Abstract
Systems biology can offer a great deal of insight into evolution by quantitatively linking complex properties such as protein structure, folding, and function to the fitness of an organism. Although the link between diseases such as Alzheimer's and misfolding is well appreciated, directly showing the importance of protein folding to success in evolution has been more difficult. We show here that predicting success during adaptation can depend critically on enzyme kinetic and folding models. We used a ‘weak link’ method to favor mutations to an essential, but maladapted, adenylate kinase gene within a microbial population that resulted in the identification of five mutants that arose nearly simultaneously and competed for success. Physicochemical characterization of these mutants showed that, although steady-state enzyme activity is important, success within the population is critically dependent on resistance to denaturation and aggregation. A fitness function based on in vitro measurements of enzyme activity, reversible and irreversible unfolding, and the physiological context reproduces in vivo evolutionary fates in the population linking organismal adaptation to its physical basis.
Keywords: adenylate kinase, enzyme kinetics, experimental evolution, fitness functions, protein folding
Introduction
The success or failure of organisms during adaptation is based on changes in molecular structure that give rise to changes in fitness. Although the conceptual link between genotype, phenotype, and fitness is clear, the ability to relate these complex adaptive landscapes in a quantitative manner remains difficult (Kacser and Burns, 1981; Dykhuizen et al, 1987; Weinreich et al, 2006). Experimental evolution has allowed the exploration of adaptation under controlled settings that often brings together both in vivo and in vitro approaches. The success of experimental evolution in examining the adaptive landscape accessible to organisms is highlighted by the work of Lenski and co-workers, where over the course of 40 000 generations, they mapped the changes in organismal fitness to glucose and citrate usage (Blount et al, 2008) as well as a series of studies that have explored adaptive landscapes in both bacteria and viruses (Bull et al, 2003; Elena and Lenski, 2003; Knies et al, 2006; Rokyta et al, 2006). In contrast, biophysical chemists have typically used more in vitro approaches such as error-prone PCR to generate libraries of molecules whose functional properties can be assessed through screening and intensive physicochemical characterization (Wintrode et al, 2003; Silberg et al, 2004; Bloom et al, 2005a; Lunzer et al, 2005; Bershtein et al, 2006; Miller et al, 2006; Fasan et al, 2008; Bloom and Arnold, 2009). Dean and Thornton (2007) noted the convergence of these approaches and coined the term ‘functional synthesis’ to capture the synergy between evolutionary and molecular biology to address important questions such as the evolution of complexity and antibiotic resistance. The ‘functional synthesis,’ in its most fully realized form, is an integrated systems biology approach to evolutionary dynamics that links physicochemical properties of molecules to evolutionary fates in a quantitative and predictive manner.
Functional synthesis flourishes in an experimental framework that allows investigators to directly link population dynamics (fitness) to changes in molecular function that result from alterations at the nucleotide level. The ‘weak link’ approach was developed to tightly couple adaptive changes within the genome to changes in fitness and provide a population-based approach that can be used to examine alterations in function and fitness at the level of atomic structure and function (Counago and Shamoo, 2005; Counago et al, 2006). A homologous recombination strategy was used to replace the chromosomal copy of the essential adenylate kinase gene (adk) of the thermophilic bacterium Geobacillus stearothermophilus with that of the mesophile Bacillus subtilis. Recombinant G. stearothermophilus cells that expressed only B. subtilis adenylate kinase (AKBSUB) were unable to grow at temperatures higher than 55°C because of heat inactivation of the mesophilic enzyme and consequent disruption of adenylate homeostasis (Counago and Shamoo, 2005). A continuously growing population of bacteria was then subjected to selection at increasing temperatures (from 55 to 70°C) that favored changes in the one gene not adapted for thermostability, adk. During the course of selection, the population was sampled and intermediates of adaptation were observed as mutations to adk and examined to determine how the mutant populations traversed the adaptive landscape to increased fitness (Counago et al, 2006). Adaptation to increased thermo-tolerance by Geobacillus in our system is analogous to adaptation of mesophiles to higher temperature niches (Berezovsky and Shakhnovich, 2005). An initial single mutation (AKBSUB Q199R), which broadens AK activity at higher temperatures, became fixed in the bacterial population (Counago et al, 2008). Subsequently, five double mutants with varying degrees of success arose nearly simultaneously within the population as the temperature increased further (Figures 1 and 4C) (Counago et al, 2006). Our initial attempts to find the physicochemical mechanism for the relative success and failure of each mutant allele based on the thermal stability and activity of the mutated enzymes were qualitatively reasonable. However, these interpretations failed to predict quantitatively the outcome of natural selection on the microbial population (Counago et al, 2006). To correctly predict the evolutionary outcomes within the population, it was necessary to accurately determine the extent and rate of both reversible and irreversible unfolding of each new mutant as well as its catalytic parameters. The resultant analysis shows that the winners and losers of evolution can be critically dependent on both protein activity and folding dynamics.
Figure 1.
Structure of AKBSUB Q199R (purple ribbons for the main chain) complexed to the transition-state inhibitor Ap5A (sticks) (PDB ID: 2eu8). The positions of the Cα atoms of the five adaptive mutations that conferred additional fitness at higher temperatures are highlighted as AKBSUB Q199R/Q16L, red (PDB ID: 2osb); AKBSUB Q199R/T179I (PDB ID: 2oo7), yellow; AKBSUB Q199R/A193V, green (PDB ID: 2ori); AKBSUB Q199R/G213E, blue (PDB ID: 2qaj); and AKBSUB Q199R/G214R, orange (PDB ID: 2p3s).
Results
Enzyme kinetics
The primary function of AK in vivo is to maintain adenylate homeostasis by catalyzing the reaction:  We determined the catalytic activity of the adapted AK mutants using an in vitro end-point assay. The rate of ADP production was measured at saturating concentrations of AMP and varying concentrations of ATP (Saint Girons et al, 1987; Glaser et al, 1992; Counago et al, 2008). To determine the dependence of catalytic rate on temperature, AK activity was measured at 55, 60, 62.5, 65, 67.5, and 70°C for each mutant. At lower temperatures, the reaction rates show a hyperbolic Michaelis–Menten dependence on [ATP] for all the mutants examined; however, at higher temperatures, distinctly non-hyperbolic kinetics are observed (Figure 2). For example, the steady-state kinetic pattern for AKBSUB Q199R/A193V activity changes markedly as a function of temperature, showing a sigmoidal dependence on substrate concentration above 65°C. At these elevated temperatures, the AK activity is close to zero at low [ATP] but is restored at higher substrate concentrations. Clearly, ATP/AMP binding stabilizes the active form of the enzyme. Substrate stabilization, along with macromolecular crowding and chaperone activity, supports our observation that many of the AK mutants are viable in vivo at temperatures in which the purified enzyme is largely denatured in vitro in the absence of ATP and AMP.
