Summary
Prevalence studies indicate that transmission of drug-resistant HIV has been rising in the adult population, but data from the perinatally infected pediatric population are limited. In this retrospective study, we sequenced the pol region of HIV from perinatally infected infants diagnosed in New York State in 2001–2002. Analyses of drug resistance, subtype diversity, and perinatal antiretroviral exposure were conducted, and the results were compared with those from a previous study of HIV-infected infants identified in 1998–1999. Eight of 42 infants (19.1%) had provirus carrying at least 1 drug-resistance mutation, an increase of 58% over the 1998–1999 results. Mutations conferring resistance to nucleoside reverse transcriptase inhibitors, nonnucleoside reverse transcriptase inhibitors, and protease inhibitors were detected in 7.1%, 11.9%, and 2.4% of specimens, respectively. Consistent with previous results, perinatal antiretroviral exposure was not associated with drug resistance (P = 0.70). Phylogenetic analysis indicated that 16.7% of infants were infected with a non–subtype B strain of HIV. It seems that drug-resistant and non–subtype B strains of HIV are becoming increasingly common in the perinatally infected population. Our results highlight the value of resistance testing for all HIV-infected infants upon diagnosis and the need to consider subtype diversity in diagnostic and treatment strategies.
Keywords: perinatal, HIV drug resistance, subtype diversity, prevalence
Over the past decade, a dramatic reduction in the rate of mother-to-child transmission (MTCT) of HIV has occurred in the United States and elsewhere in the developed world. Antiretroviral (ARV) treatment of pregnant women has evolved from the 3-part zidovudine regimen established as a result of the Pediatric AIDS Clinical Trials Group protocol 076 to the current recommendation for highly active antiretroviral treatment during pregnancy.1 Additional prevention efforts, including increased access to HIV testing and prenatal care, elective cesarean delivery, and formula feeding, have also played a major role in decreasing MTCT rates. However, despite these immense gains, MTCT continues to occur at a low frequency in the United States.2
The widespread use of highly active antiretroviral treatment has greatly reduced morbidity and mortality due to HIV infection, but it has also resulted in the emergence of ARV-resistant HIV in the treated population.3 Transmission of drug-resistant HIV in the adult population has been widely reported4–8 and has been associated with suboptimal virologic response to initial ARV therapy.5 Studies on HIV drug resistance in newly infected adults have shown that the prevalence of primary drug resistance has increased over time, and mutation patterns have evolved as well.9,10 However, data on the prevalence and patterns of drug-resistance mutations in the newly diagnosed pediatric HIV-infected population are much more limited. In a retrospective analysis of diagnostic proviral DNA specimens, drug-resistance mutations were detected in 12% of 91 perinatally infected infants born in New York State in 1998 and 1999.11 Notably, the occurrence of drug-resistant virus in this infant cohort was not associated with perinatal ARV exposure.
In addition to characterizing drug resistance in the HIV-infected infant population, it is also of interest to monitor the distribution of HIV-1 subtypes. Diagnosis of HIV infection in infants is contingent on detection of HIV proviral DNA. Several HIV DNA polymerase chain reaction (PCR) assays currently in use for infant diagnosis have not been optimized for detection of non–subtype B HIV strains. Furthermore, antiretroviral treatment efficacy and the consequences of drug resistance have predominantly been studied in subtype B HIV strains. Therefore, it is important to characterize the genetic diversity of HIV strains in the infected infant population to aid in the development of more effective diagnostic and disease management strategies for perinatal HIV infection. We undertook the current study to determine the prevalence of drug-resistant and non–subtype B strains of HIV in perinatally infected infants diagnosed in New York State in 2001 and 2002 and to investigate the association between perinatal ARV exposure and drug resistance in this population.
