Abstract
One limit on developing complex synthetic gene circuits is the lack of basic components such as transcriptional logic gates that can process combinatorial inputs. Here, we propose a strategy to construct such components based on reusable designs and convergent reengineering of well-studied natural systems. We demonstrated the strategy using variants of the transcription factor (TF) LacI and operator Olac that form specifically interacting pairs. Guided by a mathematical model derived from existing quantitative knowledge, rational designs of transcriptional NAND, NOR and NOT gates have been realized. The NAND gates have been designed based on direct protein–protein interactions in coupling with DNA looping. We demonstrated that the designs are reusable: a multiplex of logic devices can be readily created using the same designs but different combinations of sequence variants. The designed logic gates are combinable to form compound circuits: a demonstration logic circuit containing all three types of designed logic gates has been synthesized, and the circuit truthfully reproduces the pre-designed input–output logic relations.
Keywords: combinability, mathematical model, reusable designs, transcriptional logic gates
Introduction
The control of gene expression by transcription factors (TFs) and promoters can be paralleled to electronic circuits (Hasty et al, 2002). Making use of naturally existing TFs and operators, synthetic transcriptional circuits with various signal processing functions have been designed and successfully implemented in vivo, including genetic toggle switches (Atkinson et al, 2003), oscillators (Elowitz and Leibler, 2000; Stricker et al, 2008; Tigges et al, 2009), as well as bacteria ecosystems (Balagadde et al, 2008). Despite these successes, our ability to design and synthesize compound transcriptional circuits that can function exactly as desired is still limited. One major limiting factor is the lack of general designs for devices processing combinatorial inputs. Although it is routine to design and implement promoters that take input from single TFs, it is considered non-trivial to design promoters that accept compound inputs from several TFs and generate outputs predictably according to certain computational rules (Cox et al, 2007; Ellis et al, 2009). Only recently, a few examples of rationally designed synthetic logic gates (Anderson et al, 2007; Bronson et al, 2008; Sayut et al, 2009) that can process combinatorial inputs have been reported. Of significant contributions to the limited supply of basic devices is the fact that each of such non-trivially designed devices lacks reusability, or can be used at most once in a compound circuit. This is because molecular circuits use diffusing molecules as signal carriers, and devices based on the same set of molecules are not reusable or combinable as mutually insulated components in one circuit.
In this report, we propose and demonstrate a strategy to develop reusable designs of transcriptional logic gates based on well-studied natural systems. For such systems, the known physical mechanisms of cooperative transcriptional regulations (Buchler et al, 2003; Bintu et al, 2005) as well as existing experimental data facilitate the use of mathematical models to guide the design of basic devices of sophisticated functions. In addition, we carry out convergent engineering of TFs and operators to create new sets of specific TF/operator interactions without affecting those molecular interactions governing cooperative transcriptional regulation. Such kind of reengineering has been shown to be feasible through either rational design or directed evolution. For instances, the DNA-binding specificity of nucleases (Ashworth et al, 2006; Maeder et al, 2008) or TFs (Krueger et al, 2007) have been reengineered without impairing the catalytic or regulatory activities of the respective proteins.
With this approach, basic transcriptional devices are first designed as prototypes. A multiplex of basic devices, each relying on a unique set of specific DNA–protein interactions for connections, can be implemented using the same designs. Different instances of devices are combinable if their composing TF/operator pairs interact in a pairwise-specific manner.
We demonstrate the above strategy using as templates the lac repressor (LacI) and its cognate operator (Olac), whose protein–protein and protein–DNA interactions have been characterized with atomic details (Friedman et al, 1995; Lewis et al, 1996; Swigon et al, 2006). First, we engineered variants of the LacI/Olac pairs with new specific protein–DNA interactions, using experimental structures of LacI in complex with Olac (Kalodimos et al, 2002) as well as previous mutational analyses of the system (Sartorius et al, 1989; Kopke Salinas et al, 2005) as guidance. We then developed designs of transcriptional devices that can receive single or combinatorial inputs and perform designated basic logic operations, including NAND, NOR and NOT, guided by a mathematical model describing cooperative transcription regulation. We validated the reusability of the designs, namely, different instances of devices can be implemented based on the same prototypes but using different specific LacI/Olac variant pairs. Finally, to demonstrate that individual designed logic gates as separate components are combinable in compound logic gene circuits, we synthesized a circuit composed of three such gates, and showed that this in vivo circuit processes input information as designed.
