Abstract
Protein kinase D1 (PKD1), founding member of PKD protein family, is down-regulated in advanced prostate cancer (PCa). We demonstrate that PKD1 and androgen receptor (AR) are present as a protein complex in PCa cells. PKD1 is associated with a transcriptional complex which contains AR and promoter sequence of the Prostate Specific Antigen (PSA) gene. Ectopic expression of wild type PKD1 and the kinase dead mutant PKD1 (K628W) attenuated the ligand-dependent transcriptional activation of AR in prostate cancer cells and yeast cells indicating that PKD1 can affect AR transcription activity, whereas knocking down PKD1 enhanced the ligand-dependent transcriptional activation of AR. Co-expression of kinase dead mutant with AR significantly inhibited androgen-mediated cell proliferation in both LNCaP and DU145 PC cells. Our data demonstrate for the first time that PKD1 can influence AR function in PCa cells.
Keywords: Protein kinase D1, Androgen receptor, Interaction, Prostate cancer
Protein kinase D1 (PKD1), a receptor for diacylglycerol (DAG) is a family of serine/threonine protein kinases [1]. The enzyme is comprised of four structural domains: the catalytic domains, C1a and C1b at the N-terminus followed by the central pleckstrin homology (PH) domain whereas the kinase domain is located at the C-terminus [2]. PKD1 has been implicated in many cellular processes such as apoptosis, immune regulation, cell proliferation, oxidative stress signaling, adhesion, and motility [2,3]. Of particular interest is that PKD1 is down-regulated in advanced prostate cancer human specimens and influences cell adhesion and motility of prostate cancer cells in vitro [3,4]. We have demonstrated that PKD1 interacts with E-cadherin and over expression of PKD1 results in increased cellular aggregation, decreased cellular motility and promoting antiproliferative effects in prostate cancer cells [5,6]. In contrast, PKD1 expression was elevated in hyperproliferative human skin disorders and basal cell carcinoma [7]. The plurality and seemingly contrasting role of PKD1 in cells may be related to its ability to interact with diverse array of proteins which may be cell type specific. A protein that is implicated in prostate development and cancer progression is the androgen receptor (AR). Because our charter studies discovered that PKD1 is down-regulated in advanced prostate cancer human samples, we explored whether PKD1 interacted with AR and if so, the functional implications of PKD1–AR cross talk in prostate cancer cells.
Androgens play a pivotal role in prostate cancer progression and its action is mediated by androgen receptors, which belong to the nuclear receptor superfamily [8,9]. Androgen receptors do not act independently to mediate androgen action in target tissues, but requires interaction with other cell-specific or ubiquitous cofactors. There are many AR interacting proteins including coactivators, corepressors, and growth factors which can modulate AR activity in prostate cancer cells [10,11]. In this report, we provide experimental evidence that PKD1 directly interacts with AR and PKD1 can influence AR function in prostate cancer cells. Because both PKD1 and AR expression are dysregulated in prostate cancer, the potential for PKD1–AR interaction as an attractive target for biomarker evaluation or therapeutic intervention merits further study.
Materials and methods
Cell lines and reagents
The growth conditions of the prostate cancer cells and yeast strains have been described previously [5,12], Transformation of yeast cells were performed according to manufacturer's protocol (Clontech, CA). 5α-Dihydrotestosterone (5αDHT; Sigma, St. Louis, MO) or Casodex (Astra Zeneca Westborough, MA) was dissolved in DMSO to make a 10 μM stock. Anti-AR and anti-PKD1(Santa Cruz Biotechnology, CA) primary antibodies were used for Western blot analysis and fluorescein isothiocynate, or rhodamine R X conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA) were used for immunofluorescence study.
Tissue specimens
Archived prostate cancer samples were procured from the Pathology Department of Sioux Valley Hospital, now Sanford Health. The utilization of archived cancer samples used in this study was approved by the University of South Dakota Institutional Review Board (USD-IRB).
