Abstract
Spatial normalization is the process of standardizing images of different subjects into the same anatomical space. The goal of this work was to assess standard and unified methods in SPM5 for the normalization of structural magnetic resonance imaging (MRI) data acquired in mid-life/elderly subjects with diabetes. In this work, we examined the impact of different parameters (i.e. nonlinear frequency cutoff, nonlinear regularization and nonlinear iterations) on the normalization, in terms of the residual variability. Total entropy was used to assess the residual anatomical variability after spatial normalization in a sample of 14 healthy mid-life/elderly control subjects and 24 mid-life/elderly subjects with type 2 diabetes. Spatial normalization was performed using default settings and by varying a single parameter or a combination of parameters. Descriptive statistics and nonparametric tests were used to examine differences in total entropy. Statistical parametric mapping analyses were performed to evaluate the influence of parameter settings on the spatial normalization. Total entropy results and SPM analyses suggest that the best parameters for the spatial normalization of mid-life/elderly image data to the MNI template, when applying the standard approach, correspond to the default cutoff (25 mm), heavy regularization, and the default number of nonlinear iterations (16). On the other hand, when applying the unified approach, the default parameters were the best for spatial normalization of mid-life/elderly image data to the MNI priors. These findings are relevant for studies of structural brain alterations that may occur in normal aging, chronic medical conditions, neuropsychiatric disorders, and neurodegenerative disorders.
Keywords: statistical parametric mapping, spatial normalization, residual variability, total entropy, type 2 diabetes
Introduction
Magnetic resonance (MR) imaging is a technique that is widely used to generate images of brain structure and/or function. One way of analyzing imaging data consists of comparing brain images between subjects on a voxel-wise basis. These comparisons require that all brain structures occupy the same standard anatomical space in a consistent manner. To achieve this, an image-processing step known as spatial normalization is applied. This step is required to ensure that homologous regions in the brain are comparable across subjects before performing subsequent analyses.
Several techniques are available to perform spatial normalization. The most widely used normalization software in the research literature is Statistical Parametric Mapping (SPM). Most researchers choose to use the default parameter settings for spatial normalization in SPM when conducting their studies. An advantage of using the default parameters is that brain images can be compared across different studies. On the other hand, a disadvantage is that the default parameter set might not be the best choice for spatial normalization of images-of-interest to a standard space.
Several studies (e.g. Meyer et al., 1999; Sugiura, et al., 1999; Crivello et al., 2002; Gispert et al., 2003; Hellier et al., 2003; Robbins et al., 2004; Hosaka et al., 2005; Wu et al., 2006, Crinion et al., 2007) have compared and assessed different spatial normalization procedures. In addition, the precision of the spatial normalization in SPM has been assessed in terms of anatomical landmarks (Crinion et al., 2007; Salmond et al., 2002). Nearly all previous studies have focused on healthy and young subjects as opposed to diseased and elderly subjects, with the exception of Salmond et al. (2002) and Gispert et al. (2003), who respectively focused on bilateral hippocampal atrophy patients and schizophrenic patients. Crinion et al. (2007) also examined simulated brain lesions.
Spatial normalization involves applying a spatial transformation that moves and warps images into the same standard anatomical space defined by a template. The objective of normalization is to remove, to some extent, anatomical variability between the subjects’ images to allow subsequent analysis of the data. Spatial normalization is a critical step in the analysis of brain imaging data since it produces the “raw” data for the subsequent analyses. Therefore, it is important to study and understand how spatial normalization in SPM works under different conditions and parameter settings.
The purpose of this comparative study was to assess how changes in parameter settings in SPM affect the performance of spatial normalization, by examining the residual variability of normalized image data across three groups of mid-life/elderly subjects who were healthy controls, non-depressed diabetics and depressed diabetics. Structural brain alterations, such as brain atrophy and changes in ventricle size, were expected to occur to a greater extent for these subjects than for younger subjects. In particular, we were interested in how the nonlinear frequency cutoff, the nonlinear regularization and the number of nonlinear iterations, affect the residual variability of normalized structural MR imaging data. Because of the difficulty of defining a gold standard for parameter settings in SPM, we define the default parameters as the gold standard. In addition, we will compare total entropy results using two approaches: SPM5 standard method and SPM5 unified method.
Materials and Methods
Human Subjects
Fourteen healthy control (Control), sixteen type 2 diabetic (Diabetes) and eight depressed type 2 diabetic (Depressed Diabetes) subjects were recruited for a positron emission tomography imaging study (Price et al., 2002, 2003). The present work focuses only on the structural MR data that were acquired to guide region-of-interest determination and partial volume correction. Only a brief description of the participant recruitment and characteristics will therefore be provided. Table 1 describes the subject characteristics including age and gender. Subjects were recruited through university collaborations that included the University of Pittsburgh Obesity and Nutrition Research Center and Intervention Research Center for Late-life Mood Disorders, as approved by the Biomedical Institutional Review Board. None had a history of substance abuse or dependence. Exclusion criteria included medical or neurological illnesses likely to affect brain physiology or anatomy, suicidal intent, and exposure to psychotropic or other medications. Subjects were excluded if they were currently on antidepressants, taking insulin, or had major medical complications (peripheral vascular disease-CAD, PVD, CVA/TIA; history of stroke), or complex medical regimens. The glycosylated hemoglobin index of diabetes was measured for 10/14 controls (5.6±0.3%) and all diabetics (non-depressed: 7.1±1.2%; depressed: 7.5±1.5%). The Hamilton Depression Inventory averaged 1.6±1.6 for controls to 18.6±4.4 for the depressed diabetics.