We determined the catalytic activity of the adapted AK mutants using an in vitro end-point assay. The rate of ADP production was measured at saturating concentrations of AMP and varying concentrations of ATP (Saint Girons et al, 1987; Glaser et al, 1992; Counago et al, 2008). To determine the dependence of catalytic rate on temperature, AK activity was measured at 55, 60, 62.5, 65, 67.5, and 70°C for each mutant. At lower temperatures, the reaction rates show a hyperbolic Michaelis–Menten dependence on [ATP] for all the mutants examined; however, at higher temperatures, distinctly non-hyperbolic kinetics are observed (Figure 2). For example, the steady-state kinetic pattern for AKBSUB Q199R/A193V activity changes markedly as a function of temperature, showing a sigmoidal dependence on substrate concentration above 65°C. At these elevated temperatures, the AK activity is close to zero at low [ATP] but is restored at higher substrate concentrations. Clearly, ATP/AMP binding stabilizes the active form of the enzyme. Substrate stabilization, along with macromolecular crowding and chaperone activity, supports our observation that many of the AK mutants are viable in vivo at temperatures in which the purified enzyme is largely denatured in vitro in the absence of ATP and AMP.
Figure 2.
Enzyme activity of adaptive mutants AKBSUB Q199R, Q199R/A193V, and Q199R/Q16L reveal a critical function for reversible and irreversible folding. The initial velocity is defined as the concentration of ADP produced in 1 min by 1 nm of AK. ADP production was determined at 5, 10, 25, 50, 100, 250, 500, and 1000 μM ATP with constant 1400 μM AMP. Symbols represent observed data points. Plots of initial velocity versus [ATP] were fit to Equation (1) and the fitted curves are presented as solid lines. The non-hyperbolic and sigmoidal curves observed at elevated temperatures are characteristic of an irreversible-unfolding pathway. Error bars represent the s.d. of three measurements.
The distinct, sigmoidal shape of the reaction curves at higher temperatures indicates strongly that AK activity is lost by irreversible unfolding or aggregation in the absence of high concentrations of ATP. The observed sigmoidal, non-Michaelian kinetics can only be modeled assuming an irreversible reaction that directs active enzyme away from the catalytic pathway in competition with substrate binding (Scheme I) (Thomas and Scopes, 1998). The simplest mechanism that can explain the observed kinetics is shown in Scheme I below:
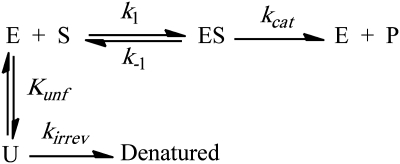 |
Scheme I
In this mechanism, the substrate-free enzyme in its active, native state, E, can reversibly and rapidly unfold into an inactive state, U. The equilibrium constant defining this unfolding reaction is Kunf, which is equal to [U]/[E]. This equilibrium ratio is unchanged throughout the reaction even though the absolute values of [U] and [E] are changing with both time and [S]. In effect, unfolding is competitively inhibiting substrate binding and can be overcome at high [S]. The net result is that increases in Kunf increase the apparent KM for the reaction (i.e. KM(obs)=KM(1+Kunf)). However, this effect alone cannot explain the sigmoidal kinetics observed at high temperatures and low [S]. In our experiments, unfolded AK aggregates and becomes irreversibly denatured at a rate proportional to the fraction of unfolded enzyme, YU, during the assay. This in vitro process was modeled as an exponential decay of the U state, that is exp(−kirrev·t·YU) (see Supplementary Information). The final initial velocity equation is
where
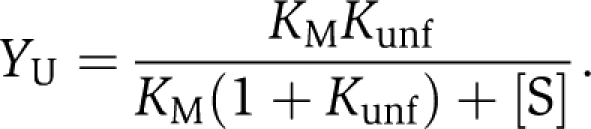 |
The amounts of ADP formation at 20, 40, and 60 s time points were fitted to a straight line and the slope was used to determine initial steady-state velocities, v. Plots of v versus [ATP] were fitted to Equation (1), allowing kcat, KM, and kirrev to vary (Figure 2). The unfolding equilibrium constants at different temperatures were determined independently by differential scanning calorimetry (DSC). As the AK reaction is a bisubstrate reaction with a ternary complex mechanism, the values of kcat and KM in Equation (1) depend on [AMP], which was fixed at 1400 μM for all reactions.
As shown in Figure 2, Equation (1) predicts simple Michaelis–Menten kinetics at low temperatures when Kunf and YU are very small and exp(−kirrev·t·YU)≈1. However, when the temperature is elevated and Kunf becomes very large, the observed activity at low [ATP] is dramatically reduced by irreversible denaturation during the time interval between initiation of the reaction and the first measurement of product formation, ∼40 s. However, at high [ATP], the fraction of unfolded protein is reduced markedly because of ES complex formation and reduction in the fraction of the U state. As a result, irreversible denaturation is inhibited, enzymatic activity is retained, and the kinetics seem sigmoidal with increasing [S]. Substrate stabilization occurs through competition for free enzyme between the catalytic and unfolding pathways in a substrate concentration-dependent manner. At high substrate concentrations, the catalytic pathway out competes the unfolding pathway. The fraction of unfolded protein, YU in Equation (1), is 0.94 for AKBSUB Q199R in the absence of substrate at 55°C, whereas in the presence of 1000 μM ATP, YU is only 0.02. This dramatic effect shows the significant influence of substrate on the ensemble of folding states and the activity of the enzyme both in vitro and presumably in vivo.
Fits of the observed data to Equation (1) are remarkably good (Figure 2), particularly considering that Kunf was determined independently, and allow determination of kcat, KM, and kirrev for the AK mutants that appear during adaptation to high temperatures (Table I). In the case of the AKBSUB Q199R/Q16L mutant, the observed kinetics showed high activity that depends almost linearly on the concentration of ATP, making it difficult to define the individual kcat, KM, and kirrev values. The linear kinetics for this mutant indicate that the reaction is effectively bimolecular with kcat≫k−1, and, therefore, the enzyme is not readily saturated at accessible concentrations of ATP.