Plasmids
The four functional domains of PKD1 containing C1a, C1b, PH (pleckstrin homology), and kinase domains [2,3] were cloned separately into the GADT7 vectors as Gal4 activation domain fusion proteins as described previously [3]. To construct the fusion protein with the kinase dead mutant (K628 W), the last 360 amino acids were amplified by PCR using full length PKD1-kinase dead mutant as a template and the following primers: 5′-CA TATGGGAACCAACTTGCACAGAGATATCTCT-3′ (forward), 5′-GGAT CCTCAGAGGATCGTGACACGCTCA-3′ (reverse). The resulting PCR fragments containing unique NdeI and BamHI restriction sites at the 5′ and 3′ ends respectively were cloned into the GADT7 expression plasmid. The full length AR was generated by PCR with unique 5′ BamHI and 3′ SalI restriction sites and cloned into the GBKT7 vector. The yeast expression and reporter plasmids for androgen receptor (AR) have been described previously [12]. To construct the yeast expression plasmids for PKD1-kinase or PKD1(K628W) for yeast transcription assays, the RARβ gene of YEpcRARβ vector [13] was replaced by PCR-mediated PKD1-kinase domains.
Co-immunoprecipitation and immunoblotting
LNCaP cells seeded in 10 cm dishes were transfected with AR and PKD1 expression plasmids as previously described. After 48 h, total cell lysate was prepared in Triton Lysis Buffer (Sigma) for each dish of cells. Aliquots (20 μl) of cell lysate were taken as inputs (5%) for PKD1 and AR in the co-IP experiment. The remaining cell lysate was incubated with 1 μg of rabbit IgG (control), 1 μg of PKD1 or 1 μg of AR antibodies and subsequently with 40 μl protein A Sepharose beads. After 3–4 h of incubation at 4 °C, the protein complex bound to the beads was washed three times with lysate buffer and the beads were resuspended in 25 μl sample buffer and subjected to SDS–PAGE. Western blot analysis was performed as described previously [5,6] using anti-AR or anti-PKD1 as primary antibodies.
Yeast two hybrid assay
The yeast two hybrid plasmids, GBKT7 fusion with PKD1-kinase, PKD1 (K628W), PH, C1a, or C1b and GADT7 fusion with AR were used to transform AH109 yeast strain according to manufacturer's protocol (Clontech, CA). These yeast transformants were grown in SD medium (-Leu-Trp-His) to score for protein–protein interaction.
Mammalian cell transfection assays
DU145 or LNCaP cells at a density of 1.5 × 104 ml−1 were seeded in 24-well plates, cells were allowed to grow for 24 h until they reached a confluence of 60–70%, vectors expressing AR (50 ng), GRE-luciferase (100 ng), PKD1 (50 ng), PKD-K628W (50 ng) or PKD1 shRNA (50 ng) were transfected into the cells using Lipofectamine Plus (Invitrogen). Concurrently, the plasmid, pRL-CMV (Renilla luciferase) (50 ng) was cotransfected into all cells as an internal control for transfection efficiency. For shRNA targeted to PKD1 Oligos: 5′-GATCCGGAAGGAA ATATCTCATGATTCAAGAGA TCATGAGATATT TCCTTCC TTTTT A and 5′-AGCTT AAAAAGGAAG GAAATATCTC ATGATCTCTTGAATC ATGAGATATTTCCTTCC G were annealed and ligated to KSU6 vector that had been digested with BamHI and HindIII [14]. The amount of transfected DNA for each well was normalized to 300 ng. After 24 h, cells were incubated with medium containing charcoal stripped serum with or without 5α-dihydrotestosterone (10 nM) and Luciferase or Renilla activities were determined after another 24 h using Dual-Glo™ Luciferase Assay (Promega, Madison WI).
Yeast transcription assays
The yeast strain carrying the androgen receptor expression and the GRE-luciferase reporter plasmids [12] was transformed with the yeast expression plasmids for PKD1-kinase (YEp-PKD1-kinase) or PKD1-kinase dead mutant, YEp-PKD1(K628W). To induce the expression of PKD1 proteins, cupric sulfate (1 μM) was added to the desired culture [13] when the cell density reached late log phase for 6 h. A ligand-dependent transcription assay was performed using the Beta-Glo Assay System (Promega, WI) as described previously [15].
