Abstract
OBJECTIVE
Variation in transcription factor 7-like 2 (TCF7L2) gene has been shown to be associated with type 2 diabetes and diabetes-related quantitative traits. We examined variation in a 0.1-Mb region surrounding marker DG10S478 for association with diabetes-related quantitative traits in 132 Mexican-American families of a proband with previous gestational diabetes mellitus (GDM).
RESEARCH DESIGN AND METHODS
Study participants were phenotyped by an oral glucose tolerance test (OGTT) and an intravenous glucose tolerance test and by a dual-energy X-ray absorptiometry scan for percentage of body fat. Of the 42 tag single nucleotide polymorphisms (SNPs) genotyped, 15 were identified.
RESULTS
On univariate analysis, none of the SNPs showed association with diabetes-related quantitative traits. However, rs12255372 showed association with 30′ Δinsulin (OGTT 30′ min fasting insulin) in an interaction with percentage of body fat (Bonferroni-corrected P = 0.027). The effect of adiposity to increase 30′ Δinsulin was greater in subjects with the T allele. This interaction was not associated with acute insulin response to intravenous glucose. rs12255372 also showed an association with β-cell compensation for insulin resistance based on 30′ Δinsulin in an interaction with percentage of body fat (Bonferroni-corrected P = 0.014). rs12255372 was also associated with GDM (odds ratio [OR] 2.49 [95% CI 1.17–5.31]; P = 0.018) in our case-control sample.
CONCLUSIONS
We conclude that variation in TCF7L2 is associated with GDM and interacts with adiposity to alter insulin secretion in Mexican Americans. Our observations partly explain the increased ORs observed in previous associated studies when analyses were restricted to lean subjects and the variability in quantitative trait association results.
Variation in transcription factor 7-like 2 (TCF7L2) gene was first shown to be associated with type 2 diabetes in the Icelandic population (1) and replicated in a variety of populations (2– 8) with some reporting association with type 2 diabetes–related quantitative traits (2,6,7). Thus, there is ample evidence supporting TCF7L2 as a type 2 diabetes susceptibility gene, possibly by altering insulin secretion. However, evidence for TCF7L2’s association with type 2 diabetes is mostly derived from Caucasian samples, and its role in other ethnic/racial groups or other forms of diabetes has not been delineated.
Mexican-American women with previous gestational diabetes mellitus (GDM) exhibit significant β-cell dysfunction and are at high risk for type 2 diabetes (9). The BetaGene Study is a family-based study to identify genetic determinants underlying differences in β-cell function in Mexican Americans. We examined genetic variation in a 0.1-Mb region surrounding DG10S478 (1) and tested variants for association with type 2 diabetes–related quantitative traits in our BetaGene cohort. We also tested whether the association between TCF7L2 and type 2 diabetes–related quantitative traits is modified by adiposity, given some suggestion that TCF7L2 variants may interact with adiposity to modify the risk of type 2 diabetes (6,8).
RESEARCH DESIGN AND METHODS
Subject recruitment
Subject recruitment for the BetaGene Study is ongoing, and for this report we describe only those clinical protocols and assays relevant to the results presented herein. Participation in the BetaGene Study is restricted to Mexican Americans from families of a proband with previous GDM. Details regarding subject recruitment can be found in the supplemental materials (located in an online appendix at http://dx.doi.org/10.2337/db06-1682). In addition, we are recruiting Mexican-American women who have gone through pregnancy without GDM but are also selected to be age-, BMI-, and parity-matched to the GDM probands. For the present report, we performed the relevant genotyping and data analysis on all control subjects, GDM probands, siblings, and cousins who had been phenotyped by the end of November 2005. All protocols for the BetaGene Study have been approved by the institutional review boards of the participating institutions.