METHODS
Specimens
In this retrospective analysis, remnant peripheral blood mononuclear cell (PBMC) DNA from HIV diagnostic specimens was analyzed. There were 51 infants diagnosed as HIV positive by the New York State Department of Health Wadsworth Center Laboratory in 2001 and 2002, representing approximately 90% of all HIV-infected infants born in New York State during this period. Of these, 47 were eligible for the study based on the following inclusion criteria: (1) residence within New York State at the time of specimen collection; (2) diagnosis of HIV confirmed by 2 consecutive, positive PCR tests; and (3) availability of a PCR-positive specimen collected when the infant was 24 weeks of age or younger. The earliest available PCR-positive specimen was obtained for analysis. Data on age of infant at diagnosis, region of residence, and antiretroviral exposure in the antepartum, intrapartum, and newborn periods were obtained by chart review, as part of the New York State monitoring program for HIV-exposed infants. A study database was established, and specimens were blinded before analysis. This study was approved by the New York State Department of Health Institutional Review Board.
PCR Amplification and Sequencing
PBMC DNA was prepared as previously described.12 The protease and reverse transcriptase (RT) regions were amplified by nested PCR using the GeneAmp XL PCR kit (Applied Biosystems, Foster City, CA). For primary PCR, 10 μL of PBMC DNA was amplified using outer primers 1828F (5′-ATGACAGCATGTCAGGGAG-3′) and 3887R (5′-AGCTGCYCCATCTACATAGAAAG-3′). Secondary PCR was identical to the primary round, except that 10 μL of the primary PCR product was amplified with inner primers 2138F (5′-AGAGCAGACCAGAGCCAACAG-3′) and 3813R (5′-AGGGAGGGGTATTGACAAAC-3′). The 1.6-kilobase product was extracted from 1% agarose by using the QIAquick gel extraction kit (Qiagen, Valencia, CA).
Codons 1 to 99 of protease and 1 to 446 of RT were bidirectionally sequenced with the Big Dye Terminator v3.1 Cycle Sequencing kit and 3700 DNA Analyzer (Applied Biosystems) using primers 2138F, 3813R, HIV2518F (5′-TG TTGACTCAGATTGGYTGCAC-3′), HIV2984R (5′-CTG ATATCTAATYCCTGGTG-3′), HIV2868F (5′-ACAGTACT GGATGTGGGTG-3′), and SR3462R (5′-CTGCCAGTTCTAGCTCTGCTTC-3′). An alternative primer, HIV2868AG-F (5′-ACAGTACTAGATGTGGGGG-3′), was used to obtain sequence from some non–subtype B strains. For 2 specimens, amplification failed with this procedure, and sequences were obtained using the TRUGENE HIV-1 Genotyping Kit and OpenGene DNA Sequencing System (Bayer Diagnostics, Inc, Tarrytown, NY) with modifications for amplifying proviral DNA.11
Sequence Analysis
Consensus sequences were assembled using Sequencher 4.2. Drug-resistance mutations were identified using the HIVdb program of the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu), in accordance with the drug-resistance mutation list of the International AIDS Society–USA.13 Reference sequences were obtained from the Los Alamos National Laboratory Database (http://hiv-web.lanl.gov/content/hiv-db/mainpage.html) and aligned with the study sequences using CLUSTAL W.14 Phylogenetic trees were constructed by the neighbor-joining method using the Kimura 2-parameter model and assessed for reliability by bootstrap sampling with a minimum of 1000 replicates. HIV subtypes were determined by phylogenetic analysis using MEGA version 3.015 and were confirmed by distance plotting and bootscanning using SimPlot v2.5 (http://sray.med.som.jhmi.edu/SCRoftware/simplot/).
Statistical Analysis
The 95% confidence intervals (CI) including continuity correction were computed using programs available at http://faculty.vassar.edu/lowry/prop1.html. The significance of association between perinatal ARV exposure and the occurrence of resistance mutations was calculated using Fisher exact test (Epi Info 2002).