Results
Engineer specifically interacting LacI and Olac variant pairs
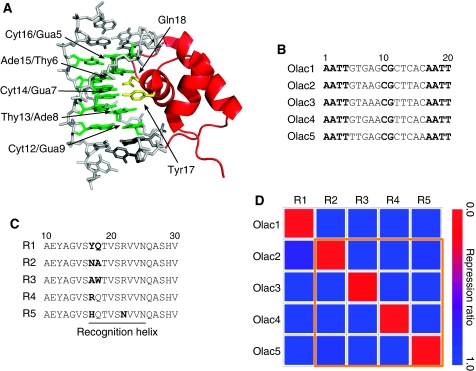
Briefly, palindromic variants of an ideal Olac1 derived from the natural lac O1 operator (Sadler et al, 1983), Olac2–4 in Figure 1B, are designed. Sequence variants (R2, R3 and R4 in Figure 1C) of the wild-type LacI (R1 in Figure 1C) specifically recognizing these variant operators have been selected from a sequence library. An additional pair R5/Olac5 has been selected from the literature (Sartorius et al, 1989). These TF/operator pairs have been chosen based on an experimentally measured ‘repression matrix' tabulating the repression effects of each LacI variant against each Olac variant. Figure 2D shows the diagonal repression matrix formed by the selected LacI/Olac variant pairs, indicating relatively stringent pairwise specificity. More details of the directed evolution and characterization of the LacI variants are given in Supplementary information.
Figure 1.
(A) The N-terminal headpiece of LacI in complex with its operator (PDB: 1L1M). The backbone of the headpiece is shown in red. The side chains of Tyr17 and Gln18 are shown as yellow sticks. Base pairs in direct contacts with the repressor in the DNA major groove are shown in green and the rest of the DNA are shown in gray. This graph has been generated using PyMol. (B) The nucleotide sequences of the five operator variants considered in this work. Conserved nucleotides are shown in bold. (C) Amino acid sequences around the DNA recognition helix in LacI wild type (R1) and mutants (R2–R5). Residues 17 and 18 in R1 and the mutated residues in R2–R5 are shown in bold. (D) A graphical representation of the repression matrix associated with R1–R5 and Olac1–Olac5. The values represent the repression ratios, defined as the promoter activity at the presence of the input LacI variant over that in absence of the input LacI, measured for promoters each containing a single-operator element at the position +10. The yellow boundary encloses the subgroup associated with a diagonal repression matrix, suggesting within-subgroup pairwise specificity.
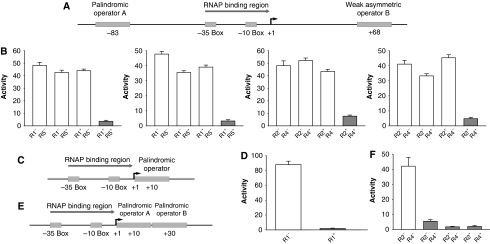
Figure 2.
(A) The promoter design for the NAND gates. (B) Fluorescent outputs of four NAND gates in response to different inputs. Different NAND gates share the promoter design shown in (A) but use different Olac variants for operators A and B, respectively. From left to right, the promoters are PU83Olac1+D68Oweak5, PU83Olac5+D68Oweak1, PU83Olac2+D68Oweak4 and PU83Olac4+D68Oweak2, respectively. Here, OweakX, where X is one of 1–5, indicates that the downstream operator has been modified as described in the main text, with the unmodified half coming from one of Olac1–Olac5. (C) The promoter design for the NOT gates. (D) Fluorescent outputs of a NOT gate constructed using R1/Olac1 in response to different inputs. (E) The promoter design for the NOR gate. (F) Fluorescent outputs of the NOR gate constructed using the R2/Olac2 and R4/Olac4 pairs in response to difference inputs. (A, D, F) White bars correspond to expected ‘ON' outputs and gray bars correspond to expected ‘OFF' outputs. Activities are reported as slopes obtained from linearly fitting fluorescence versus OD600. Error ranges are root mean square residuals of the respective fittings. Source data is available for this figure at www.nature.com/msb.