Clinical protocols
Phenotyping is performed on two separate visits to the University of Southern California General Clinical Research Center. Visit 1 consists of a physical examination, DNA collection, and a 75-g 2-h oral glucose tolerance test (OGTT) with 30-min blood sampling. Participants with fasting glucose <126 mg/dl are invited for a second visit, which consists of a dual-energy X-ray absorptiometry scan for determination of percentage of body fat and an insulin-modified intravenous glucose tolerance test (IVGTT).
Molecular analysis
Single nucleotide polymorphisms (SNPs) in all four HapMap (release no. 19) populations were, whenever possible, selected at ~2.5-kb intervals across a 0.1-Mb region surrounding DG10S478. Forty-two SNPs were selected and genotyped using the Applied Biosystems TaqMan system (10).
Data analysis
We calculated two measures of insulin response to glucose; the difference between the 30′ and fasting plasma insulin concentrations from the OGTT (30′ Δinsulin) and the incremental area under the insulin curve for the first 10 min of the IVGTT (acute insulin response [AIR]). IVGTT glucose and insulin data were analyzed by minimal model (MINMOD Millennium version 5.18). The disposition index (DI), a measure of β-cell compensation for insulin resistance, was computed as the product of the insulin sensitivity index (SI) and early insulin response (DI = SI × AIR from the IVGTT [11,12]; DI30 = SI × 30 min Δinsulin from the OGTT [13]).
Genotype data were tested for deviation from the Hardy-Weinberg equilibrium and for non-Mendelian inheritance using PEDSTATS version 0.6.4 (14), and allele frequencies were estimated using SOLAR version 2.1.4 (15). Tagging SNPs were selected from among the genotyped SNPs using TAGGER as implemented in Haploview version 3.32 (16,17). Linkage disequilibrium (LD) and haplotype block structure were also assessed using Haploview (18). The measured genotypes approach under a variance components and a likelihood ratio framework was used to test SNP associations with continuous phenotypes and implemented using SOLAR (15) with ascertainment correction.
Each SNP was first tested for a trend for association with quantitative traits under an additive genetic model. The reference allele was the minor allele, and models were adjusted for age, sex, and, where appropriate, percentage of body fat. A significant trend was defined as a nominal P value of 0.1, corrected for the number of SNPs tested (n = 15; P = 0.0067). SNPs showing a trend were then tested for association under dominant and/or recessive genetic models. SNPs significantly associated with any type 2 diabetes–related quantitative trait were also tested for a multiplicative interaction with percentage of body fat. The three SNPs that tagged previously associated SNPs (rs7901695, rs7100927, and rs12255372; Table 1) were tested for an interaction with adiposity regardless of their univariate association results.
TABLE 1.
Tag SNP characteristics
| SNP | Position (kb)* | Minor allele | Minor allele frequency | SNPs tagged |
|---|---|---|---|---|
| rs7901695† | 114744078 | C | 0.24 | rs7903146† |
| rs7896811 | 114756707 | T | 0.07 | |
| rs6585202, rs7077247, rs11196208, rs6585200, rs10885405, rs10885409, rs11196205†, rs7895340†, rs12265291, rs7924080 | ||||
| rs7100927 | 114786038 | G | 0.28 | |
| rs11196203 | 114795850 | A | 0.08 | |
| rs12255372† | 114798892 | T | 0.19 | rs12243326 |
| rs7087006 | 114813416 | G | 0.26 | rs11196213, rs7085989 |
| rs10885410 | 114814463 | A | 0.20 | |
| rs7919409 | 114814966 | C | 0.21 | |
| rs7100388 | 114815803 | G | 0.09 | |
| rs7908486 | 114824488 | G | 0.37 | rs911769 |
| rs11196218 | 114830484 | A | 0.23 | rs12768310, rs3750804 |
| rs7082458 | 114836639 | G | 0.10 | |
| rs3814572 | 114837713 | G | 0.07 | |
| rs10749127 | 114839343 | T | 0.29 | |
| rs4639863 | 114844373 | T | 0.32 |
Based on NCBI build 36.1.