Table 1. Kinetic and folding parameters with s.d. for AKBSUB mutants at various temperatures determined by non-linear fits of reaction rates as a function of ATP concentrationa.
| Enzyme | T (°C) | Kinetics |
Folding |
||||
|---|---|---|---|---|---|---|---|
| kcat (s−1) | KM (μM) | kcat/KM (μM−1 s−1) | vi for [S]=1000 μM (μM min–1 nM–1 AK) | K unf b | kirrevc (s−1) | ||
| aThe kinetic parameters for B. subtilis adenylate kinase mutants were fit to triplicate data with a quasi-steady state approximation (Equation (1)). | |||||||
| bKunf was calculated using parameters determined by DSC. | |||||||
| ckirrev is fit as one value shared across temperatures. | |||||||
| dThe enzyme has no measureable activity at the specified condition. | |||||||
| eA value of 0.01 was used as a constant in the fitting scheme. | |||||||
| fValue could not be determined. | |||||||
| Q199R | 55 | 14.2±0.2 | 1.4±0.3 | 9.9±1.8 | 13.2±1.0 | 16.6±3.3 | 0.07±0.01 |
| 60 | 17.0±0.7 | 0.9±0.2 | 18.7±4.1 | 12.2±0.8 | 103±34 | ||
| 62.5 | 29.9±12.5 | 2.0±1.1 | 14.6±10.0 | 7.1±0.3 | 269±105 | ||
| 65 | 13.2±224 | 2.0±26.9 | 6.6±140 | 1.3±0.4 | 701±324 | ||
| 67.5 | d | d | d | d | 1880±1030 | ||
| 70 | d | d | d | d | 5120±3150 | ||
| Q199R/G213E | 55 | 10.0±0.2 | 2.2±0.4 | 4.5±0.9 | 9.7±0.7 | 8.1±2.5 | 0.06±0.01 |
| 60 | 11.2±0.3 | 1.0±0.2 | 11.6±2.4 | 10.0±0.6 | 36.5±21.5 | ||
| 62.5 | 12.9±0.6 | 1.4±0.3 | 9.3±2.2 | 9.1±0.9 | 79.9±62.3 | ||
| 65 | 23.9±9.2 | 3.1±1.7 | 7.7±5.1 | 6.4±0.7 | 175±159 | ||
| 67.5 | d | d | d | d | 392±441 | ||
| 70 | d | d | d | d | 893±1140 | ||
| Q199R/T179I | 55 | 9.9±0.2 | 1.8±0.2 | 5.7±0.8 | 9.7±0.2 | 6.02±1.20 | 0.09±0.01 |
| 60 | 13.7±0.2 | 0.7±0.1 | 20.5±2.8 | 12.6±0.9 | 32.4±14.7 | ||
| 62.5 | 20.9±0.6 | 0.8±0.1 | 26.6±4.0 | 16.4±0.1 | 76.4±43.5 | ||
| 65 | 17.1±1.2 | 0.9±0.2 | 19.3±3.8 | 8.8±0.5 | 181±127 | ||
| 67.5 | 49.6±198 | 2.5±6.9 | 20.2±98.2 | 3.8±1.2 | 429±355 | ||
| 70 | d | d | d | d | 1020±987 | ||
| Q199R/G214R | 55 | 9.5±0.2 | 3.5±0.7 | 2.7±0.5 | 9.3±1.0 | 4.57±0.21 | 0.06±0.01 |
| 60 | 12.5±0.3 | 1.3±0.3 | 9.8±2.0 | 11.2±1.1 | 30.0±3.6 | ||
| 62.5 | 14.5±0.9 | 1.5±0.4 | 9.7±2.4 | 10.0±0.5 | 77.5±14.0 | ||
| 65 | 39.4±39.9 | 4.1±4.3 | 9.6±13.9 | 6.5±0.9 | 207±55 | ||
| 67.5 | d | d | d | d | 527±156 | ||
| 70 | d | d | d | d | 1390±488 | ||
| Q199R/A193V | 55 | 15.1±0.3 | 8.2±0.9 | 1.9±0.2 | 14.4±1.2 | 2.36±0.29 | 0.08±0.01 |
| 60 | 18.6±0.3 | 1.6±0.2 | 11.3±1.3 | 17.1±0.6 | 17.1±5.2 | ||
| 62.5 | 23.9±0.5 | 1.3±0.2 | 18.7±2.4 | 19.0±0.7 | 47.3±19.1 | ||
| 65 | 33.7±2.5 | 1.6±0.3 | 21.3±3.9 | 16.1±0.4 | 132±67 | ||
| 67.5 | 70.9±229 | 3.1±7.7 | 22.7±92.0 | 6.2±1.0 | 381±236 | ||
| 70 | d | d | d | d | 1100±788 | ||
| Q199R/Q16L | 55 | 57.1±1.3 | 561.9±24.9 | 0.10±0.01 | 34.5±1.1 | 0.13±0.06 | 0.01e |
| 60 | 60.6±1.5 | 372.5±16.6 | 0.16±0.01 | 34.7±0.7 | 0.72±0.10 | ||
| 62.5 | 65.0±2.4 | 249.6±16.2 | 0.26±0.02 | 34.9±1.6 | 1.74±0.08 | ||
| 65 | 69.3±2.7 | 169.1±10.2 | 0.41±0.03 | 31.2±0.8 | 4.37±0.91 | ||
| 67.5 | 128.0±13.4 | 248.3±30.7 | 0.52±0.08 | 24.5±0.6 | 11.0±4.1 | ||
| 70 | f | f | f | 7.6±0.4 | 28.4±15.4 | ||
A continuous function for activity with respect to temperature for AK that incorporates both reversible and irreversible protein folding is given in Equation (2). A temperature-dependent function for activity is favored over an interpolation between measured activities because it provides a model that can be tested against evolutionary outcomes under various temperature regimens and provides a continuum of values that can be referenced by a fitness function (Figure 3). The temperature dependence of the catalytic constants and unfolding equilibria are expressed as:
Figure 3.
The temperature-dependent activity of the mutants AKs are very well reproduced by a continuous function that includes reversible and irreversible folding (Equation (2)). The modeled reaction rates for various mutant AKs as a function of temperature are shown as solid lines and were used in the population simulation. Experimentally measured enzyme activities are represented by circles with s.d.
Linear fits of Eyring plots (1/T versus ln(kcat/T)) of the catalytic constants (kcat)) of each mutant were used to construct a temperature-dependent function for catalytic constants:
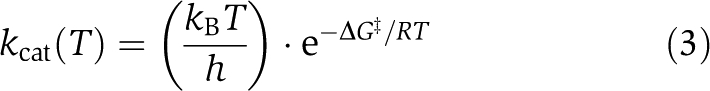 |
where kB is Boltzmann's constant, h is Plank's constant, R is the gas constant, and ΔG‡ is the free energy of activation. The kcat values at the highest temperature in which activity is often barely detectable were not used in these analyses because of the large uncertainties associated with the fitted values.