Cell proliferation assays
In order to determine the effects of PKD1 and/or AR expression on prostate cancer cell growth, MTS growth and viability assays were used as described previously [15]. DU145 or LNCaP cells were plated at 4 × 103/well in 200 μl DMEM medium in 96-well plates and transfected with different combinations of PKD1, PKD1 shRNA or AR expression plasmids. After 24 h, medium was removed and replaced with fresh medium containing 10 nM of DHT. Following 72 h incubation, medium was aspirated and the cells were washed once with 200 μl of Hanks balanced salt solution prior to MTS assays.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation assays were performed according to the instruction manual of ChIP-IT™ from Active Motif (Carlsbad, CA). The primary antibodies (2 μg each) used to bind protein/DNA complexes were IgG, androgen receptor and PKCμ (PKD1) (Santa Cruz, CA). Eluted DNA was purified and used as templates for PCR analysis. The two primers to amplify the 100 bp fragment of the PSA promoter were: 5′-CCCTCCCCTTCCACAGCTCTGGGT-3′ and 5′-CCGCCCCTG CCCTGC TGGCACCC-3′.
Immunofluorescence microscopy
AR/PKD1-transfected DU145 cells were cultured on glass cover slips (40,000 cells/well) until subconfluence and processed for immunofluorescence study. The coverslips were incubated with primary antibodies in 10% goat serum in PBS. After washing with PBS, coverslips were incubated with fluorescein isothiocynate, or rhodamine R X conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA) for 1 h. The cells were washed in PBS and mounted on slides with SlowFade Antifade reagent (with DAPI) by Molecular Probes.aqueous antifade medium followed by analysis with a Olumpus Flow view 1000 confocal laser scanning microscope and Imaris software was used for image analysis.
Results
Protein kinase D1 (PKD1) interacts with the human androgen receptor and forms a complex in mammalian cells
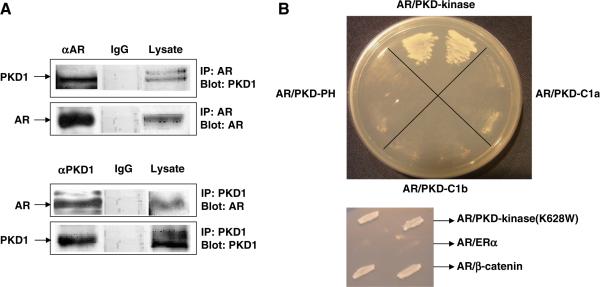
Co-immunoprecipitation assays were performed using whole cell lysate prepared from LNCaP cells transfected with AR and PKD1 expression plasmids. Protein complexes immunoprecipitated by AR antibody indicated the presence of full length PKD1, which was also present in the whole cell lysate (Fig. 1A top). The specificity PKD1–AR interaction in vivo in PCa cells was demonstrated by the absence of PKD1 protein when αAR was replaced by rabbit IgG. When the blot was stripped and reprobed with AR antibody, full length AR was present only in crude lysate and the sample immunoprecipitated by αAR. In the reciprocal co-immunoprecipitation experiment, the full length AR was detected when the lysate was immunoprecipitated by αPKD1 but not rabbit IgG. Full length PKD1 was also detected in cell lysate and the sample immunoprecipitated by αPKD1 (Fig. 1A bottom). We subsequently carried out PKD1 domain specific AR interaction studies using four main structural PKD1 domains (C1a, C1b, PH, and kinase domains) in the yeast two hybrid assay. As shown in Fig. 1B, only the kinase domain of PKD1 was able to interact with the full length AR. Interestingly, the mutated PKD1-kinase domain (K628W), with attenuated kinase activity also interacted with the AR, demonstrating that total PKD1-kinase activity may not be necessary for interaction with AR. We used known AR coactivator β-catenin and estrogen receptor α as positive and negative controls, respectively (Fig. 1B).
Fig. 1.
(A) PKD–AR protein complex detected by co-immunoprecipitation assays. LNCaP cells were transfected with AR and PKD1 expression plasmids. Cell lysate was prepared for co-IP experiments as described in materials and method. The amount of input protein in these experiments was 5% of those in the total lysates. (B) PKD–AR interaction in yeast two hybrid assays. The four functional domains of protein kinase D1 (PKD1), i.e. C1a, C1b, PH, and kinase domains were fused to Gal4 activation domain plasmid (GADT7) whereas the androgen receptor (AR) was fused to the Gal4 binding domain plasmid (GBKT7). Positive protein–protein interaction is indicated by the growth of yeast cells.