Previously reported by Grant et al. (1).
Linear modeling results are reported as age-, sex-, and percentage of body fat–adjusted medians and interquartile ranges. For results from the interaction analysis that included percentage of body fat, trait values are reported as age-and sex-adjusted medians and interquartile ranges. In all cases, reported P values are Bonferroni corrected for multiple comparisons unless otherwise specified.
RESULTS
We report results from 537 individuals in 132 families (Table 2). Control subjects were slightly younger and less obese compared with GDM probands, reflecting recruitment of control subjects lagging slightly behind GDM probands to allow for matching as described above. Pairwise LD and haplotype blocks (Fig. 1) were estimated using 40 of 42 genotyped SNPs (rs7904519 and rs7907632 failed Hardy-Weinberg equilibrium), resulting in a density of ~2.57 kb. Seven haplotype blocks were identified. The largest block included three previously associated SNPs (rs7895340, rs1196205, and rs12255372) (1), and two other previously associated SNPs (rs7901695 and rs7903146) (1) formed an independent block. Table 2 shows the characteristics of the 15 tag SNPs. None of the tag SNPs showed a trend for association with quantitative traits under an additive genetic model; the two strongest associations were with traits related to insulin secretion: rs10885410 was associated with 30′ Δinsulin (uncorrected P = 0.010), and rs11196218 was associated with AIR (uncorrected P = 0.008).
TABLE 2.
Subject characteristics
| GDM probands | Siblings | Cousins | Control subjects | |
|---|---|---|---|---|
| n | 94 | 241 | 179 | 58 |
| Male/female | 0/94 | 88/153 | 80/99 | 0/58 |
| Age (years) | 35.0 (8.6) | 34.5 (11.0) | 32.8 (11.8) | 33.4 (7.6) |
| BMI (kg/m2) | 30.9 (8.9) | 29.1 (6.3) | 27.4 (6.6) | 27.2 (6.6) |
| Body fat (%) | 39.1 (7.3) | 33.5 (13.9) | 30.7 (12.8) | 36.0 (7.0) |
| Fasting glucose (mmol/l) | 5.4 (0.9) | 5.1 (0.6) | 5.1 (0.6) | 4.9 (0.4) |
| 2-h glucose (mmol/l) | 8.4 (3.4) | 7.3 (2.6) | 6.4 (2.2) | 6.9 (2.1) |
| Fasting insulin (pmol/l) | 69 (48) | 42 (42) | 36 (36) | 30 (36) |
| 2-h insulin (pmol/l) | 522 (462) | 372 (378) | 300 (336) | 252 (312) |
| 30′ Δinsulin (pmol/l) | 372 (252) | 321 (252) | 366 (342) | 294 (324) |
| SG (×10−2 per min−1) | 1.29 (0.41) | 1.55 (0.71) | 1.74 (0.88) | 1.84 (0.94) |
| SI (×10−3 per min−1 per pmol/l) | 3.63 (2.06) | 4.32 (3.28) | 4.85 (3.17) | 5.67 (3.01) |
| AIR (pmol/l × 10 min) | 1,990 (2,648) | 2,572 (2,788) | 3,044 (3,610) | 2,974 (2,348) |
| DI | 780 (780) | 1,141 (1,145) | 1,339 (1,106) | 1,714 (972) |
Data are unadjusted median (interquartile range) unless otherwise indicated.
FIG. 1.

TCF7L2 pairwise LD structure. Pairwise LD and haplotype block structure as determined by the method of Gabriel for the 40 SNPs genotyped in our Mexican-American families. LD is displayed as pairwise r2 values (values within boxes), where white indicates r2 = 0, varying shades of gray indicate 0 < r2 < 1, and black indicates r2 = 1.