Table 1.
Subject Demographics
| Group | Age (years) | Gender(M:F) |
|---|---|---|
|
Control n = 14 |
62.14 ± 13.00 | 6:8 |
|
Diabetes n = 16 |
55.94 ± 9.33 | 9:7 |
|
Depressed Diabetes n = 8 |
62.75 ± 10.61 | 3:5 |
Magnetic Resonance Imaging
Magnetic resonance imaging was performed on a 1.5 Tesla G.E. Signa system. All subjects were positioned in a standard head coil and a brief scout T1-weighted image was obtained. The axial series was acquired (oriented to the anterior and posterior commissures) to screen subjects for unexpected pathology: fast spin-echo T2-weighted (effective TE=102, TR=2500, NEX=1, slice thickness=5mm/1mm interslice) and proton density weighted images (effective TE=17, TR=2000, NEX=1, slice thickness=5 mm/1mm interslice). A field of view of 24 cm and image matrix of 256×192 pixels were used for all axial MR series. A volumetric spoiled gradient recall (SPGR) sequence with parameters optimized for maximal contrast among gray matter, white matter, and CSF was acquired in the coronal plane (TE=5, TR=25, flip angle=40 degrees, NEX=1, slice thickness=1.5 mm/0mm interslice). All MR image data analyzed were skull stripped manually using ANALYZE AV.
Spatial Normalization
SPM5 Standard Method
The standard normalization method in SPM5 minimizes the sum of squared differences between the subject’s image and the template, while maximizing the prior probability of the transformation. This spatial normalization begins by determining the optimum twelve-parameter affine transformation to account for differences in position, orientation and overall brain size. After affine transformation, a nonlinear transformation is applied to correct for gross differences in head shape that were not accounted for by the affine transformation. The nonlinear deformations are described by the lowest frequency components of a three-dimensional discrete cosine transform basis functions (Ashburner and Friston, 1999).
For the standard method, the user has the ability to select parameter estimation settings for the nonlinear transformation. We will focus on three parameter estimation settings: nonlinear frequency cutoff, nonlinear regularization and the number of nonlinear iterations.
Nonlinear Frequency Cutoff represents the cutoff (mm) of the period of the cosine basis functions. SPM will only estimate warps of the order of the specific “cutoff” or larger. Smaller cutoff values represent the use of a larger number of basis functions that result in greater warping along each axis. The default for nonlinear transformation in SPM is 25 mm.
Nonlinear Regularization is the inclusion of a log-likelihood penalty term for unlikely deformations. Greater regularization provides smoother deformations. The smoothness measure is determined by the bending energy of the deformations. The default value for degree of regularization in SPM is medium regularization (1).
Nonlinear Iterations is the number of iterations performed during the parameter estimation process. The default number for nonlinear iterations in SPM is 16.
SPM5 Unified Method
The new SPM5 method combines segmentation, spatial normalization and bias correction in a unified model approach (Ashburner and Friston, 2005). Spatial normalization begins with an affine transformation to achieve an approximate alignment. Then, deformation of the tissue probability maps is performed to achieve a better model fit to the data. Similar to the standard method, the unified model option allows the user to choose different values of the warp frequency cutoff and warping regularization.
Segmentation
The SPM segmentation of structural MR brain images generates probability maps for the following tissue types: gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF). The segmentation method in SPM5 is based upon a Gaussian mixture model, provides an intensity non-uniformity (or bias) correction and implements deformable tissue probability maps (Ashburner and Friston, 2005).
Total Entropy
Total entropy is a measure that can be used to assess the amount of uncertainty associated with a random variable and has been used by others to assess the residual variability remaining after spatial normalization (Warfield et al., 2001; Robbins et al., 2004).
Assume V is a discrete random variable that represents the distribution of tissue types for a given voxel ν. Let p(ν) = Pr{V = ν}, t ∈ T, where ν denotes the voxel, T is the set of possible tissue types that define the probability distribution at voxel ν, and pt (ν) is the probability that voxel ν is of tissue type t (i.e., GM, WM or CSF) based upon the segmentation of the normalized MR images. The entropy of a random variable V (Cover and Thomas, 2006) is defined by
| (1) |
where 0·log2·0 is defined to be zero. The total entropy is thus defined over all voxels as:
| (2) |
For the purpose of this work, the entropy of V is measured in base b units that will correspond to logarithm base 2 (i.e. H2), and the measurement of entropy in bits. Entropy was calculated for each subject’s normalized segmented image in a voxel-wise manner, based upon Equation (1):
| (3) |
According to equation (1), a large amount of uncertainty can reflect more information and hence a greater entropy. Total entropy is zero when a spatial normalization achieves complete matching of homologous regions of a subject’s brain to the template; or when there is a distribution where each label is equally likely. Ideally, one would like to find a combination of parameter settings in SPM that minimizes the total entropy. That is, smaller total entropy is assumed to generally reflect less uncertainty and therefore better spatial normalization.
Study Design
Spatial normalization (standard and unified methods) was performed using SPM5 software (Wellcome Department of Imaging Neuroscience, London, UK, 2007). All normalized images were written out using the template-bounding box and voxel size of 2mm. Prior to calculation of total entropy, each normalized segmented MR image was smoothed using an 8-mm FWHM Gaussian isotropic kernel.