Functional association of PKD1 with AR in the promoter region of androgen responsive prostate specific antigen (PSA) gene (ChIP assays)
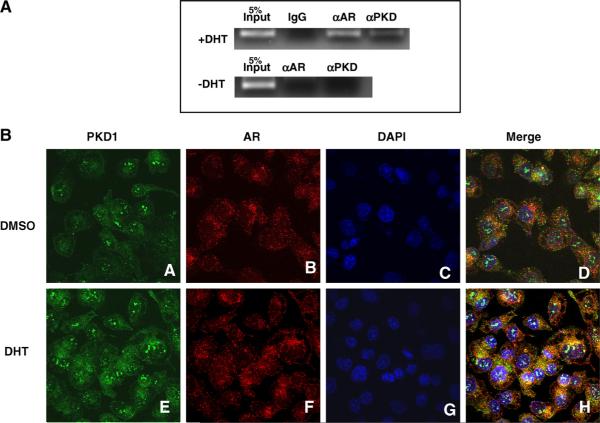
In the chromatin immunoprecipitation (ChIP) assays, IgG and αAR, were used as negative and positive controls, respectively. In the presence of androgen (+DHT), a distinct PCR fragment with the expected size of 100 bp in the PSA promoter was detected when αAR was used to immunoprecipitate the protein/DNA complex (Fig. 2A). A similar size of PCR fragment was also apparent when αPKD1 was used in the assay indicating that PKD1 is functionally associated with AR in the promoter region of the PSA gene. In contrast, this 100 bp PCR fragment could not be detected when the cells were not incubated with DHT, confirming that PKD1 association with PSA promoter region is androgen responsive.
Fig. 2.
(A) Chromatin immunoprecipitation assays. Cells were fixed with 37% formaldehyde and lysed; chromatin was sheared using optimized conditions according to the protocol of Active Motif ChIP-IT kit. The cell extracts were incubated with 1 μg of IgG, antibodies (1 μg each) for AR or PKD1. Eluted DNA was purified and used as templates for PCR analysis. Input DNA (5%) was served as a control with the expected size of the PCR fragment (100 bp). (B) Localization of PKD1 and AR in prostate cancer cells. DU145 cells were transfected with PKD1 or AR expression plasmids and were grown in charcoal stripped media in the absence or presence of DHT (10 nM). Confocal Microscopy of DU145 shows the localization of PKD1 (A), AR (B), and the nucleus (DAPI) (C), and a merge of the three (D) in DMSO treated cells. In DHT treated DU145 cells, PKD1 (E) is located in both the cytoplasm and the nucleus. AR (F) is located primarily in the nucleus. The merge of images (H) is indicated by whitish blue color in the nucleus. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Localization of PKD1 and AR in PCa cells
Confocal immunofluorescence microscopy experiments (Fig. 2B) demonstrate cytoplasmic and nuclear localization of PKD1 (A), AR (B), DAPI (C), and merge of these images (D) in DMSO treated DU145 cells in the same confocal plane. When DU145 cells were treated with DHT (10 nM), we observed cytoplasmic and nuclear staining of PKD1 (E: green) whereas AR (F: red) was localized exclusively in the nucleus. Merger of images shown in green (PKD1), red (AR) and blue (nucleus) suggests co-localization of PKD1 and AR in the nucleus (H: white).
PKD1 co-localizes with AR in human prostate cancer tissues
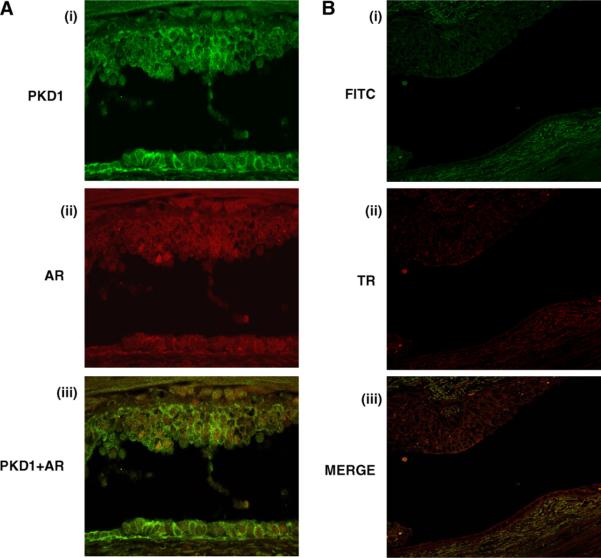
Confocal images of prostate tissue samples stained for PKD1 and AR demonstrated co-localization of PKD1 and AR at cytoplasm and nucleus. PKD1 also showed junctional and nuclear localization (Fig. 3A i) and AR was localized to cytoplasm and nucleus in human prostate cancer tissue (Fig. 3A ii). Merging of images 1 and 2 demonstrated that PKD1 and AR co-localized mostly in nuclei of prostatic glandular epithelial cells (Fig. 3A iii). Specificity of expression was confirmed by using isotype controls (Fig. 3B).