We then tested whether previously associated SNPs (rs7100927, rs7901695, and rs12255372) interacted with adiposity to alter variation in quantitative traits. rs12255372 under an additive genetic model showed a significant interaction with percentage of body fat to alter 30′ Δinsulin (uncorrected P = 0.009), and rs7901695 interacted with percentage of body fat to alter SI (uncorrected P = 0.027). Only the interaction with rs12255372 remained significant after correcting for multiple comparisons. The interaction remained significant, assuming a dominant genetic model (P = 0.016). The interaction between rs12255372 and adiposity was not associated with AIR or SI and showed marginal association with fasting insulin (uncorrected P = 0.157, P = 0.744, and P = 0.067, respectively).
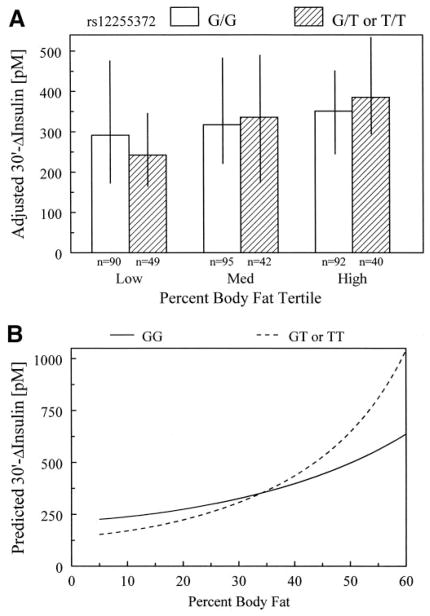
We stratified 30′ Δinsulin by rs12255372 genotype assuming a dominant genetic model and by percentage of body fat tertiles within each genotype group (Fig. 2). 30′ Δinsulin increases with increasing adiposity, with the effect stronger among individuals with a T allele (Fig. 2A). The model-predicted 30′ Δinsulin concentration (Fig. 2B) revealed that within the range of percentage of body fat up to ~34%, subjects with a T allele had lower 30′ Δinsulin, whereas they had higher 30′ Δinsulin in the higher percentage of body fat range when compared with G homozygotes.
FIG. 2.
Interaction between rs12255372 and percentage of body fat on 30′ Δinsulin concentrations. A: Median age- and sex-adjusted 30′ Δinsulin concentration and interquartile range stratified by genotype and percentage of body fat tertiles. Within the lowest body fat tertile, individuals homozygous for the G allele have higher adjusted 30′ Δinsulin compared with individuals with at least one copy of the T allele. However, within the highest body fat tertile, individuals homozygous for the G allele have lower adjusted 30′ Δinsulin concentrations. The effect of adiposity to alter 30′ Δinsulin is greater among subjects with a T allele compared with those homozygous for the G allele. B: Interaction based on the model parameter estimates and covering the range of body fat observed in the BetaGene Study.
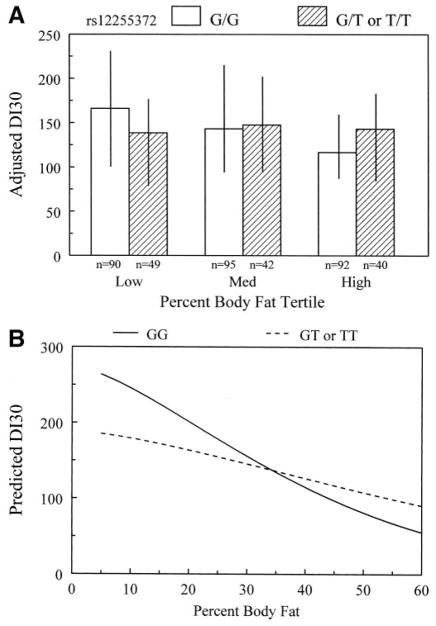
Examination of the covariate-adjusted, genotype-specific median SI for rs12255372 stratified by percentage of body fat tertiles revealed a pattern of increasing insulin resistance with adiposity regardless of genotype (data not shown). Such differences in SI could confound patterns in 30′ Δinsulin, which changes reciprocally with changes in SI (11,12). Therefore, we tested whether an index of β-cell compensation based on the 30′ Δinsulin (DI30 = SI × 30′ Δinsulin) was associated with the interaction between rs12255372 and adiposity. Assuming a dominant genetic model, the interaction was significantly associated with DI30 (P = 0.014). Increasing adiposity had a modest modifying effect on DI30 among individuals with a T allele but a dramatic effect in reducing DI30 among G homozygotes (Fig. 3A). The model-predicted DI30 (Fig. 3B) shows that at low percentages of body fat, DI30 was higher in G homozygotes than those with a T allele. DI30 fell with increasing percentages of body fat in both groups, but the fall was less steep in individuals with a T allele. DI from IVGTTs was not associated with the interaction between rs12255372 and percentage of body fat (uncorrected P = 0.152).