The calculated Gibbs-free energy of unfolding (ΔGunf) determined for temperatures between 55 and 70°C were fit to a linear function in temperature. Kunf was calculated as:
The temperature dependence of the fraction of unfolded enzyme (YU) was computed as:
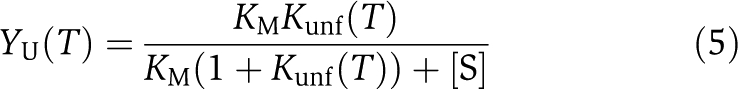 |
KM is a complex term that can vary non-linearly with temperature because of the temperature dependence of three constituent rate constants: k1, k−1, and kcat. The measured KMs were relatively independent of temperature and thus averaged across experimental temperatures for a given mutant and used as a constant in the continuous temperature-dependent activity function. The linearity seen by Arrhenius plots (1/T versus ln(KM(obs))) in our earlier studies can be accounted for by the temperature dependence of the unfolding equilibrium constant (Kunf) in the expression for the observed Michaelis constant (KM(obs)=KM(1+Kunf)) (Counago et al, 2008). kirrev is considered as a constant over the temperature range examined.
Thermal stabilities of mutant AKs
Experimental parameters obtained by DSC (midpoint, enthalpy change at Tm, and heat capacity change at Tm) were used to calculate the Gibbs-free energy change for unfolding (ΔGunf) at each temperature (Figure 4A; Table II) (Becktel and Schellman, 1987). ΔGunf was then used to calculate the unfolding equilibrium constants (Kunf=exp(−ΔGunf/RT)) for use in analyzing the steady-state kinetic data with Equation (1) (Table I).
Figure 4.
In vitro activity and stability can be used to accurately reproduce experimentally determined evolutionary outcomes during natural selection. (A) Stabilities of B. subtilis wild type and mutant AK as determined by DSC are presented here as the heat capacity as a function of temperature. The double mutants have midpoints or Tm values at higher temperatures compared with the AKBSUB Q199R single mutant, which itself is ∼3°C more stable than wild type (Tm values are given in parentheses in panel A). The DSC data were used in determining Kunf for each mutant at various temperatures. (B) The kinetic (kcat and KM) and folding (kirrev and Kunf) terms were used to fit ADP production rates for each mutant at 1000 μM ATP as a function of temperature. (C) The cumulative frequency of alleles obtained by experimental evolution within an exponentially growing population as a function of temperature. (D) The frequency of alleles determined from numerical modeling of the exponentially growing population using in vitro measurements. Above an activity threshold of 8.75 μM ADP min–1 nM–1, relative increases in activity do not correspond to increases in growth rate and increased fitness. Below the threshold, there is a direct correspondence between growth rate and the AK activity presented in panel B. The colors used in panels A–D correspond to the color scheme used to identify the positions of the mutations in Figure 1.
Table 2. Calculated Gibbs-free energy change of unfolding, ΔGunf (kcal mol–1), at respective temperatures for each mutant with s.d.a.
| Enzyme | Temperature (°C) |
|||||
|---|---|---|---|---|---|---|
| 55 | 60 | 62.5 | 65 | 67.5 | 70 | |
| aΔG was calculated for each mutant at the listed temperature using Equation (S4) and the parameters listed in Supplementary Table SI. The s.d. is determined for three calculated values of ΔG based on three independent DSC measurements for each mutant. | ||||||
| Q199R | −1.83±0.13 | −3.07±0.22 | −3.73±0.26 | −4.40±0.31 | −5.10±0.37 | −5.82±0.42 |
| Q199R/G213E | −1.36±0.20 | −2.38±0.39 | −2.92±0.52 | −3.47±0.61 | −4.04±0.76 | −4.63±0.87 |
| Q199R/T179I | −1.17±0.13 | −2.30±0.30 | −2.89±0.38 | −3.49±0.47 | −4.10±0.56 | −4.72±0.66 |
| Q199R/G214R | −0.99±0.03 | −2.25±0.08 | −2.90±0.12 | −3.58±0.18 | −4.24±0.20 | −4.93±0.24 |
| Q199R/A193V | −0.56±0.08 | −1.88±0.20 | −2.57±0.27 | −3.28±0.34 | −4.02±0.42 | −4.77±0.49 |
| Q199R/Q16L | 1.35±0.30 | 0.22±0.09 | −0.37±0.03 | −0.99±0.14 | −1.62±0.25 | −2.28±0.37 |
AK unfolding can be well modeled as an apparent two-state transition, although re-folding is hampered by aggregation and partial loss of an intrinsic zinc ion (Bae and Phillips, 2004; Miller et al, 2010). The thermodynamic parameters for each protein were determined as a function of concentration and scan rate to assess the extent to which AK and the associated mutants can be reasonably modeled as an apparent two-state transition (Supplementary Information). All of the isolated mutants have increased stability with shifts in Tm to higher temperatures relative to wild-type AK (Figure 4A). As temperature is increased further, the instability or unfolding constants of the mutant proteins correlate well with the order in which their respective allelic frequencies decrease. The first mutant to appear, AKBSUB Q199R, has a modest 2.8°C increase in Tm compared with the original wild-type enzyme (Tm=44.0°C). Subsequent double mutants, AKBSUB Q199R/G213E, AKBSUB Q199R/T179I, AKBSUB Q199R/G214R, and AKBSUB Q199R/A193V, provide 1–6° of additional stabilization, with Tm values equal to 48.0, 49.5, 51.0, and 52.8°C, respectively. The very successful AKBSUB Q199R/Q16L mutant is dramatically more stable, with a Tm=60.9°C, which is 10°C greater than that for its AKBSUB Q199R progenitor.
Stabilization of AK by ligand binding
Enzyme kinetics and calorimetric data fitted to Scheme I (Table I) can be used to estimate the extent of stabilization that should be induced when 50% of the protein is in complex with substrate. As a test of these estimates, DSC was used to experimentally measure the amount of stabilization induced by substrate binding (Figure 5). To preclude enzyme activity during the experiment, the transition-state analog Ap5A was used to measure substrate stabilization. DSC measurements at 0.25:1, 0.5:1, and 1:1 stoichiometric ratios of Ap5A to AKBSUB Q199R/A193V (Figure 5A) were performed. Under very tight binding conditions, two populations with two melting temperatures corresponding to unbound protein (To) and the protein Ap5A complex (Tm) are observed (Figure 5A) (Brandts and Lin, 1990). Ap5A affinity was estimated to be about 60 nM at 64.3°C (Supplementary Information). Ap5A increased AKBSUB Q199R/A193V stability by 12.3±1°C. AKBSUB Q199R/A193V showed little change in Tm as a function of Ap5A concentration for the bound state (as expected from Brandts and Lin, 1990). Knowing the affinity of Ap5A for AK and the Tm (Supplementary Table S4) allowed us to compare the measured stabilization to that predicted from Supplementary Equation (S8) and is in excellent agreement with the observed data (Table III). Conversely, this data can be used to predict the amount of Ap5A that is required to shift the Tm of AKBSUB Q199R/A193V to 64.3°C. The predicted concentration of Ap5A would be 12 μM in excellent agreement with the experimental value of 10 μM Ap5A.
Figure 5.