Fig. 3.
Immunostaining was performed on paraffin-embedded prostate tissue sections using antibodies for PKD1 and AR antibodies. (3A) PKD1 localized to perinuclear and cell junctions (i: green) whereas AR localized to nucleus (ii: red). Following merging of green and red channel images, co-localization of PKD1 and AR (iii: yellow) was observed in human prostate glandular epithelium. Specificity of expression was confirmed by using isotype controls (3B). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
PKD1 attenuates AR transcriptional activity in prostate cancer cells
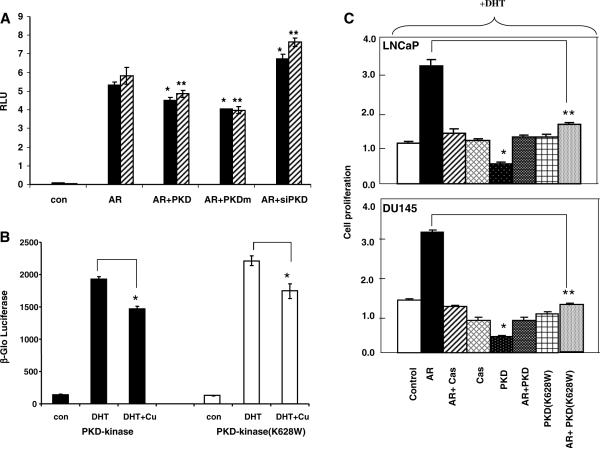
When DU145 or LNCaP cells were transfected with AR and the reporter gene, GRE-luciferase, there was a significant induction (>5-fold) of the reporter gene activity in a ligand-dependent manner (Fig. 4A). This AR transcriptional activity was significantly attenuated with the co-expression of PKD1 or PKD1-kinase dead mutant in both cell lines. Our observation suggests that the inhibition of AR transcription by PKD1 may not be related to kinase activity. As a corollary, down regulation of PKD1 by shRNA could enhance the ligand-dependent AR transcription. The specificity of the shRNA directed against PKD1 has been confirmed previously by comparing with a scrambled sequence of the shRNA [6,13].
Fig. 4.
(A) AR transcription assays. DU145 (Black bars) or LNCaP (Hatched bars) cells at a density of 1.5 × 104 ml−1 were seeded in 24-well plates. Vectors expressing AR (50 ng), GRE-luciferase (100 ng), PKD1 (50 ng), PKD1-kinase dead mutant, PKDm (50 ng) or PKD1 shRNA (50 ng) were transfected with different combinations into the cells using Lipofectamine Plus (Invitrogen). After 24 h, cells were incubated with medium with or without 5α-dihydrotestosterone (10 nM). Luciferase versus Renilla activities were determined as Relative Luciferase Units (RLU) with SEM (n = 3) after another 24 h. (*) Indicates statistical significant as compared to the control (student's t-test: p < 0.05). Similar results were obtained from two separate experiments. (B) Yeast transcription assays. Triple transformant yeast cells harboring the expression plasmids for AR, GRE-luc reporter and PKD-kinase or PKD (K628W), a kinase dead mutant, were grown in SD medium (-Leu-Trp-Ura) in the absence or presence of dihydrotestosterone (DHT, 10 nM). Cupric sulfate (+Cu) was added to desired culture when the cell density reached late log phase to induce the expression of PKD1 protein. After 6 h incubation at 30 °C. Cells were collected and analyzed for β-gal induction as β-Glo luciferase activity with SEM (n = 3). (*) Indicates statistical significant as compared to the control (student's t-test: p < 0.05). Similar results were obtained from two separate experiments. (C) Cell proliferation assays. DU145 or LNCaP cells were plated at 4 × 103/well in 200 μl DMED medium in 96-well plate. Cells were transfected with different combinations of PKD, AR and shPKD expression plasmids. After 24 h, medium was removed and replaced with fresh medium containing 10 nM of DHT. MTS assays were performed after 72 h incubation as described previously [6].(*) Indicates statistical significant as compared to the control (student's t-test: p < 0.05).