Design NAND, NOR and NOT gates
We designed these gates based on the previously revealed repression mechanisms of LacI (Friedman et al, 1995; Lewis et al, 1996; Swigon et al, 2006). The promoters in these devices share the basic structure of a 37-bp RNA polymerase binding region (Figure 2A, C and E) that has the same sequence as the promoter PlacUV5 between the −35 box and the +1 transcription starting site. This region is separated from one or two Olac variants by randomly selected non-sense sequences. The various LacI variants would bind to these operators as a tetramer, or dimer of dimers, one dimer binding to one operator, inducing DNA looping when two dimers in one tetramer simultaneously bind to two operators (Friedman et al, 1995).
A mathematical model has been constructed to link the transcriptional activity of such promoters to the input repressor signals (see Materials and methods). This model containing mostly experimentally derived parameters integrated existing knowledge about the natural LacI/Olac system as well as characteristics of engineered LacI/Olac variants, including the experimentally characterized effects of factors like positions of the operators (Elledge and Davis, 1989), DNA looping induced by TF binding (Oehler et al, 1990; Muller et al, 1996), varying association constants between variants of repressors and different operator sequences, as well as equilibriums between different forms of repressor tetramers when a multiplex of repressor variants are present.
More details of the mathematically guided design are given in Supplementary information. The final NAND gate design (Figure 2A) consists of one palindromic operator recognizable by one TF variant at the position −83 (the number refers to the position of the 10th base of the 20-bp long operator sequence) and another non-palindromic operator recognizable by a different TF variant (noted as Weak Asymmetric Operator B in Figure 2A) with deliberately weakened interactions with its cognate repressor to achieve a satisfactory NAND performance (see Supplementary information). This design uses DNA looping. There can be other types of designs that have combinatorial effects, such as a single operator composed of two half-sites each recognized by a different TF variants. Compared with the latter, the DNA looping-based design has two main advantages concerning reusability and logic performance. First, it uses the combination between full sites, thus avoids possible influences between contiguous half-sites in a design that uses the combination between half-sites. Such influences may lead to sequence variant-dependent input–output relationships, reducing the reusability of designs. Second, it uses the absolute and relative positions of operators as the main design parameters (see the mathematical model in Materials and methods) to achieve good logic performance, being tolerant to variations in unselected binding constants such as those between different TF heterodimers to a particular operator. On the other hand, the mathematical model for a single-hybrid operator indicates that the main parameters are the relative binding constants associated with different TF homo- and heterodimers to the hybrid operator. Good logic performance may appear only by chance, as these binding constants can be highly variable between different combinations of sequence variants that have been selected based on specific binding between the TF homodimers and palindromic operators.
In the final NOR gate design (Figure 2C), two different Olac variants are put at the downstream positions +10 and +30, respectively. In the NOT gate (Figure 2E), one Olac variant is put at the downstream position +10.
Performance, reusability and combinability of the designs
The mathematical model predicts that the above designs are of good logic performance (Supplementary Figures S2–S4). Experimental tests of the designs in vivo confirmed the predictions, and the results are shown in Figures 2B, D and F. In Supplementary Figure S5, the outputs of an alternative design based on combining two half-sites of different TF specificity are shown, indicating combinatorial regulation but only with suboptimal logic performance.
The designs are reusable in the sense that different LacI/Olac variant pairs can be substituted into the same design to obtain different instances of devices. Figure 2B shows the test results for four different in vivo NAND gates constructed based on the same design but using different combinations of LacI/Olac variant pairs. The results confirm the reusability of the NAND design. Similar substitutions have been performed on the NOR gate and NOT gate designs. Results (not shown) lead to the same conclusion.