The extent of ligand induced stabilization monitored by DSC for AKBSUB Q199R/A193V corresponds to values predicted by Scheme I (Supplementary Equation (S6)). (A) Tight binding of the transition-state analog Ap5A shows distinct peaks with Tms corresponding to the unbound and bound states in the expected stoichiometric ratios. (B) In the weaker binding case of nucleotides ATP and AMP, protein stabilization is observed as a single peak that has an increase in Tm that is dependent on ligand concentration (Brandts and Lin, 1990). Greater extent of stabilization is achieved with ATP relative to AMP because of a greater affinity for this substrate.
Table 3. Comparison of Tm values for AKBSUB Q199R/A193V as determined by DSC in the presence of various ligands and those predicted by Scheme I and Supplementary Equation S8 (Supplementary Information).
| Ligand | KBa (M−1) | [S]b (M) |
TMc (°C) |
|
|---|---|---|---|---|
| Observed | Predicted | |||
| aKB is the binding constant determined at 20 μM protein at a scan rate of 90°C h–1. | ||||
| bEstimation of the concentration of free ligand in the presence of 20 μM protein. | ||||
| cTM is the thermal midpoint of a transition in which approximately 50% of the ensemble is folded and bound in the presence of ligand. | ||||
| Protein alone | — | — | 52.0 | — |
| +Ap5A | 1.6 × 107 | 1 × 10−5 | 64.3 | 65.4 |
| +AMP | 7.6 × 102 | 1 × 10−3 | 53.4 | 54.4 |
| +ATP | 1.7 × 104 | 1 × 10−3 | 59.1 | 60.0 |
A study of stabilization of AKBSUB Q199R/A193V by ATP and AMP showed comparable results, although these measurements should only be regarded as estimates because of enzyme activity (Figure 5B; see Supplementary Information for further discussion). Again after Scheme I, a solution of 20 μM AKBSUB Q199R/A193V, incubated with either 1 mM ATP or 1 mM AMP, would be expected to be stabilized by 8.0 and 2.4°C, respectively (Table III). The extent of stabilization measured by DSC for AKBSUB Q199R/A193V in the presence of 1 mM ATP or 1 mM AMP was 7.1 and 1.4°C in very good agreement with the expected extent of stabilization.
Fitness function and population dynamics
The biochemical and structural properties of the AKBSUB mutants provide the molecular basis for different physiological and metabolic phenotypes that are tightly linked to the fitness of the organism within the population. A single mutant and five double mutant alleles of adk were isolated during experimental evolution of a continuous microbial population under exponential growth conditions (Figure 4C) (Counago et al, 2006). Relative growth rates (fitnesses) are reflected by changes in frequencies of mutants within the polymorphic population over time. A fitness function consisting of temperature-dependent kinetic and folding parameters was constructed and provides a biochemical description that can be used to describe quantitatively the experimentally observed population frequencies. This function also includes the particular properties of the turbidostat, provides a function for mutation rate in seeding the mutant subpopulations, and uses the product formation expression in Equation (2) as a fitness proxy for relative growth rates.
A numerical solution with discrete time steps was used to simulate the population dynamics of an exponentially growing polymorphic population as described in Supplementary Information. The growth of each mutant subpopulation occurring at each generation time is given by:
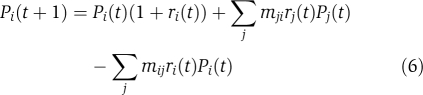 |
Pi is the frequency of a subpopulation i growing at a rate of ri at generation t. The model includes terms whereby subpopulation i increases when mutations occur in the j populations forming more of the i population with a rate mji and decreases by drift away from i population by at a rate mij:
 |
The relative growth rate (ri) is defined as a value between 0, no growth, and 1, entire population replicates, and is the parameter that determines fitness selection. In our model, ri is proportional to AK activity up to a specific threshold value, vthreshold, and is defined by v(T) in Equation (2). Variation in the relative growth rates comes from the temperature dependences of Kunf and kcat, which are measured independently in vitro (Table I). The fraction of each subpopulation i is calculated as fi(t)= Pi(t)/Ptotal(t).
A fitness function based on enzyme activity and stability that includes an activity threshold, vthreshold, can accurately reproduce the frequencies and the order in which alleles are observed within the polymorphic, experimental population (Figure 4D). In this model, fitness is only proportional to v(T) when activity falls below a critical physiological level (Equation (7)). Increases in activity levels above the threshold have no corresponding increases in fitness and are not advantageous during competition between subpopulations. If activity or stability were directly or largely proportional to fitness, AKBSUB Q199R/Q16L would be the only double mutant observed at any of the elevated temperatures because this mutant is both highly active and very stable. However, significant transient subpopulations of AKBSUB Q199R/T179I, Q199R/G213E, and Q199R/G214R do appear with increasing temperature, implying that only a threshold of activity is needed for viability. Thus, the very favorable stability and kinetic attributes of AKBSUB Q199R/Q16L serve to eliminate alternative models and help to define the critical physiological threshold for AK performance.
For polymorphism to persist within the rapidly growing population, the fitness differences between single nucleotide substitutions at the adk locus must be relatively small. Even the transient appearance of AKBSUB Q199R/G213E at 63°C corresponds to a substantial subpopulation of several billion cells that was reached within a total population of ∼100 billion between the time it became more fit than AKBSUB Q199R and less fit than the remaining double mutants. Under the condition in which the relative fitness values of the mutants are very close, mutational frequency can influence the speed with which one strain is able to replace another. AKBSUB Q199R/Q16L is one of the first three double mutants isolated at 62°C, but, despite its much more favorable kinetic and stability parameters, does not reach a majority status until 66°C, which occurs 8 days or roughly 400 generations later (Figure 4C). AKBSUB Q199R/Q16L is the only AK mutant that was the result of a transversion and suggests that AKBSUB Q199R/Q16L could have been constrained by clonal interference if transitions occurred at a rate 10 times greater than that for the transversion leading to AKBSUB Q199R/Q16L. As shown in Equation (6), mutation rates are included within the general population model. The prevalence of base transitions relative to transversions is well documented and has been observed for many organisms including B. subtilis and may certainly contribute to the observed evolutionary outcome (Wakeley, 1994, 1996; Sasaki and Kurusu, 2004).
Substrate stabilization of AKBSUB contributes to the population dynamics observed in the numerical solution and presumably during natural selection. Increases in the concentration of ATP leads to prolonged persistence of Q199R, shifting the transition between allelic dominance to later generations (see Supplementary Figure S3). Marked increases in substrate concentration increase the activity of all the AK enzymes, eventually making them equally competitive when their activities surpass the threshold. The unfolding midpoints of almost all mutants in the absence of ATP/AMP (with the exception of Q199R/Q16L) are below the beginning experimental temperature. Q199R has a Tm of 47°C, but the mutant strain was isolated in the culture up to 62°C. Thus, substrate stabilization is almost certainly a major contributor to the activity of the mutant AKs at the temperatures above the Tm values of the apoproteins. Additional mechanisms of stabilization such as macromolecular crowding and chaperones may also be occurring. In vivo, a combination of these factors will produce additional stability, but in the in vitro assay increases in [S] clearly produce marked stabilization and retention of AK activity, which is reproduced quantitatively by our model.