PKD1 can directly inhibit AR transcriptional activation
Since PKD1 is known to interact with many other proteins such as β-catenin, E-cadherin and metallothionein IIA [3,4], it is unclear whether PKD1 could directly inhibit AR transcriptional activity in the absence of other adaptor mammalian proteins. Therefore, we examined the effects of PKD1 on AR transcription in yeast cells, which do not have the mammalian factors that interact with PKD. Since the expression of full length PKD1 is lethal in yeast cells (unpublished observation), therefore we circumvent this problem by using an inducible promoter (CUP1) [13] for PKD1 expression so that yeast cells can grow normally in the absence of cupric sulfate induction. As shown in Fig. 4B, while AR transcriptional activity was clearly inducible by treatment with DHT, the effect was significantly attenuated by induction of PKD1(+Cu) with or without total kinase activity. Expression of PKD1 alone with the reporter plasmid had no effect on AR transcription in yeast (data not shown). The data demonstrates that PKD1 can directly repress AR transcription in absence of other mammalian proteins.
PKD1 inhibits AR mediated effect on cell proliferation in PC cells
The effect of PKD1 expression on prostate cancer cell proliferation was analyzed in AR transfected DU145 or LNCaP cells in the absence or presence of PKD1 or PKD1-kinase mutant (K628W) expression plasmids. As shown in Fig. 4C, in the presence of DHT (10 nM), there was a 3-fold increase of cell proliferation in both cell lines. This effect was abolished by incubating with the AR antagonist Casodex (10 μM) indicating that the growth promoting effect of DHT on PC cells is specifically mediated through AR. While expression of PKD1 alone could inhibit cell proliferation, Co-expression of PKD1 or kinase dead PKD1 (K628W) with AR could abolish the ligand-dependent AR mediated growth demonstrating that PKD1 inhibits AR mediated cell proliferation, which may be independent of PKD1-kinase activity.
Discussion
In the present study, we clearly demonstrate that PKD1-kinase domain can be physically associated with AR and forms a protein complex in mammalian cells. More importantly, PKD1 is associated with a transcriptional complex which contains AR and promoter sequence of the PSA gene in a ligand-dependent manner. In resting DU145 cells without DHT stimulation, both PKD1 and AR are present mainly in the cytoplasm. However, our confocal study indicated that these two proteins were localized mainly in the nucleus after DHT stimulation. AR is an important signaling molecule involved in early and late stages of prostate cancer and is regulated by a combination of tissue specific coactivators or corepressors [11]. Our transcription and cell proliferation assays clearly demonstrate that PKD1 could attenuate AR transcription in the two prostate cancer cell lines. Our data demonstrates that PKD1 can directly attenuate AR transcription in yeast cells, which do not have any mammalian factors, further confirming the significance of PKD1-AR novel protein–protein interaction in regulating AR function. This observation also suggests that the binding of PKD1 to AR might affect the formation of a productive transcription initiation complex at the promoter region of target genes.
The exact mechanism of PKD1 mediated AR transcriptional activity remains to be investigated. One potential mechanism is that PKD1 can increase β-catenin expression, which can sensitize AR to DHT. Indeed, we have recently shown that down regulation of PKD1 can increase cell proliferation in prostate cancer cells with a concomitant increase of β-catenin expression [6]. Cell proliferation studies support the notion that AR can promote prostate cancer cell growth in the presence of DHT [16]. The growth inhibitory effect of PKD1 alone on both prostate cancer cells appears to be kinase dependent since the kinase dead mutant has no effect on cell proliferation. This observation provides us an ideal condition to test whether PKD1–AR interaction is an essential pathway to inhibit prostate cancer cell growth. Co-expression of PKD1-kinase dead mutant and AR was able to suppress androgen-mediated cell proliferation further suggesting that this inhibitory effect is due to PKD1–AR interaction and may be independent of kinase activity.
In conclusion, the current study demonstrates that PKD1 is a novel interacting protein of AR in PCa cells. Because both PKD1 and AR are dysregulated in prostate cancer, PKD1–AR interaction provides us a novel signaling pathway worth exploring further to understand prostate cancer progression. Further examination and correlation in the expression of PKD1 and AR in normal and different Gleason grades and stages of PCa tissues can provide novel insights into this newly discovered PKD1–AR signaling pathway in prostate cancer.