SPM5 Standard Method
Firstly, each subject’s MR data was spatially normalized to the standard MNI (Montreal Neurological Institute) template in SPM using the standard approach, varying a single parameter at a time while setting all other parameters to the default. The normalization parameter settings were nonlinear frequency cutoff (affine only, 25mm, 45mm and 70mm), nonlinear regularization (light, medium and heavy) and nonlinear iterations (3, 8 and 16). Secondly, the standard method (with the MNI template) was applied to each subject’s MR by varying a combination of parameters for nonlinear frequency cutoff and nonlinear regularization to determine how the relationship of these two parameters affects the performance of the normalization. After each spatial normalization, segmentation was applied to the normalized images and total entropy (see Equations 1–3) was calculated using the GM, WM and CSF image data.
SPM5 Unified Method
The unified segmentation was also applied to each subject’s MR data (default tissue priors in SPM), varying a single parameter at a time while setting all other parameters to the default. The parameter settings were: warp frequency cutoff (affine only, 25mm, 45mm and 70mm) and warping regularization (light, medium and heavy). In contrast to the standard method, the unified segmentation produces modulated normalized segmented images of GM, WM and CSF. In addition, the unified method was applied to each subject’s MR using a combination of parameters for warp frequency cutoff and warping regularization to determine how the relationship of these two parameters affects the performance of spatial normalization.
Statistical Analysis
Total entropy data were analyzed for each subject group in SPSS 14 (SPSS Inc.) to provide descriptive statistics (including box plots) and to perform nonparametric testing for two-related or three-related samples (e.g. comparison across three different frequency cutoffs). Each box plot indicates minimum value, lower quartile (lowest 25% of data), median, upper quartile (highest 25% of data) and maximum value. Open circles indicate outliers that were either 1.5 fold less than the first quartile or 1.5 fold greater than the third quartile. Stars indicate outliers that were either 3 fold less than the first quartile or 3 fold greater than the third quartile. The nonparametric tests were used to examine differences in total entropy that might arise from different levels of frequency cutoff, regularization or number of nonlinear iterations. If the three-related samples test indicated statistically significant differences, a paired t-test was performed to compare the “minimum” and “default” total entropy results for that parameter set. To assess the extent to which differences in the normalization results could influence subsequent analyses of the MR data, paired t-tests were performed in SPM. The paired t-tests compared normalized MR data obtained by using the default parameter settings to the normalized MR data for which the total entropy was lowest (contrast [−1 1]). The SPM analyses were only performed for the control MR data. The SPM significance threshold was set to a FDR corrected p-value of p < 0.001 and an extent threshold of 50 voxels.
Results
Total Entropy
SPM5 Standard Method (SM)
Spatial normalization performed using affine transformation resulted in smaller total entropy, relative to all nonlinear transformation results (Figure 1A, bottom). The difference between the mean total entropy values determined using an affine transformation and a nonlinear transformation with default parameters was approximately 6% for all three-subject groups. The mean total entropy for affine transformation for controls, diabetics and depressed diabetics using the standard method was 1.93×105 (s.d.=0.10), 1.98×105 (s.d.=0.10) and 2.05×105 (s.d.=0.05) bits respectively.
Figure 1.
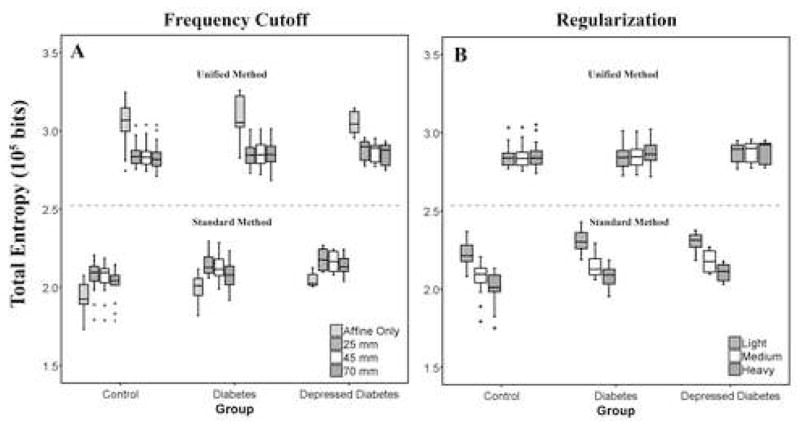
Residual variability (i.e., total entropy) for normalizations performed by varying a single parameter at a time, while all other parameters were fixed to their default value, for both SPM5 methods (top: unified, bottom: standard). (A) Affine transformation and nonlinear transformations that correspond to nonlinear frequency cutoffs of 25mm*, 45mm or 70mm. (B) Normalizations were performed using light, medium* or heavy regularization. For the standard method, an affine transformation resulted in the lowest total entropy relative to the nonlinear. In addition, a larger frequency cutoff or heavy regularization provided significantly lower residual variability for nonlinear normalization when applying the standard approach. For the unified method, a nonlinear transformation resulted in the lowest total entropy relative to an affine transformation. The unified method was relatively insensitive to the choices of frequency cutoff or regularization. In addition, the unified method provided greater values of total entropy relative to the standard method. One outlier was excluded from the figure (unified method) because the total entropy for this subject was consistently more than three fold lower than the first quartile of the diabetic group. (* denotes default parameters for nonlinear transformation in SPM).
Nonlinear Frequency Cutoff (SM)
A frequency cutoff of 70 mm tended to give lower total entropy, when compared to smaller cutoff values. This finding held true for all groups. Total entropy results for different levels of the frequency cutoff are shown in Figure 1A (bottom). The mean entropy for the smallest frequency cutoff of 25 mm (default) for controls, diabetics and depressed diabetics was 2.06×105 (s.d.=0.11), 2.13×105 (s.d.=0.10) and 2.18×105 (s.d.=0.07) bits respectively, while that for the largest cutoff setting (70mm) was 2.01×105 (s.d.=0.10), 2.07×105 (s.d.=0.11) and 2.14×105 (s.d.=0.06) bits, respectively. The difference in the mean total entropy value between 25 mm and 70 mm was only about 2–3 percent.