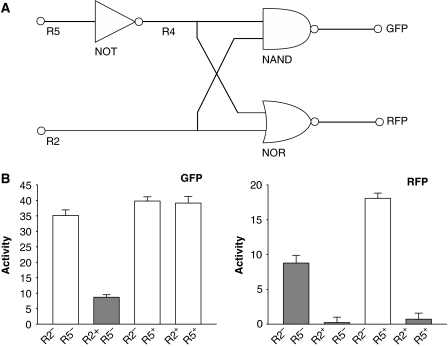
To demonstrate the combinability of the engineered logic devices or the feasibility of implementing compound transcriptional circuits using these devices, we assembled a demonstration system that includes all the three types of logic gates designed in this work (Figure 3A). Figure 3B shows the experimental results of testing the designed circuit in vivo in association with the theoretical ones. The in vivo circuit reproduced the theoretical expectations faithfully, indicating not only that the individual logic devices operated as designed, but also that interferences between the different connecting wires or the TF/operator pairs, if present, are acceptably weak to not significantly distort the major signal flows in the circuit, in consistence with mathematical predictions such as Supplementary Figure S6. We note that in Figure 3B, the repression of RFP corresponding to the ‘RFP-off' state at input R2−R5− seems to fall short of ideal as compared with the other ‘RFP-off' states. This result is likely to be caused by that R4, the internal repressor leading to this particular ‘off' state through the NOR gate, has been produced from a low copy number plasmid (see Materials and methods). Despite this, the repression is still significant enough to be considered as an ‘off' output as compared with the ‘RFP-on' output that is achieved through the repression of R4 under the input R2−R5+ (see also Supplementary Figure S7).
Figure 3.
(A) Diagram of the demonstration circuit. Labels directly under the wires represent repressors carrying the respective input and intermediate signals. GFP and RFP refer to the green and red fluorescence proteins reporting outputs, respectively. (B) Theoretical and experimental outputs of the circuit in response to different inputs. White bars correspond to ‘ON' outputs expected by theory and gray bars correspond to ‘OFF' outputs expected by theory. Activities are reported as slopes obtained from linearly fitting fluorescence versus OD600. Error ranges are root mean square residuals of the respective fittings. Source data is available for this figure at www.nature.com/msb.
Discussion
We have demonstrated a strategy to create reusable and combinable designs of basic logic devices at the transcriptional level. In terms of creating a multiplex of mutually insulating components based on reusable designs, the approach can be generally applied to systems in which the parts responsible for the specificity of intermolecular interactions can be reengineered without impairing the overall regulatory function. Such modularity commonly exists in natural regulatory systems. For an increasing number of these systems, knowledge about key specificity-determining sequence positions can be or have been provided by sequence, structural and/or functional analyses, facilitating such engineering.
Our study also showed the use of mathematical models in developing synthetic gene circuits. Recently, Ellis et al have demonstrated the use of mathematical models to guide the selection of components from a randomly diversified component library to achieve predictable network dynamics (Ellis et al, 2009). As compared with that work, mathematical models used in our work are of a different type and complementary values. The model used here is not purely a mathematical formulation of kinetics, but incorporates physical mechanisms underlying cooperative transcriptional controls, thus can guide the developments of synthetic components of extended or new functionalities as compared with their natural counterparts. The convergent engineering of TF and operator sequences in this work is also different but complementary to the diversification of components in the earlier studies (Cox et al, 2007; Ellis et al, 2009). The purpose here is to create a range of new molecular specificities, but not to introduce components of diverse dynamics. We note that programmable protein–DNA interaction, a much desired property for designed transcriptional devices, although not yet achievable for the DNA-binding module in lacI, can be achieved by using zinc finger-based DNA-binding modules (Ashworth et al, 2006; Maeder et al, 2008). On the other hand, the lacI-based designs reserve a nature-selected molecular scaffold that integrates different types of regulations, including protein–protein and protein–small molecule interactions besides protein–DNA interactions and DNA looping. In this work, we have considered reengineering only the latter two. Using the same protein–protein interface in different repressor variants causes undesired repressor hetero-oligomers to coexist with the desired ones. In this sense, the components having the same protein interface are not completely free from mutual interferences, although the undesired oligomers should be inactive against the target operators and the interferences may have insignificant effects in small-scale systems (see also Supplementary Figure S6). To eradicate such interferences, the reengineering can be extended to include the specificity of protein–protein interactions (Schnappinger et al, 1998; Spott et al, 2000). In addition, reengineering the protein–protein as well as the protein–small molecule (Lewis, 2005; Tang et al, 2008) interactions may potentiate the design of more extended and more sophisticated transcriptional controls.