References
- [1].Valverde AM, Sinnett-Smith J, Van Lint J, Rozengurt E. Molecular cloning and characterization of protein kinase D: a target for diacylglycerol and phorbol esters with a distinctive catalytic domain. Proc. Natl. Acad. Sci. USA. 1994;91:8572–8576. doi: 10.1073/pnas.91.18.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang QJ. PKD at the crossroads of DAG and PKC signaling. Trends Pharmacol. Sci. 2006;27:317–323. doi: 10.1016/j.tips.2006.04.003. [DOI] [PubMed] [Google Scholar]
- [3].Jaggi M, Du C, Zhang W, Balaji KC. Protein kinase D1: a protein of emerging translational interest. Front. Biosci. 2007;12:3757–3767. doi: 10.2741/2349. [DOI] [PubMed] [Google Scholar]
- [4].Jaggi M, Roa PS, Smith DJ, Hemstreet GP, Balaji KC. Protein kinase C mu is down-regulated in androgen independent prostate cancer. Biochem. Biophys. Res. Commun. 2003;307:254–260. doi: 10.1016/s0006-291x(03)01161-6. [DOI] [PubMed] [Google Scholar]
- [5].Jaggi M, Rao PS, Smith DJ, Wheelock MJ, Johnson KR, Hemstreet GP, Balaji KC. E-cadherin phosphorylation by protein kinase D1/protein kinase C{mu} is associated with altered cellular aggregation and motility in prostate cancer. Cancer Res. 2005;65:483–492. [PubMed] [Google Scholar]
- [6].Syed V, Mak P, Du C, Balaji KC. β-Catenin mediates alterations in cell proliferation, motility and invasion of prostate cancer cells by differential expression of E-cadherin and Protein Kinase D1. J Cell. Biochem. 2008;104:82–95. doi: 10.1002/jcb.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ristich VL, Bowman PH, Dodd ME, Bollag WB. Protein kinase D distribution in normal human epidermis, basal cell carcinoma and psoriasis. Br. J. Dermatol. 2006;154:586–593. doi: 10.1111/j.1365-2133.2005.07073.x. [DOI] [PubMed] [Google Scholar]
- [8].Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gelmann EP. Molecular biology of the androgen receptor. J. Clin. Oncol. 2002;20:3001–3015. doi: 10.1200/JCO.2002.10.018. [DOI] [PubMed] [Google Scholar]
- [10].Agoulnik IU, Vaid A, Bingman WE, III, Erdeme H, Frolov A, Smith CL, Ayala G, Ittmann MM, Weigel NL. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65:7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- [11].Chmelar R, Buchanan G, Need EF, Tiley W, Greenberg NM. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. Int. J. Cancer. 2006;120:719–733. doi: 10.1002/ijc.22365. [DOI] [PubMed] [Google Scholar]
- [12].Mak P, Young CY-F, Tindall DJ. A novel yeast expression system to study androgen action. Recent Prog. Horm. Res. 1994;49:47–352. doi: 10.1016/b978-0-12-571149-4.50023-4. [DOI] [PubMed] [Google Scholar]
- [13].Salerno AJ, He Z, Goos-Nilsson A, Ahola H, Mak P. Differential transcriptional activation of the apoAI gene by retinoic acid receptor homo and heterodimers in yeast. Nucleic Acids Res. 1996;24:566–572. doi: 10.1093/nar/24.4.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Du C, Ge B, Liu Z, Fu K, Chan WC, McKeithan TW. PCR-based generation of shRNA libraries from cDNA. BMC Biotechnol. 2006;6:28. doi: 10.1186/1472-6750-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mak P, Leung YK, Tang WY, Harwood C, Ho SM. Apigenin suppresses cancer cell growth through ERβ. Neoplasia. 2006;8:896–904. doi: 10.1593/neo.06538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xu Y, Chen SY, Ross KN, Balk SP. Androgens induce prostate cancer cell proliferation through mammalian target of Rapamycin activation and posttranscriptional increase in cyclin D proteins. Cancer Res. 2006;66:7783–7792. doi: 10.1158/0008-5472.CAN-05-4472. [DOI] [PubMed] [Google Scholar]