.4161/cc.6.2.3718. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Patel K, Wilkinson DG. Methods in Cell Biology. New York: Academic Press, Inc; 1996. In situ hybridization analysis of chick embryos in whole mount and tissue sections. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Oates AC, Pratt SJ, Vail B, Yan Y, Ho RK, Johnson SL, Postlethwait JH, Zon LI. The zebrafish klf gene family. Blood. 2001;98:1792–1801. doi: 10.1182/blood.v98.6.1792. [DOI] [PubMed] [Google Scholar]
- Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Kruppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Patterson M, Farrell E, Munsterberg A. Dynamic expression of Lef/Tcf family members and beta-catenin during chick gastrulation, neurulation, and early limb development. Dev Dyn. 2004;229:703–707. doi: 10.1002/dvdy.20010. [DOI] [PubMed] [Google Scholar]
- Skromne I, Stern CD. Interactions between Wnt and Vg1 signalling pathways initiate primitive streak formation in the chick embryo. Development. 2001;128:2915–2927. doi: 10.1242/dev.128.15.2915. [DOI] [PubMed] [Google Scholar]
- Streit A, Lee KJ, Woo I, Roberts C, Jessell TM, Stern CD. Chordin regulates primitive streak development and the stability of induced neural cells, but is not sufficient for neural induction in the chick embryo. Development. 1998;125:507–519. doi: 10.1242/dev.125.3.507. [DOI] [PubMed] [Google Scholar]
- Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Aizawa K, Matsumura T, Nagai R. Vascular implications of the Kruppel-like family of transcription factors. Arterioscler Thromb Vasc Biol. 2005;25:1135–1141. doi: 10.1161/01.ATV.0000165656.65359.23. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]