Summaries of the stability and kinetic properties of the AK mutants are shown in Figure 4A and B, and the observed and calculated relative frequencies of each mutant population are shown in Figure 4C and D. Although there are many factors that determine the effective concentration of intracellular proteins, the fitness function used to compute the theoretical populations in Figure 4D only required the relative in vitro stabilities and kinetic properties of the mutant enzymes to capture the general features of the in vivo evolutionary outcomes for the three major mutants, AKBSUB Q199R/A193V, AKBSUB Q199R/T179I and AKBSUB Q199R/Q16L, and even for the limited transient success of AKBSUB Q199R/G213E and AKBSUB Q199R/G214R. Remarkably, at a given substrate concentration, the only parameter that needed to be varied to accurately reproduce the experimentally observed population time courses was the activity threshold, vthreshold (see Supplementary Information).
Growth rates of AK mutants
Our physiological threshold model predicts equivalent growth rates between mutant strains at temperatures in which AK activities meet or surpass the threshold requirement. If the threshold is met, growth rates, that is fitness, will no longer be dependent on increases in activity of AK, but on other limiting aspects of the metabolic pathways shared by NUB3621-R and the mutant strains, which differ only at the adk locus. Growth rates were determined for NUB3621-R along with mutants AKBSUB Q199R and AKBSUB Q199R/Q16L from 50 to 70°C (Figure 6). Relative to the original G. steraothermophilus strain NUB3621-R, AKBSUB Q199R and AKBSUB Q199R/Q16L have equivalent growth rates within the error of the experiment at low temperatures (50–55°C) as predicted by the model. It is only at higher temperatures, between 55 and 57.5°C, AKBSUB Q199R becomes less fit than the double mutant. AKBSUB Q199R/Q16L mirrors the growth curve of NUB3621-R over the entire temperature range. This result is consistent with a recent experiment in which a starting population of the mutant AKBSUB Q199R/Q16L was grown at 50°C for 10 days (∼500 generations) to test whether new mutants would be fixed within the turbidostat at low temperature. An AK mutant with a 5% or greater fraction of the total population would have been readily detected; however, none were observed (not shown), further confirming the predictions of the physiological threshold model and the growth rate study. A significant difference between growth rate of Q199R and Q199R/Q16L at 57.5°C would suggest that the progenitor would be less fit before reaching 60°C. In the original experiment of Counago et al (2006), fixation of the double mutant strains is significantly slowed by clonal interference in the turbidostat population (Counago et al, 2006).
Figure 6.
Equivalent growth rates at low temperatures support a physiological threshold for AK activity in vivo. Only at higher temperatures in which the activity of AKBSUB Q199R decreases below a threshold because of thermal denaturation do the growth rates, that is fitness, diverge from NUB3621-R.
Discussion
The successful adaptation of an organism during natural selection depends on improvements in fitness that result from changes in molecular structure and function. In our experiments, AK activity is tightly linked to the fitness of the organism and allows us to highlight the critical function of enzyme kinetics as well as reversible and irreversible protein folding to molecular adaptation. We have constructed a fitness function that incorporates classic Michaelis–Menten kinetics and protein-folding properties to successfully reproduce the population dynamics of a microbial population during adaptation to higher temperature. As shown in Scheme I, the fraction of active, folded protein is dependent on both substrate stabilization and the probability that the protein becomes irreversibly lost from the system by aggregation or protein misfolding.
Misfolding and aggregation of proteins in vivo can have a substantial effect on fitness. As shown in Figure 2, it is essential to include a pathway for irreversible protein loss that reflects protein aggregation or misfolding as temperature increases. The irreversible unfolding step is analogous to the in vivo tendency of proteins to aggregate and can be an important factor in the evolutionary outcomes for a population with a particular mutant allele. The rate of this process, kirrev, is required to appropriately account for enzyme loss in the in vitro assay and, when incorporated into the fitness function, represents the in vivo loss of physiological activity associated with aggregation or degradation. In vivo rates of irreversible unfolding will be affected by many factors including temperature, osmolytes, and chaperones, each of which could have a function in adaptation (Tokuriki and Tawfik, 2009a). Even at lower temperatures, proteins sample unfolded or partially unfolded states and thus stability and denaturation kinetics are important and selectable properties. AKBSUB Q199R/Q16L has the largest Tm among the selected mutants and is the only variant that is predominantly folded in vitro at 55°C in the absence of substrate (∼90%). In comparison, <10% of AKBSUB Q199R ensemble occupies the native state at 55°C. In vivo, AKBSUB Q199R is sufficiently folded and active to extend the viability of the organism by 9°C and shows a critical function for the in vivo folding environment.
An appealing aspect of our fitness function is that it permits an evaluation of specific and quantitative aspects of protein stability and activity to evolutionary fates. As shown in Figure 4, in vivo population dynamics can be reproduced using in vitro physicochemical measurements. Although in vitro measurements are made under relatively non-physiological buffer conditions, the relative activities and stabilities of the mutant AKs with respect to one another in vivo seem to be well captured as judged by the close correspondence of the observed and predicted population fractions in Figure 4C and D.
Our work shows the importance of physiological context in defining fitness. Cellular energy charge (EC) is maintained by AK and is defined as [ATP]+1/2[ADP]/[ATP+ADP+AMP] (Atkinson, 1968). When deficiencies in adenylate homeostasis limit growth rate, improvements in enzyme function can result in increased fitness, but only to the extent that cellular needs are met, which is included as a physiological threshold in the fitness function for AK. In G. stearothermophilus, the EC during exponential growth is 0.4–0.5, but falls dramatically and irrevocably once AK activity is lost (Counago and Shamoo, 2005). AK alleles that are able to maintain EC do not limit cell growth because AK is no longer the ‘weak link’ in the cell. We believe this physiological threshold effect to be a general feature that defines and circumscribes the adaptive fitness landscape of protein evolution.