Nonlinear Regularization (SM)
The total entropy results show that increases in the regularization for the nonlinear transformation (when using the standard method in SPM5) reduces the residual variability (Figure 1B, bottom). Heavy regularization yielded lower residual variability when compared to medium regularization (default). This finding held true for all groups. The mean entropy for heavy regularization for controls, diabetics and depressed diabetics was 2.00×105 (s.d.=0.11), 2.06×105 (s.d.=0.09) and 2.11×105 (s.d.=0.06) bits respectively, while the mean entropy for medium regularization for controls, diabetics and depressed diabetics was 2.06×105 (s.d.=0.11), 2.13×105 (s.d.=0.10) and 2.18×105 (s.d.=0.07) bits respectively. The difference in mean entropy value between smallest and largest regularization was approximately 3% for all groups.
Combination of Nonlinear Frequency Cutoff and Nonlinear Regularization (SM)
Figure 2 (top) shows that when combining different levels of nonlinear cutoff and nonlinear regularization the total entropy changes. As the frequency cutoff increases, the effect of nonlinear regularization on the spatial normalization decreases. That is, regularization of a larger nonlinear frequency cutoff had little mean effect. This was seen for all three cutoffs evaluated (including 45 mm), although results for two are shown in Figure 2 for clarity.
Figure 2.
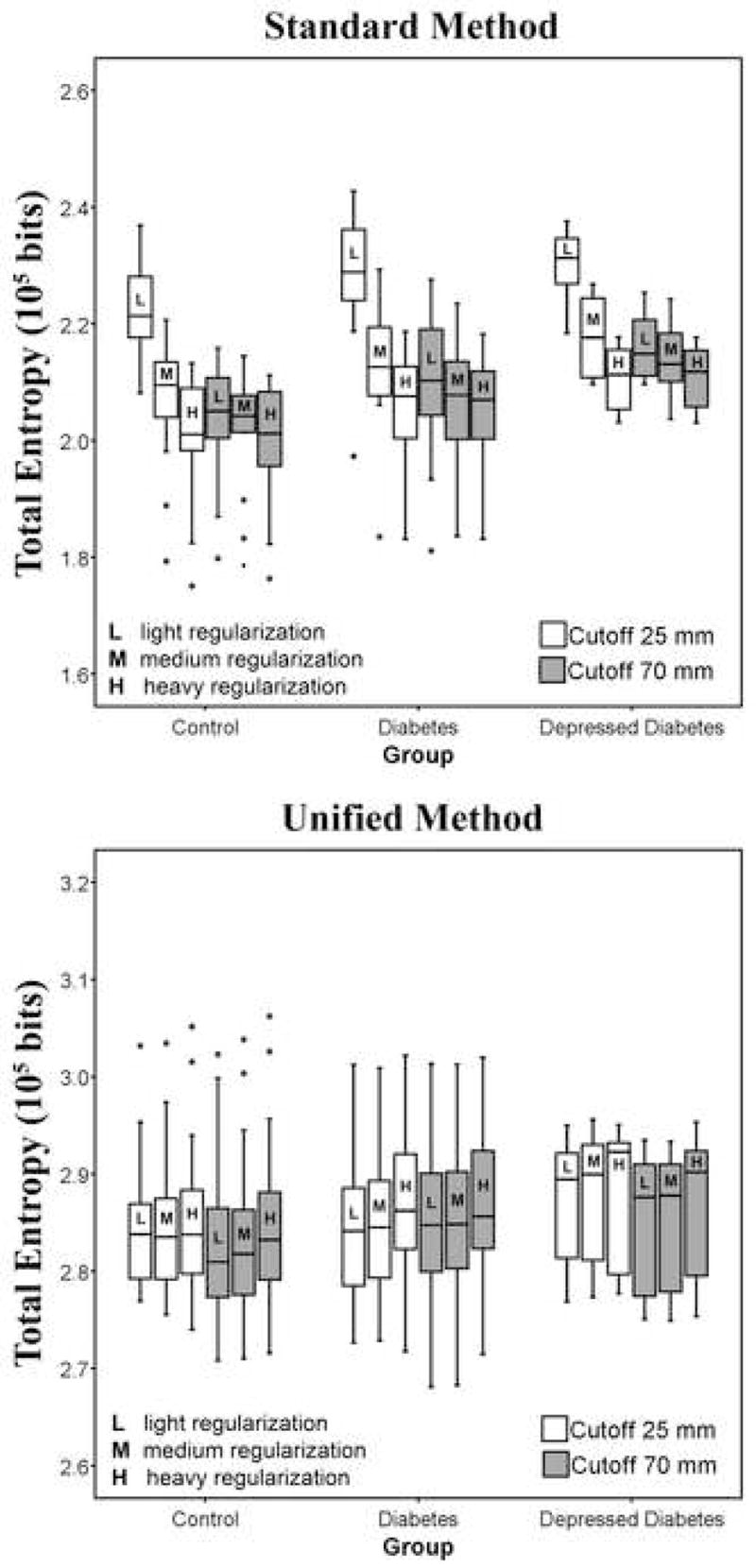
Residual variability (i.e., total entropy) for normalizations performed by varying more than one parameter at a time: frequency cutoff (25 mm* or 70 mm) and regularization (light, medium* or heavy), for both SPM5 methods (top graph: standard, bottom graph: unified). Regularization had little effect when applying a larger frequency cutoff. The unified model was relatively insensitive to the choice of frequency cutoff or regularization. The unified method provided greater values of total entropy relative to the standard method. One outlier was excluded from the figure (unified method) because the total entropy for this subject was consistently more than three fold lower than the first quartile of the diabetic group. (* denotes default parameters for nonlinear transformation in SPM).