Materials and methods
Mathematical modeling
We consider the transcriptional activity (designated as A) of a single promoter containing two operator sequences, noted as Oα and Oβ, respectively. Their positions in the promoter are designated as Pα and Pβ, respectively. Each of their sequences is viewed as composed two halves that are either palindromic or asymmetric. The promoter activity is controlled or influenced by three species of repressor, RA and RB that may recognize the entire or half-operator sequences in Oα or Oβ, and RC that does not recognize either Oα or Oβ. RA and RB correspond to the true input signals and RC the interfering signals, probably collections of designated inputs for other logic gates.
We assume that the various tetramer forms of the repressors, Ti, are far more stable than the dimer or monomer forms, and the stability of different tetramers corresponding to different combinations of RA, RB and RC are the same. Then the concentrations of various tetramers, [Ti], which are dimers of dimers such as (RA)2(RA)2, (RA)2(RARB), (RA)2(RARC),… and so on, can be easily determined from the concentrations [RA], [RB] and [RC]. Extending from the models proposed by Hwa and coworkers (Buchler et al, 2003; Bintu et al, 2005), the transcription activity of the promoter, which parametrically depends on Oα, Oβ, Pα and Pβ and functionally depends on [RA], [RB] and [RC], can be written as,
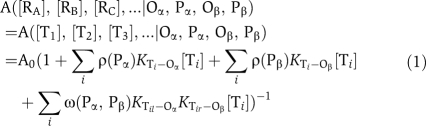 |
Here, A0 is the activity of unrepressed promoter. The summations are over all possible tetramers.
The middle two summations in the parentheses in equation (1) correspond to contributions of repressor binding at mono-operator site. The function ρ (Px), x corresponding to either α or β, denotes the dependence on operator position. The KTi−Ox is the effective association constant between Ti with Ox. A tetramer can uses either of its two composing dimers (designated as Til and Tir, respectively) to bind to Ox. We assume KTi−Ox=KTil−Ox+KTir−Ox, KTil−Ox and KTir−Ox corresponding to dimer-operator association constants.
The last term in the parentheses in equation (1) corresponds to simultaneous binding of Til to Oα and of Tir to Oβ. The function ω (Pα, Pβ) denotes the inter-operator distance dependence of DNA looping accompanied by repressor binding (Oehler et al, 1990; Muller et al, 1996).
Equation (1) describes a promoter containing two operators with selected binding specificity to different TF variants to achieve combinatorial effects. The main design parameters are the absolute and relative positions of the operators. If a single operator composed of two half-sites with different TF specificity is used, the terms involving Oβ and (Oα, Oβ) in equation (1) disappear, leaving the unselected binding constants associated with different TF homo- and heterodimers to the hybrid operator as the main design parameters.
Details of parameterizing the model based on experimentally derived knowledge about the LacI/Olac system as well as final parameters are provided in Supplementary information. Briefly, the function ρ (Px) has been determined by measuring experimentally the repression strength, putting a single operator at different positions (Supplementary Figure S1). The function ω (Pα, Pβ) has been determined by fitting to experimental date reported in Sayut et al (2009). Three categories of dimer-operator association constants have been discriminated in the model: strong associations, in which both half-sequences in Ox are recognized by the repressor dimer; weak associations, in which only one half-sequence of the Ox is recognized by a monomer in the dimer; and negligible associations, in which neither half-sequence of Ox is recognized by any monomer in the dimer.
Simulations using equation (1) with different promoter structures and repressor concentrations have been performed (Supplementary Figures S2–S5). Especially, comparisons between Supplementary Figures S3 and S4 show the effects of assymetrizing one operator sequence in the NAND gate, and Supplementary Figure S6 shows the influences of the interfering signal RC, on the performance of the NAND gate.
The bacteria strains and plasmids
The in vivo experiments have been performed using the Escherichia coli strain TOP10 for screening the sequence variants and the E. coli lac− strain MC4100 for testing the designed components. Three types of plasmids have been constructed in this work. The first type, including pUC-lacI, pDR-Ri and pDR-RiRj, have been used to express the repressor variants. Each of these plasmids contains a sequence coding for one or two lacI variants controlled by a constitutively activated tet promoter. In pUC-lacI, the coding sequence is from the gene library of LacI mutants. In pDR-Ri or pDR-Ri-Rj, the coding sequence Ri or Rj corresponds to one of the LacI variants from R1 to R6. The second type, including those derived from pZS*-lacZα and pDPv2F, have been used to report the activity of designed promoters. In each of those reporter plasmids derived from pZS*-lacZα, a designed promoter controls the expression of a lacZ α fragment used as a reporter. In those derived from pDPv2F, a designed promoter additionally controls the expression of a reporter green fluorescence protein gene GFPmut3b. The third type of plasmids, including pDEMO-4, have been used in the demonstration compound logic circuit. More details of the plasmids and their construction processes can be found in the Supplementary information.