Adaptability is facilitated by increasing the diversity of the population and can be accomplished by the accumulation of near neutral or even modestly destabilizing mutations that provide more possibilities for success. Chaperones have an exceptionally important function in buffering biological systems against these destabilizing mutations as well as mistakes in translation that lead to polymorphic populations and have been shown to increase rates of adaptation (Rutherford, 2003; Drummond and Wilke, 2008; Tokuriki and Tawfik, 2009a). An increase in the diversity of the molecular ensemble permits accumulation of ‘global suppressor’ mutations that act largely on protein stability increasing the robustness of the molecule to subsequent adaptive changes (Bloom et al, 2005b; Bershtein et al, 2008). Thus, adaptation through protein evolution is circumscribed by protein stability. As most mutational events will be destabilizing (Tokuriki and Tawfik, 2009b), higher mutation rates can lead to decreases in fitness eventually leading to extinction (Zeldovich et al, 2007; Chen and Shakhnovich, 2009). We would predict that increases in substrate concentrations or global suppressors of unfolding such as chaperones or specific osmolytes could shift the stability of the protein ensemble and ameliorate adaptive mutations that increase activity at the cost of stability. It would be interesting to increase adenylate flux or alter the in vivo concentration of substrate to further test the function of substrate or chaperone stabilization in evolutionary dynamics within a population under selection. This would be difficult in our system as the adenine nucleotides are stabilizing molecules for the protein as well as substrates that are used not only by AK, but a host of other critical proteins such as kinases and crucial energy metabolites for maintaining cellular functions such as DNA replication. Thus, the activities directly related to AK stabilization by substrate cannot be decoupled from their critical function in the overall fitness of the cell. However, it should be possible to recover or engineer new AK mutants with physicochemical properties that could be used to further probe the function of substrates in evolutionary outcomes.
Taken together, these studies and others suggest that the balance of molecular diversity and mutation rate is critical to understanding adaptation. Although our system links the physicochemical properties of adaptive changes that increase stability, the principles apply equally to those changes that might decrease stability of the ensemble either through mutation or translational errors (Drummond and Wilke, 2008). Thus, regardless of how protein diversity is generated, evolutionary dynamics will likely be strongly coupled to stability and function.
Materials and methods
Measuring rates of forward reaction: ATP+AMP → 2ADP
Rates of ADP production for AK mutants were measured using an end-point-coupled assay (Saint Girons et al, 1987; Glaser et al, 1992; Counago et al, 2008). The assay mixture contained buffer (25 mM phosphate buffer pH 7.2, 5 mM MgCl2, 65 mM KCl), 1.4 mM AMP, and various ATP concentrations (5, 10, 25, 50, 100, 250, 500, and 1000 μM). The reaction was started by the addition of AK to a final concentration of 5 nM. The final reaction volume was 1.0 ml in a polyethylene tube. The reaction mixtures were incubated in a water bath at 55, 60, 62.5, 65, 67.5, and 70°C for 5 min before the addition of AK. The reactions were quenched 20, 40, and 60 s after initiation by rapidly adding 300 μl of the mixture to 100 μl of ice-cold, 1.0 mM P1, P5-di(adenosine-5′) pentaphosphate (Ap5A).
The samples were maintained on ice until the amount of ADP could be measured. ADP was quantitated by a secondary assay in which five units of pyruvate kinase, 0.3 mM β-nicotinamide adenine dinucleotide (NADH), and 0.5 mM phosphoenol pyruvate (PEP) were added to the quenched reaction. Absorbance at 340 nm was measured before and after the addition of five units of lactate dehydrogenase. The conversion of NADH to NAD+ was used as a measure of the ADP produced by AK at various temperatures, substrate concentrations, and reaction durations. The slope of [ADP] versus time for these three time points was used as the rate of product formation, v, at the middle time point. All measurements were performed in triplicate.
Plots of v versus [ATP] were fit to Equation (1) using Prism (GraphPad, La Jolla, CA) with t=40 s for the exponential term describing irreversible denaturation. Calculated values for Kunf at each respective temperature for each mutant were used as constants in the fit and kirrev was shared across all temperatures for each individual mutant to constrain the fits. AMP, ADP, ATP, Ap5A, NADH, PEP, pyruvate kinase, and lactate dehydrogenase were all purchased from Sigma-Aldrich (St Louis, MO).
Differential scanning calorimetry
Measurements were performed using VP-DSC differential scanning microcalorimeter (MicroCal, LLC, Northampton, MA) at a scan rate of 90°C h–1. Data could be well modeled as an apparent two-state transition under the conditions of the DSC experiment. To test this approximation, a study of the concentration and scan rate-dependent properties of wild type and all the mutants was performed (Supplementary Information). Protein concentration was 40 μM in 10 mM HEPES pH 7.0. Before measurements, sample and buffer were degassed for 10 min at room temperature. A pressure of 2 atm was kept in the cells throughout the heating cycles to prevent degassing. A background scan collected with the buffer in both cells was subtracted from each scan. Data were analyzed with Origin 7.0 software (MicroCal) to obtain the transition temperature (Tm), the calorimetric enthalpy of thermal unfolding (ΔH), the heat capacity change at the Tm (ΔCp), and the Gibbs-free energy change (ΔGunf) at each temperature T.
Numerical simulation with MATLAB
Numerical integration was used to calculate the population size at time t+1 based on the population size at time t, the mutation rate, and a growth rate (0⩽r⩽1) specified by the AK enzyme activity computed from a continuous function for activity, Equation (2). The numerical solution requires inputs of initial frequencies of each allele and the rate at which the two-substitution alleles are formed from the progenitor strain (AKBSUB Q199R) (Equation (6)). A mutation rate of 5E-10 was used as seeding rate for each double mutation involving a simple base transition, with the exception of the AKBSUB Q199R/Q16L mutation, which involves a transversion and was assigned a rate at one-tenth of this value. The mutation rates were kept constant for the course of the simulation. Enzyme activity was modeled between 55 and 70°C for each enzyme, and this activity was converted to a relative growth rate as described in Equation (7). If the activity was above or equal to the threshold, the growth rate was 1. However, if the activity fell below the threshold, the growth rate was equal to the fraction of the activity divided by the threshold value. The numerical solution was calculated for 1500 generations, with each 100 generations corresponding to 1°C temperature span in the experiment.