FIG. 3.
Interaction between rs12255372 and percentage of body fat on β-cell compensation, computed as the product of SI × 30′ Δinsulin concentration (DI30). A: Median age- and sex-adjusted DI30 and interquartile range stratified by genotype and percentage of body fat tertiles. Within the lowest body fat tertile, individuals homozygous for the G allele have higher adjusted DI30 compared with individuals with at least one copy of the T allele. However, within the highest body fat tertile, individuals homozygous for the G allele have lower adjusted DI30. The effect of adiposity to alter DI30 is greater among subjects homozygous for the G allele compared with those with at least one copy of the T allele. B: Interaction based on the model parameter estimates and covering the range of body fat observed in the BetaGene Study.
Given the trait associations, we tested rs12255372 for association with previous GDM. The frequency of the T allele among probands with previous GDM (n = 94) was 39.4% compared with 20.7% in control subjects without previous GDM (n = 58). The association, assuming a dominant genetic model, was significant (odds ratio [OR] 2.49 [95% CI 1.17–5.31]; P = 0.018) and remained significant when adjusting for age and percentage of body fat (2.62 [1.13– 6.11]; P = 0.025). rs7901695 and rs7100927 showed no evidence for association with GDM (P = 0.601 and P = 0.627, respectively).
DISCUSSION
There are two novel findings from this study. First, rs12255372 in TCF7L2 was significantly associated with GDM under a dominant genetic model. This is the first evidence that TCF7L2 is associated with forms of pre-diabetes. We did not genotype ancestrally informative markers, so it is possible that population substructure may confound our results. However, given that the identical marker also shows association with type 2 diabetes–related quantitative traits, it is likely that this is a true association with GDM.
Second, rs12255372 was associated with 30′ Δinsulin in an interaction with percentage of body fat. This finding could explain some of the inconsistencies in association between TCF7L2 and quantitative traits among studies. It may also explain the increased OR for association with type 2 diabetes (1.89 vs. 1.69) reported by Cauchi et al. (8) when their analysis was restricted to lean subjects (BMI <30 kg/m2). The lack of association between this interaction and AIR from the IVGTT is consistent with an effect mediated through an enteroendocrine mechanism (e.g., glucagon-like peptide 1), as proposed by Grant et al. (1).
Since insulin secretion normally varies as a function of insulin resistance (11,12), associations with insulin secretion could simply reflect an underlying difference in insulin resistance to which β-cells are responding. Two lines of evidence argue against this. First, the interaction between adiposity and rs12255372 was not associated with SI. Second, the interaction was associated with DI30, which tended to be relatively low in lean individuals with a T allele but to fall relatively little with increasing adiposity compared with G homozygotes. The biological significance of this finding in relation to diabetes risk remains to be determined, but β-cell compensation at any point in time reflects dual influences of acute regulatory stimuli and cumulative effects of chronic factors such as insulin resistance. We have reported that chronic insulin resistance and high levels of insulin secretion may be detrimental to long-term β-cell compensation (19,20). It is possible that TCF7L2 variants have dual effects, limiting β-cell compensation for insulin resistance at any given time through acute effects (e.g., incretins) but minimizing deterioration of insulin secretion related to obesity and insulin resistance over time. This mechanism would be analogous to the effects of MODY2 variants, where glucose levels are elevated but tend not to deteriorate over time, unlike most forms of diabetes (A. Hattersley, personal communication).