Nonlinear Iterations (SM)
Total entropy results show that smaller number of nonlinear iterations led to smaller residual variability for the standard approach in SPM5. A nonlinear iteration number of three tended to provide smaller residual variability relative to the default (16 iterations). These results are shown in Figure 3. The mean entropy for three iterations for controls, diabetics and depressed diabetics was 2.04×105 (s.d.=0.10), 2.10×105 (s.d.=0.10) and 2.15×105 (s.d.=0.07) bits respectively and 2.06×105 (s.d.=0.11), 2.13×105 (s.d.=0.10) and 2.18×105 (s.d.=0.07) bits respectively, for the default setting. However, the difference in the mean total entropy value was only about 1% for all three groups.
Figure 3.
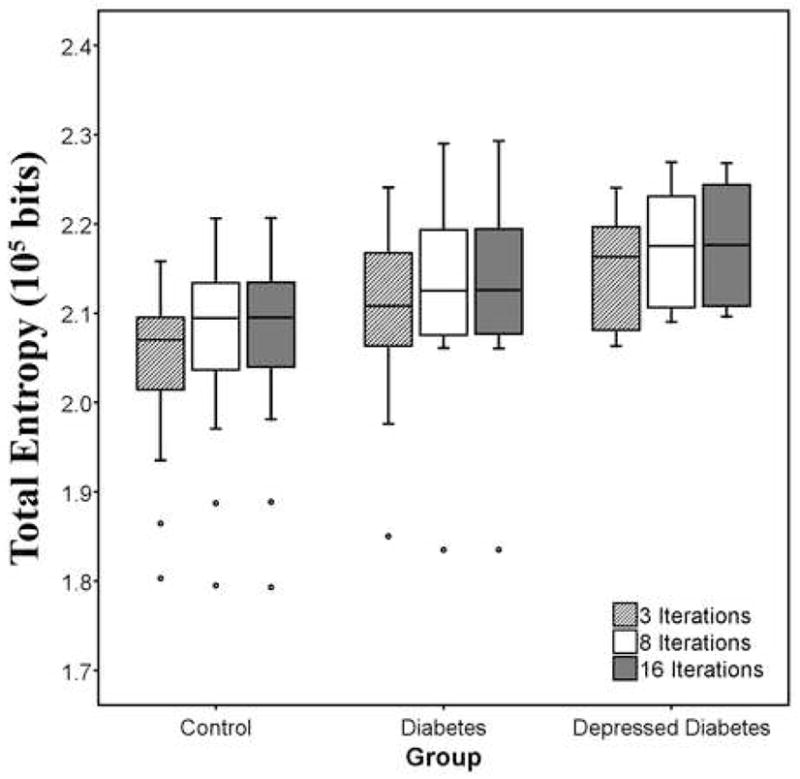
Residual variability (i.e., total entropy) for normalization performed by varying the number of nonlinear iterations (3, 8 or 16* iterations), while setting all other parameters to the default using the SPM5 standard approach. Fewer number of nonlinear iterations can provide significantly lower residual variability but a greater number may be needed to achieve finer matching of more complex deformations. (* denotes default parameters for nonlinear transformation in SPM5).
Nonparametric Statistical Results
Our results indicate that there are differences in residual variability when parameter settings change in the nonlinear transformation using the standard approach for this mid-life/elderly data set. Nonparametric testing for differences in entropy across parameter settings were statistically significant for each of the three groups (controls, diabetics and depressed diabetics), with a maximum observed significance of p = 0.008 for choice of nonlinear frequency cutoff, p < 0.001 for choice of nonlinear regularization and p = 0.013 for nonlinear iterations. In addition, individually optimal parameters: larger nonlinear frequency cutoff, higher nonlinear regularization and fewer nonlinear iteration provide significantly lower residual variability than the default parameters using the standard approach in SPM5. Table 2 summarizes nonparametric results obtained for comparison of the spatial normalization determined for the minimum total entropy data relative to the default values. All comparisons were statistically significant. It should be noted that the p-values from the nonparametric tests, comparing minimum total entropy to the default value, should be interpreted with caution since these values are not corrected for multiple comparisons.
Table 2.
Nonparametric tests comparing lowest total entropy with default parameter for the standard method in SPM5
| SPM5 Standard Method Default Parameter versus Parameter with Smallest Total Entropy |
|||
|---|---|---|---|
| Nonlinear Frequency Cutoff | Nonlinear Regularization | Nonlinear Iterations | |
| 25mm* versus 70mm | medium* versus heavy | 16* versus 3 | |
| Control | |||
| p-value | p = 0.001 | p = 0.001 | p = 0.002 |
| Diabetes | |||
| p-value | p = 0.001 | p < 0.001 | p = 0.013 |
| Depressed Diabetes | |||
| p-value | p = 0.012 | p = 0.012 | p = 0.012 |
Denotes default parameters in SPM
SPM5 Unified Method (UM)
Nonlinear transformation gave lower total entropy when compared to affine transformation when using the unified approach. This finding held true for all three groups. The difference in mean total entropy value between an affine and nonlinear transformation for the unified method was between 6–9 percent.