Determination of temperature-dependent growth rates
G. stearothermophilus strain NUB3621-R and recombinant strains expressing B. subtilis adk with Q199R or Q199R/Q16L were plated on modified LB (mLB) with rifampicin (5 μg ml–1) and chloramphenicol (7 μg ml–1) overnight at 55°C. Single colonies were used to inoculate 10 ml of mLB with rifampicin at 55°C for approximately 3 h with shaking at 225 r.p.m. (Abs600≈0.5). A total of 50 μl of the 3 h culture was used to inoculate 50 ml of mLB and incubated at the desired temperature. Growth was monitored as changes in optical density. Growth curves were determined by fitting to an exponential function. All LB media were modified with the addition of 1.05 mM nitrilotriacetate, 0.59 mM MgSO4, 0.91 mM CaCl2, and 0.04 mM FeSO4.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation grant 0641792 (YS), NIH Grants HL047020 and GM35649 (JSO) and Welch Foundation Grants C1584 (YS) and C0612 (JSO). We also thank the three anonymous referees for their valuable comments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Atkinson DE (1968) The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry 7: 4030–4034 [DOI] [PubMed] [Google Scholar]
- Bae E, Phillips GN Jr (2004) Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. J Biol Chem 279: 28202–28208 [DOI] [PubMed] [Google Scholar]
- Becktel WJ, Schellman JA (1987) Protein stability curves. Biopolymers 26: 1859–1877 [DOI] [PubMed] [Google Scholar]
- Berezovsky IN, Shakhnovich EI (2005) Physics and evolution of thermophilic adaptation. Proc Natl Acad Sci USA 102: 12742–12747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Goldin K, Tawfik DS (2008) Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol 379: 1029–1044 [DOI] [PubMed] [Google Scholar]
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS (2006) Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 444: 929–932 [DOI] [PubMed] [Google Scholar]
- Bloom JD, Arnold FH (2009) In the light of directed evolution: pathways of adaptive protein evolution. Proc Natl Acad Sci USA 106 (Suppl 1): 9995–10000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom JD, Meyer MM, Meinhold P, Otey CR, MacMillan D, Arnold FH (2005a) Evolving strategies for enzyme engineering. Curr Opin Struct Biol 15: 447–452 [DOI] [PubMed] [Google Scholar]
- Bloom JD, Silberg JJ, Wilke CO, Drummond DA, Adami C, Arnold FH (2005b) Thermodynamic prediction of protein neutrality. Proc Natl Acad Sci USA 102: 606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE (2008) Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA 105: 7899–7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandts JF, Lin LN (1990) Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry 29: 6927–6940 [DOI] [PubMed] [Google Scholar]
- Bull JJ, Badgett MR, Rokyta D, Molineux IJ (2003) Experimental evolution yields hundreds of mutations in a functional viral genome. J Mol Evol 57: 241–248 [DOI] [PubMed] [Google Scholar]
- Chen P, Shakhnovich EI (2009) Lethal mutagenesis in viruses and bacteria. Genetics 183: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counago R, Chen S, Shamoo Y (2006) In vivo molecular evolution reveals biophysical origins of organismal fitness. Mol Cell 22: 441–449 [DOI] [PubMed] [Google Scholar]
- Counago R, Shamoo Y (2005) Gene replacement of adenylate kinase in the gram-positive thermophile Geobacillus stearothermophilus disrupts adenine nucleotide homeostasis and reduces cell viability. Extremophiles 9: 135–144 [DOI] [PubMed] [Google Scholar]
- Counago R, Wilson CJ, Pena MI, Wittung-Stafshede P, Shamoo Y (2008) An adaptive mutation in adenylate kinase that increases organismal fitness is linked to stability-activity trade-offs. Protein Eng Des Sel 21: 19–27 [DOI] [PubMed] [Google Scholar]
- Dean AM, Thornton JW (2007) Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet 8: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO (2008) Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134: 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE, Dean AM, Hartl DL (1987) Metabolic flux and fitness. Genetics 115: 25–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Lenski RE (2003) Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet 4: 457–469 [DOI] [PubMed] [Google Scholar]
- Fasan R, Meharenna YT, Snow CD, Poulos TL, Arnold FH (2008) Evolutionary history of a specialized p450 propane monooxygenase. J Mol Biol 383: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser P, Presecan E, Delepierre M, Surewicz WK, Mantsch HH, Barzu O, Gilles AM (1992) Zinc, a novel structural element found in the family of bacterial adenylate kinases. Biochemistry 31: 3038–3043 [DOI] [PubMed] [Google Scholar]
- Kacser H, Burns JA (1981) The molecular basis of dominance. Genetics 97: 639–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies JL, Izem R, Supler KL, Kingsolver JG, Burch CL (2006) The genetic basis of thermal reaction norm evolution in lab and natural phage populations. PLoS Biol 4: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunzer M, Miller SP, Felsheim R, Dean AM (2005) The biochemical architecture of an ancient adaptive landscape. Science 310: 499–501 [DOI] [PubMed] [Google Scholar]
- Miller C, Davlieva M, Wilson CJ, White K, Counago R, Wu G, Myers JC, Wittung-Stafshede P, Shamoo Y (2010) Experimental evolution of adenylate kinase reveals contrasting strategies towards protein thermostability. Biophys J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SP, Lunzer M, Dean AM (2006) Direct demonstration of an adaptive constraint. Science 314: 458–461 [DOI] [PubMed] [Google Scholar]
- Rokyta DR, Beisel CJ, Joyce P (2006) Properties of adaptive walks on uncorrelated landscapes under strong selection and weak mutation. J Theor Biol 243: 114–120 [DOI] [PubMed] [Google Scholar]
- Rutherford SL (2003) Between genotype and phenotype: protein chaperones and evolvability. Nat Rev Genet 4: 263–274 [DOI] [PubMed] [Google Scholar]
- Saint Girons I, Gilles AM, Margarita D, Michelson S, Monnot M, Fermandjian S, Danchin A, Bârzu O (1987) Structural and catalytic characteristics of Escherichia coli adenylate kinase. J Biol Chem 262: 622–629 [PubMed] [Google Scholar]
- Sasaki M, Kurusu Y (2004) Analysis of spontaneous base substitutions generated in mutator strains of Bacillus subtilis. FEMS Microbiol Lett 234: 37–42 [DOI] [PubMed] [Google Scholar]
- Silberg JJ, Endelman JB, Arnold FH (2004) SCHEMA-guided protein recombination. Methods Enzymol 388: 35–42 [DOI] [PubMed] [Google Scholar]
- Thomas TM, Scopes RK (1998) The effects of temperature on the kinetics and stability of mesophilic and thermophilic 3-phosphoglycerate kinases. Biochem J 330 (Part 3): 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS (2009a) Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459: 668–673 [DOI] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS (2009b) Stability effects of mutations and protein evolvability. Curr Opin Struct Biol 19: 596–604 [DOI] [PubMed] [Google Scholar]
- Wakeley J (1994) Substitution-rate variation among sites and the estimation of transition bias. Mol Biol Evol 11: 436–442 [DOI] [PubMed] [Google Scholar]
- Wakeley J (1996) The excess of transitions among nucleotide substitutions: new methods of estimating transition bias underscore its significance. Trends Ecol Evol 11: 158–162 [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL (2006) Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312: 111–114 [DOI] [PubMed] [Google Scholar]
- Wintrode PL, Zhang D, Vaidehi N, Arnold FH, Goddard WA III (2003) Protein dynamics in a family of laboratory evolved thermophilic enzymes. J Mol Biol 327: 745–757 [DOI] [PubMed] [Google Scholar]
- Zeldovich KB, Chen P, Shakhnovich EI (2007) Protein stability imposes limits on organism complexity and speed of molecular evolution. Proc Natl Acad Sci USA 104: 16152–16157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