Tests of the logic devices and the compound logic circuit
The input signals have been generated by plasmids expressing the respective LacI variants. To test the response of a given device or circuit to given inputs, the reporting plasmid carrying both the designed promoter and the downstream reporter genes was first transformed into MC4100. Then, the plasmid expressing the respective input LacI variants was transformed into the same strain. For example, to test the NAND response of the promoter containing an upstream Olac2 operator at −83 and a downstream weakened Olac4 operator at +68, the input plasmids pDR-NUL (input=00), pDR-R2 (input=01), pDR-R4 (input=10) and pDR-R2R4 (input=11) were, respectively, transformed into the corresponding strain carrying the reporting plasmid pDPV2F-PU83Olac2+D68Oweak4. The activities of the promoter in each strain were determined separately. An alternative strategy is to test the responses to the four different inputs in the same strain. Then one would have to put the expression of R2 and R4 under different types of controls. More importantly, these controls must lead to the same expression level for R2 and R4 at both of their ON and OFF states, respectively. To achieve and confirm the fulfillment of this condition is, although possible, non-trivial and out of focus of the current report.
Fluorescence measurements
The outputs of the designed devices and circuit have been measured as cellular fluorescence strengths. Cells were grown in M9 medium supplemented with 0.5% glucose, 0.1% casamino acids, 1 mM thiamine hydrochloride, 50 μg/ml ampicillin and 20 μg/ml kanamycin. Each output has been obtained by growing cells in four tubes in parallel, taking samples at 3 or 4 time points from each tube for immediate fluorescence readings (much more points have been sampled on the demo systems to determine the growth rates, see source data of the figures), performing a linear fit of fluorescence readings versus OD600 using all data points with OD600 between 0.2 and 0.9, and reporting the slope as the observed transcriptional activity in figures presenting the results. Error ranges are reported as root mean square residuals of the fitting. The OD600 was determined using an Eppendorf BioPhotometer. The fluorescence intensity was measured using a spectrofluorophotometer (RF-5301PC, SHIMADZU). We used a 488-nm excitation filter in combination with a 512-nm emission filter to measure GFP fluorescence, and a 584-nm excitation filter in combination with a 607-nm emission filter to measure RFP fluorescence. Under our experimental conditions, we observed cell density-dependent ‘background,' namely, non-zero ‘fluorescence' readings for cells free of GFP or RFP expressions. The experimentally determined background values using a fluorescent protein-free strain can be represented well as linear functions of OD600. They have been subtracted from the actual readings before the linear fit.