Warp Frequency Cutoff (UM)
Nonlinear transformation with a frequency cutoff of 70 mm resulted in smaller total entropy for all three groups, when compared to the default 25 mm (top, Figure 1A). However, the difference in mean total entropy value between a smaller and larger frequency cutoff was negligible (0.14–0.80 percent). The mean entropy for a frequency cutoff of 25 mm for controls, diabetics and depressed diabetics was 2.85×105 (s.d.=0.08), 2.75×105 (s.d.=0.41) and 2.88×105 (s.d.=0.07) bits, respectively and for a cutoff of 70 mm was 2.84×105 (s.d.=0.10), 2.75×105 (s.d.=0.38) and 2.85×105 (s.d.=0.07) bits, respectively. A smaller mean entropy value for the diabetics was because of an outlier (2.85×105 bits without the outlier for both frequency cutoffs). This outlier was excluded from Figure 1A (top) because the total entropy for this subject was consistently more than three fold lower than the first quartile of the diabetic group.
Warping Regularization (UM)
The total entropy results show that the light regularization gave smaller residual variability for all three groups. Results are shown on the top of Figure 1B. The mean entropy for medium regularization for controls, diabetics and depressed diabetics was 2.85×105 (s.d.=0.08), 2.75×105 (s.d.=0.41) and 2.88×105 (s.d.=0.07) bits, respectively and the mean entropy for light regularization for controls, diabetics and depressed diabetics was 2.85×105 (s.d.=0.07), 2.71×105 (s.d.=0.54) and 2.87×105 (s.d.=0.07) bits, respectively. A smaller mean entropy value for the diabetics was because of an outlier (2.85×105 and 2.84×105 bits without the outlier for medium and light regularization respectively). This outlier was excluded from Figure 1B (top) because the total entropy for this subject was consistently more than three fold lower than the first quartile of the diabetic group.
Combination of Nonlinear Frequency Cutoff and Nonlinear Regularization (UM)
Figure 2 (bottom) shows that spatial normalization in the unified model is generally insensitive to the choice of parameter settings, when combining different levels of nonlinear frequency cutoff and nonlinear regularization. One outlier was excluded because the total entropy for this subject was consistently more than three fold lower than the first quartile of the diabetic group.
Nonparametric Statistical Results
There were no differences detected in residual variability when parameter settings change in the nonlinear transformation using the unified approach, with the exception of different levels of regularization for the diabetics (p = 0.01). Results are not shown.
Standard Method versus Unified Method
An affine transformation yielded the smallest total entropy for the standard approach, when compared to a nonlinear transformation. In contrast, nonlinear normalization provided the lowest residual variability for the unified method. The total entropy results suggest that the unified method provides larger total entropy relative to the standard method. Both methods yielded similar number of outliers; however, they identified different subjects with the exception of one subject. The reason that both methods identified the same outlier was because the subject was not optimally positioned in the field of view. Total entropy results also show that the unified method is insensitive to parameter variation.
SPM maps
Our results indicate that the use of different parameter settings for the standard approach may alter the SPM maps. The comparison of frequency cutoffs (25 mm and 70 mm) yielded statistically significant differences in cortical white matter (primarily gyri) with few gray matter differences (insula and temporal gyri). Comparison of medium and heavy regularization indicated significant differences, about equally, in white matter (fusiform and frontal gyri) and gray matter (frontal gyri, anterior cingulate, claustrum and lentiform nucleus). Finally, the comparison of iteration number (16 vs. 3) yielded significant differences in gyral areas of the frontal, temporal, and occipital lobes and the only difference noted for CSF (lateral ventricle/sub-lobar). SPM analyses that compared different levels of regularization for the unified method did not detect statistically significant differences.
Discussion
In this comparative study, we used total entropy as a performance measure to assess the effects of different parameter settings on the residual variability associated with the spatial normalization of mid-life/elderly MR imaging data (healthy controls or type 2 diabetics). Overall, we found the standard method of normalization in SPM5 to be sensitive to changes in nonlinear frequency cutoff, nonlinear regularization and number of nonlinear iterations, while the unified method was relatively insensitive to such parameter variations.
For the standard method in SPM5, the total entropy results indicated lower residual variability for the affine transformation than for all nonlinear transformations. It is possible for a larger frequency cutoff to yield less residual variability in the nonlinear normalization, relative to a smaller cutoff. This is consistent with the notion that as more basis functions are used, it is possible to match more distortions because higher frequency deformations can be modeled (Salmond et al., 2002). As expected, as the regularization increases, the total entropy decreases and is closer in value to the entropy results for the affine transformation. It is possible that a greater regularization could allow better matching because the penalty increases for quickly changing transformations. However, regularization of larger frequency cutoff had little effect on the residual variability of the normalization. The entropy results for the standard method also indicated that good spatial normalization might be achieved with only a few iterations of the nonlinear warping process. The lowest residual variability for the standard nonlinear spatial normalization of mid-life/elderly image data was achieved by using “heavy” regularization, with all other parameters set at the default value. Analogous comparisons were performed in SPM2 and these results were very similar to those reported herein for the SPM5 standard method (results not shown).
For the unified method in SPM5, the total entropy results indicated lower residual variability for the nonlinear transformations than for the affine transformation. In contrast to the standard method, variations in frequency cutoff and/or regularization had little effect on the normalization and the residual variability determined using the SPM5 unified approach. The unified method performed well for the mid-life/elderly image data, when using the default parameters, in a manner consistent with Crinion et al. (2007). These findings are consistent with those of Hellier et al. (2003) who reported better spatial normalization of skull-stripped images using nonlinear normalizations over affine.