It is noteworthy that many reports for TCF7L2 association with type 2 diabetes come from Caucasian populations in whom rs7901695 is in moderate to strong LD (r2 = 0.7) with rs12255372 and partly accounts for why both SNPs show association with type 2 diabetes (1,3–6,8), although other Caucasian populations report weaker LD between these SNPs (2), and there is a near absence of LD in African Americans (6). However, in our Mexican-American population, both were in relatively low LD (r2 = 0.55), resulting in their selection as tag SNPs. This may account for why the interaction between rs7901695 and percentage of body fat was not associated with type 2 diabetes–related quantitative traits and suggests that other functional variants of TCF7L2 may be distal to rs7901695.
In summary, we observed a strong association between TCF7L2 rs12255372 and GDM, along with an interaction between rs12255372 and adiposity to alter insulin secretion in Mexican Americans from families with GDM. The interaction with adiposity led to relatively poorer β-cell compensation in relatively lean individuals but better preservation of compensation with increasing adiposity. This pattern could explain prior observations that relative risks of type 2 diabetes associated with variants in TCF7L2 are highest in relatively lean individuals. The observed interaction also suggests a dual role of rs12255372 to downregulate acute β-cell compensation but limit the impact of obesity and chronic insulin resistance to damage β-cells and their function.
Acknowledgments
This work was supported by National Institutes of Health Grant DK-61628 and an American Diabetes Association Distinguished Clinical Scientist Award to T.A.B. A portion of this work was conducted in a facility constructed with support from Research Facilities Improvement Program Grants C06 (RR10600-01, CA62528-01, and RR14514-01) from the National Center for Research Resources.
We thank the families who participated in the BetaGene Study and the support of the University of Southern California General Clinical Research Center (M01-RR-00043). We also acknowledge the efforts of our recruiting and technical staff.
- AIR
acute insulin response
- GDM
gestational diabetes mellitus
- IVGTT
intravenous glucose tolerance test
- LD
linkage disequilibrium
- OGTT
oral glucose tolerance test
Footnotes
Additional information for this article can be found in an online appendix at http://dx.doi.org/10.2337/db06-1682.
References
- 1.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 2.Damcott CM, Pollin TI, Reinhart LJ, Ott SH, Shen H, Silver KD, Mitchell BD, Shuldiner AR. Polymorphisms in the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in the Amish: replication and evidence for a role in both insulin secretion and insulin resistance. Diabetes. 2006;55:2654–2659. doi: 10.2337/db06-0338. [DOI] [PubMed] [Google Scholar]
- 3.Scott LJ, Bonnycastle LL, Willer CJ, Sprau AG, Jackson AU, Narisu N, Duren WL, Chines PS, Stringham HM, Erdos MR, Valle TT, Tuomilehto J, Bergman RN, Mohlke KL, Collins FS, Boehnke M. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes. 2006;55:2649–2653. doi: 10.2337/db06-0341. [DOI] [PubMed] [Google Scholar]
- 4.Groves CJ, Zeggini E, Minton J, Frayling TM, Weedon MN, Rayner NW, Hitman GA, Walker M, Wiltshire S, Hattersley AT, McCarthy MI. Association analysis of 6,736 U.K. subjects provides replication and confirms TCF7L2 as a type 2 diabetes susceptibility gene with a substantial effect on individual risk. Diabetes. 2006;55:2640–2644. doi: 10.2337/db06-0355. [DOI] [PubMed] [Google Scholar]