Supplementary Material
Acknowledgments
We thank Drs Zhonghuai Hou, Lingchong You, Xiaolian Gao and Lianhong Sun for discussions and suggestions. We also thank Jichao Wang for technical support. The plasmid pZS*21-MCS1 was kindly provided by H Bujard. The plasmid pSB3K5 and pSB1A3, and the BioBricks parts R0011, R0040, B0034, E0040 and E1010 were from the iGEM 2007 Parts Kit provided by the Registry of Standard Biological Parts. This work has been supported by grant 2006AA02Z303 from the Chinese Ministry of Science and Technology and grant 30670485 from the Chinese Natural Science Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson JC, Voigt CA, Arkin AP (2007) Environmental signal integration by a modular AND gate. Mol Syst Biol 3: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ Jr, Stoddard BL, Baker D (2006) Computational redesign of endonuclease DNA binding and cleavage specificity. Nature 441: 656–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson MR, Savageau MA, Myers JT, Ninfa AJ (2003) Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113: 597–607 [DOI] [PubMed] [Google Scholar]
- Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L (2008) A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol 4: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R (2005) Transcriptional regulation by the numbers: models. Curr Opin Genet Dev 15: 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson JE, Mazur WW, Cornish VW (2008) Transcription factor logic using chemical complementation. Mol Biosyst 4: 56–58 [DOI] [PubMed] [Google Scholar]
- Buchler NE, Gerland U, Hwa T (2003) On schemes of combinatorial transcription logic. Proc Natl Acad Sci USA 100: 5136–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RS III, Surette MG, Elowitz MB (2007) Programming gene expression with combinatorial promoters. Mol Syst Biol 3: 14510.1038/msb4100187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ, Davis RW (1989) Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev 3: 185–197 [DOI] [PubMed] [Google Scholar]
- Ellis T, Wang X, Collins JJ (2009) Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat Biotechnol 27: 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S (2000) A synthetic oscillatory network of transcriptional regulators. Nature 403: 335–338 [DOI] [PubMed] [Google Scholar]
- Friedman AM, Fischmann TO, Steitz TA (1995) Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science 268: 1721–1727 [DOI] [PubMed] [Google Scholar]
- Hasty J, McMillen D, Collins JJ (2002) Engineered gene circuits. Nature 420: 224–230 [DOI] [PubMed] [Google Scholar]
- Kalodimos CG, Bonvin AM, Salinas RK, Wechselberger R, Boelens R, Kaptein R (2002) Plasticity in protein-DNA recognition: lac repressor interacts with its natural operator 01 through alternative conformations of its DNA-binding domain. EMBO J 21: 2866–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopke Salinas R, Folkers GE, Bonvin AM, Das D, Boelens R, Kaptein R (2005) Altered specificity in DNA binding by the lac repressor: a mutant lac headpiece that mimics the gal repressor. Chembiochem 6: 1628–1637 [DOI] [PubMed] [Google Scholar]
- Krueger M, Scholz O, Wisshak S, Hillen W (2007) Engineered Tet repressors with recognition specificity for the tetO-4C5G operator variant. Gene 404: 93–100 [DOI] [PubMed] [Google Scholar]
- Lewis M (2005) The lac repressor. C R Biol 328: 521–548 [DOI] [PubMed] [Google Scholar]
- Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P (1996) Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science 271: 1247–1254 [DOI] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Muller-Lerch F, Fu F, Pearlberg J, Gobel C, Dassie JP, Pruett-Miller SM, Porteus MH, Sgroi DC et al. (2008) Rapid ‘open-source' engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 31: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Oehler S, Muller-Hill B (1996) Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J Mol Biol 257: 21–29 [DOI] [PubMed] [Google Scholar]
- Oehler S, Eismann ER, Kramer H, Muller-Hill B (1990) The three operators of the lac operon cooperate in repression. EMBO J 9: 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler JR, Sasmor H, Betz JL (1983) A perfectly symmetric lac operator binds the lac repressor very tightly. Proc Natl Acad Sci USA 80: 6785–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorius J, Lehming N, Kisters B, von Wilcken-Bergmann B, Muller-Hill B (1989) lac repressor mutants with double or triple exchanges in the recognition helix bind specifically to lac operator variants with multiple exchanges. EMBO J 8: 1265–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayut DJ, Niu Y, Sun L (2009) Construction and enhancement of a minimal genetic and logic gate. Appl Environ Microbiol 75: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnappinger D, Schubert P, Pfleiderer K, Hillen W (1998) Determinants of protein-protein recognition by four helix bundles: changing the dimerization specificity of Tet repressor. EMBO J 17: 535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spott S, Dong F, Kisters-Woike B, Muller-Hill B (2000) Dimerisation mutants of Lac repressor. II. A single amino acid substitution, D278L, changes the specificity of dimerisation. J Mol Biol 296: 673–684 [DOI] [PubMed] [Google Scholar]
- Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J (2008) A fast, robust and tunable synthetic gene oscillator. Nature 456: 516–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigon D, Coleman BD, Olson WK (2006) Modeling the Lac repressor-operator assembly: the influence of DNA looping on Lac repressor conformation. Proc Natl Acad Sci USA 103: 9879–9884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SY, Fazelinia H, Cirino PC (2008) AraC regulatory protein mutants with altered effector specificity. J Am Chem Soc 130: 5267–5271 [DOI] [PubMed] [Google Scholar]
- Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M (2009) A tunable synthetic mammalian oscillator. Nature 457: 309–312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.