It is acknowledged that the lowest total entropy may not necessarily be indicative of the best normalization result. In this work, the standard method provided lower entropy values (1.7 – 2.4 × 105 bits) than the unified method (2.7 – 3.3 × 105 bits), but the latter method provided more stable results across parameter settings. The larger total entropy observed for the unified method, compared to the standard method (Figures 1 and 2) appeared to result from greater entropy in gray and white matter but not in CSF, based upon the examination of segmentation results across several default settings (data not shown).
We chose to use this measure because it reflected residual variability based upon individual tissue types. Warfield et al. (2001) proposed the use of total entropy to assess image alignment for affine and nonlinear registration algorithms. Robbins et al. (2004) utilized total entropy to assess performance, to tune, and to compare spatial normalization methods. The total entropy measure could be readily applied in this work because all imaging was performed on the same MR scanner using the same protocol. Another measure that has been very useful for assessing differences in registration or deformations is the root mean squared displacement that is based upon differences in voxel distances relative to anatomical landmarks (Crinion et al., 2007). Hellier, et al. (2003) also proposed several global and local measures to assess the performances of different nonrigid registration methods. The precision of the spatial normalization in SPM has also been assessed in terms of anatomical landmarks by calculating the standard deviation from the mean position of the landmark (Salmond et al., 2002). An advantage of the total entropy measure is that it does not require anatomical landmark definition, and therefore can be applied by a user who does not have a strong background in neuroanatomy, and no inter-rater reliability assessments of landmark identification is needed.
Technical factors, such as poor positioning of the subject in the scanner or subject movement, can influence the spatial normalization of the standard method and the spatial normalization and segmentation of the unified method, and thus the total entropy. Extreme segmentation failure (extreme outlier) caused the total entropy to be dramatically underestimated. The dependence of the total entropy on the segmentation process was further evaluated by examining the segmented image data (gray matter, white matter and CSF) for both standard and unified SPM5 methods (several default settings). Pearson’s correlations were performed to explore potential relationships between age (degree of atrophy) and total entropy for the three tissue types. The results were ambiguous across the subject groups with the only significant result indicated for diabetics for which greater age was associated with greater CSF entropy (more CSF is expected with greater atrophy) but less gray matter and white matter entropy (data not shown). Finally, as stated above, gray and white matter entropies were greater for the unified method than for the standard method, while CSF entropies were similar for both methods.
The SPM analyses of the data processed using the standard approach indicated that the frequency cutoff resulted in normalization differences most often observed in cortical white matter (frontal and temporal gyri) and less often in gray matter (temporal gyri). Differences in regularization warping influenced both gray and white matter of frontal and temporal cortices. The SPM analysis also indicates that the number of iterations might significantly affect the warping of gyral areas in cortex and along the lateral ventricle (areas adjacent to and including CSF).
Total entropy results and SPM analyses suggest that the best parameters for nonlinear spatial normalization of mid-life/elderly image data to the MNI template, when applying the standard approach, correspond to a smaller cutoff (25 mm), heavy regularization, and the default number of nonlinear iterations (16). A smaller frequency cutoff may allow for better matching of deformations, heavy regularization could limit distortions that might occur when warping mid-life/elderly image data to the young template, and sixteen iterations because these may improve the normalization in areas adjacent to or containing CSF. On the other hand, when applying the unified approach, the default parameters were the best for spatial normalization of mid-life/elderly image data to the MNI priors, given the apparent lack of sensitivity of the method to variations in the parameter settings. In contrast to the standard method, it is possible that the total entropy measure may not be a sensitive indicator for the assessment of parameter variations in the unified method.
For clinical research applications, it can be important to try to match the normalization method with the research question. The preferred normalization would be one that provides a good compromise between the extent to which an individual’s MR image is adjusted and optimal matching of the brain to the standard template. The normalization-of-choice would allow for the detection of pathology-related changes rather than differences that may exist between the individual’s MR and the template. If age-related changes are of interest, then one would want to apply a normalization that would modify the individual’s MR image least, while providing the best normalization in areas where age-related group differences are expected (e.g., frontal cortex or ventricles/CSF).
These findings are relevant for studies of structural brain alterations that may occur in normal aging, chronic medical conditions, neuropsychiatric disorders, and neurodegenerative disorders. Future studies will extend this work in the context of subject- or disease-specific templates/tissue priors.