- 5.Zhang C, Qi L, Hunter DJ, Meigs JB, Manson JE, van Dam RM, Hu FB. Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of U.S. women and men. Diabetes. 2006;55:2645–2648. doi: 10.2337/db06-0643. [DOI] [PubMed] [Google Scholar]
- 6.Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PIW, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D the Diabetes Prevention Program Research Group. TCF7L2 polymorphisms and progression to diabetes in the diabetes prevention program. N Engl J Med. 2006;355:242–250. doi: 10.1056/NEJMoa062418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saxena R, Gianniny L, Burtt NP, Lyssenko V, Giuducci C, Sjögren M, Florez JC, Almgren P, Isomaa B, Orho-Melander M, Lindblad U, Daly MJ, Tuomi T, Hirschhorn JN, Ardlie KG, Groop LC, Altshuler D. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduced insulin response to glucose in nondiabetic individuals. Diabetes. 2006;55:2890–2895. doi: 10.2337/db06-0381. [DOI] [PubMed] [Google Scholar]
- 8.Cauchi S, Meyre D, Dina C, Choquet H, Samson C, Gallina S, Balkau B, Charpentier G, Pattou F, Stetsyuk V, Scharfmann R, Staels B, Frühbeck G, Froguel P. Transcription factor TCF7L2 genetic study in the French population: expression in human β-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55:2903–2908. doi: 10.2337/db06-0474. [DOI] [PubMed] [Google Scholar]
- 9.Kim C, Newton KM, Knopp RH. Gestational diabetes and the incidence of type 2 diabetes. Diabetes Care. 2002;25:1862–1868. doi: 10.2337/diacare.25.10.1862. [DOI] [PubMed] [Google Scholar]
- 10.Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14:143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 11.Bergman RN, Phillips LS, Cobelli C. Physiologic evaluation of factors controlling glucose tolerance in man: measurement of insulin sensitivity and β-cell glucose sensitivity from the response to intravenous glucose. J Clin Invest. 1981;68:1456–1467. doi: 10.1172/JCI110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahn SE, Prigeon RL, McCulloch DK, Boyko EJ, Bergman RN, Schwartz MW, Neifing JL, Ward WK, Beard JC, Palmer JP, Porte D., Jr Quantification of the relationship between insulin sensitivity and B-cell function in human subjects: evidence for a hyperbolic function. Diabetes. 1993;42:1663–1672. doi: 10.2337/diab.42.11.1663. [DOI] [PubMed] [Google Scholar]
- 13.Buchanan TA, Xiang AH, Peters RK, Kjos SL, Berkowitz K, Marroquin A, Goico J, Ochoa C, Azen SP. Response of pancreatic β-cells to improved insulin sensitivity in women at high risk for type 2 diabetes. Diabetes. 2000;49:782–788. doi: 10.2337/diabetes.49.5.782. [DOI] [PubMed] [Google Scholar]
- 14.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 15.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.de Bakker PIW, Yelensky R, Pe’er D, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 18.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 19.Peters RK, Kjos SL, Xiang A, Buchanan TA. Long-term diabetogenic effect of single pregnancy in women with previous gestatoinal diabetes mellitus. Lancet. 1996;347:227–230. doi: 10.1016/s0140-6736(96)90405-5. [DOI] [PubMed] [Google Scholar]
- 20.Xiang AH, Peters RK, Trigo E, Kjos SL, Lee WP, Buchanan TA. Multiple metabolic defects during late pregnancy in women at high risk for type 2 diabetes. Diabetes. 1999;48:848–854. doi: 10.2337/diabetes.48.4.848. [DOI] [PubMed] [Google Scholar]
- 21.Buchanan TA, Xiang AH, Kjos SL, Trigo E, Lee WP, Peters RK. Antepartum predictors of the development of type 2 diabetes in Latino women 11–26 months after pregnancies complicated by gestational diabetes. Diabetes. 1999;48:2430–2436. doi: 10.2337/diabetes.48.12.2430. [DOI] [PubMed] [Google Scholar]