Acknowledgments
This work was supported by NIH grants (K01 MH01976, P30 DK046205, P30 MH52247, R01 DK39629, R01 MH59945, MO1-RR000056, R01 MH070729). The authors are deeply indebted for the mentorship and recruitment support provided by the University of Pittsburgh Obesity and Nutrition Research Center (Dr. David E. Kelley), Intervention Research Center for Late-life Mood Disorders (Dr. Charles F Reynolds, III), Dr. Christopher M. Ryan, and Dr. Sigrid A. Hagg. We thank Dr. Carolyn C. Meltzer for sharing MR data of 4 elderly controls. We also thank the MR and PET research staff, M. Geckle, M. Bechtold, and S. Hulland for their help with these studies. We also thank Drs. Steven DeKosky and William Klunk for their helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Bedda L. Rosario, Department of Biostatistics, University of Pittsburgh, Graduate School of Public Health, 130 DeSoto Street, Pittsburgh, PA 15261, E-mail: blr5@pitt.edu, Phone Number: (412) 563-0352, Fax: (412) 624-2183
Scott K. Ziolko, Department of Radiology, University of Pittsburgh School of Medicine, Presbyterian University Hospital, 200 Lothrop Street, Pittsburgh, PA 15213
Lisa A. Weissfeld, Department of Biostatistics, University of Pittsburgh, Graduate School of Public Health, 130 DeSoto Street, 326 Parran Hall, Pittsburgh, PA 15261, E-mail: lweis@pitt.edu
Julie C. Price, Department of Radiology, University of Pittsburgh School of Medicine, Presbyterian University Hospital, B-938, 200 Lothrop Street, Pittsburgh, PA 15213, E-mail: pricejc@upmc.edu
References
- Ashburner J, Friston KJ. Nonlinear Spatial Normalization using Basis Functions. Human Brain Mapping. 1999;7:254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Unified Segmentation. NeuroImage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Cover TM, Thomas JA. Elements of Information Theory. 2. John Wiley & Sons, Inc; 2006. [Google Scholar]
- Crinion J, Ashburner J, Leff A, Brett M, Price C, Friston C. Spatial normalization of lesioned brains: Performance evaluation and impact on fMRI analyses. NeuroImage. 2007;37:866–875. doi: 10.1016/j.neuroimage.2007.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivello F, Schormann T, Tzourio-Mazoyer N, Roland PE, Zilles K, Mazoyer BM. Comparison of spatial normalization procedures and their impact on functional maps. Human Brain Mapping. 2002;16(4):228–250. doi: 10.1002/hbm.10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gispert JD, Pascau J, Reig S, Martinez-Lazaro R, Molina V, Garcia-Barreno P, Desco M. Influence of the normalization template on the outcome of statistical parametric mapping of PET scans. NeuroImage. 2003;19:601–612. doi: 10.1016/s1053-8119(03)00072-7. [DOI] [PubMed] [Google Scholar]
- Hellier P, Barillot I, Corouge I, Gibaud B, Le Goualher G, Collins DL, Evans A, Malandain G, Ayache N, Christensen GE, Johnson HJ. Retrospective Evaluation of Intersubject Brain Registration. IEEE Transactions on Medical Imaging. 2003;22(9):1120–1130. doi: 10.1109/TMI.2003.816961. [DOI] [PubMed] [Google Scholar]
- Hosaka K, Ishii K, Sakamoto S, Sadato N, Fukuda H, Kato T, Sugimura K, Senda M. Validation of anatomical standardization of FDG PET images of normal brain: comparison of SPM and NEUROSTAT. European Journal of Nuclear Medicine and Molecular Imaging. 2005;32(1):92–97. doi: 10.1007/s00259-004-1576-z. [DOI] [PubMed] [Google Scholar]
- Meyer JH, Gunn RN, Myers R, Grasby PM. Assessment of Spatial Normalization of PET Ligand Images Using Ligand-Specific Templates. NeuroImage. 1999;9:545–553. doi: 10.1006/nimg.1999.0431. [DOI] [PubMed] [Google Scholar]
- Price JC, Kelley DE, Ryan CM, Meltzer CC, Drevets WC, Mathis CA, Mazumdar S, Reynolds CF. Evidence of Increased Serotonin-1A Receptor Binding in Type 2 Diabetes: A Positron Emission Tomography Study. Brain Research. 2002;927(1):97–103. doi: 10.1016/s0006-8993(01)03297-8. [DOI] [PubMed] [Google Scholar]
- Price JC, Kelley DE, Hagg SA, Meltzer CC, Ryan CM, Mazumdar S, Mathis CA, Reynolds CF. Measurement of serotonin-1A receptor binding in diabetes and depression. Journal of Nuclear Medicine. 2003;44(5):S896. [Google Scholar]
- Robbins S, Evans AC, Collins DL, Whitesides S. Tuning and comparing spatial normalization methods. Medical Image Analysis. 2004;8:311–323. doi: 10.1016/j.media.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Salmond CH, Ashburner J, Vargha-Khadem F, Connelly A, Gadian DG, Friston KJ. Technical Note: The Precision of Anatomical Normalization in the Medial Temporal Lobe Using Spatial Basis Functions. NeuroImage. 2002;17:507–512. doi: 10.1006/nimg.2002.1191. [DOI] [PubMed] [Google Scholar]
- Sugiura M, Kawashima R, Sadato N, Senda M, Kanno I, Oda K, Sato K, Yonekura Y, Fukuda H. Anatomic Validation of Spatial Normalization Methods for PET. Journal of Nuclear Medicine. 1999;40(2):317–322. [PubMed] [Google Scholar]
- Warfield SK, Rexilius J, Huppi PS, Inder TE, Miller EG, Wells WM, III, Zientara GP, Jolesz FA, Kikinis R. A binary entropy measure to assess nonrigid registration algorithms. Medical Image Computing and Computer-Assisted Intervention. Lecture Notes in Computer Science Proceedings of the 4th International Conference on Medical Image Computing and Computer-Asisted Intervention, Springer; 2001. pp. 266–274. [Google Scholar]
- Wellcome Department of Imaging Neuroscience. Statistical Parametric Mapping Software, London, UK. 1994–2007 http://www.fil.ion.ucl.ac.uk/spm/
- Wu M, Carmichael O, Lopez-Garcia P, Carter C, Aizenstein HJ. Quantitative Comparison of AIR, SPM, and the Fully Deformable Model for Atlas-Based Segmentation of Functional and Structural MR Images. Human Brain Mapping. 2006;27:747–754. doi: 10.1002/hbm.20216. [DOI] [PMC free article] [PubMed] [Google Scholar]