RESULTS
Drug-Resistance Data
The protease and RT regions of proviral HIV DNA from 42 of 47 eligible infants were amplified and sequenced successfully. At least 1 major drug-resistance mutation was detected in provirus from 8 of 42 infants (19.1%; CI: 9.1% to 34.6%) (Table 1). Five of these infants resided in New York City, and 3 resided elsewhere in New York State at the time of delivery. Three infants (7.1%; CI: 1.9% to 20.6%) had pro-virus carrying mutations conferring resistance to a nucleoside RT inhibitor (NRTI). Despite zidovudine exposure to 66.7% of infants in the antepartum or intrapartum periods and to 92.9% during the newborn period, none of the NRTI mutations observed were thymidine analogue mutations (TAMs), a subclass of NRTI mutations selected predominantly by zidovudine and stavudine. Provirus from 5 infants had the RT mutation G333E, which has been associated with dual resistance to zidovudine and lamivudine in a background of NRTI resistance mutations.16 In each case, the G333E mutation was detected in the absence of additional mutations and, therefore, was considered a naturally occurring polymorphism and not a resistance mutation for this analysis. Five infants (11.9%; CI: 4.5% to 26.4%) had provirus carrying a mutation conferring resistance to a nonnucleoside RT inhibitor (NNRTI), including 3 with the K103N mutation. A major protease resistance mutation was detected in one (2.4%; CI: 0.1% to 14.1%) specimen. Provirus from one (2.4%; CI: 0.1% to 14.1%) infant had mutations predictive of dual-class resistance (Table 1).
TABLE 1.
Mutation Profiles and Perinatal ARV Exposure of Infants With Predicted ARV Resistance
| Study ID | Genotyping Results |
Perinatal ARV Exposure |
|||||
|---|---|---|---|---|---|---|---|
| NRTI | NNRTI | PI | Resistance Interpretation* | Antepartum | Intrapartum | Newborn | |
| NYS158 | M184V | K103N | — | 3TC, FTC, ABC, DDI, DLV, EFV, NVP | ZDV, 3TC, NVP | ZDV | ZDV |
| NYS146 | — | K103N | — | DLV, EFV, NVP | ZDV, 3TC, IDV, NFV | ZDV | ZDV |
| NYS106 | — | K103N | — | DLV, EFV, NVP | ZDV, 3TC, ABC | ZDV | ZDV |
| NYS124 | A62V, M184V | — | — | 3TC, FTC, ABC, DDI | ZDV, 3TC, NVP | ZDV | ZDV |
| NYS154 | — | V108I | — | DLV, EFV, NVP | — | — | ZDV |
| NYS150 | — | Y181C | — | DLV, EFV, NVP | — | — | ZDV |
| NYS145 | — | — | M46L | ATV, NFV, APV, IDV, LPV | — | — | ZDV |
| NYS127 | M184V | — | — | 3TC, FTC, ABC, DDI | — | — | ZDV |
Bold type, drugs with predicted high-level resistance; light type, drugs with predicted low-level or potential low-level resistance; underline, drugs that were administered in a perinatal period.
PI indicates protease inhibitors; DLV, delavirdine; EFV, efavirenz; NVP, nevirapine; 3TC, lamivudine; FTC, emtricitabine; ABC, abacavir; DDI, didanosine; ATV, atazanavir; NFV, nelfinavir; APV, amprenavir; IDV, indinavir; LPV, lopinavir.
ARV Exposure Analysis
ARV exposure occurred in the antepartum period for 17 (40%), the intrapartum period for 24 (57%), and both periods for 13 (31%) of the 42 infants analyzed. Forty of the 42 infants (95.2%) received ARV chemoprophylaxis during the 6-week newborn period after birth. Most infants with resistance mutations had no recorded exposure to a corresponding ARV. Of the 8 babies predicted to have drug-resistant strains of HIV, 4 (50.0%) were exposed to ARV drugs in the antepartum and intrapartum perinatal periods (Table 1). Of these 4 infants, only 2 had mutations associated with resistance to an ARV administered in a perinatal period (Table 1). Antepartum ARV exposure was not significantly associated with the occurrence of detectable drug-resistance mutations (P = 0.70).
Phylogenetic Analysis
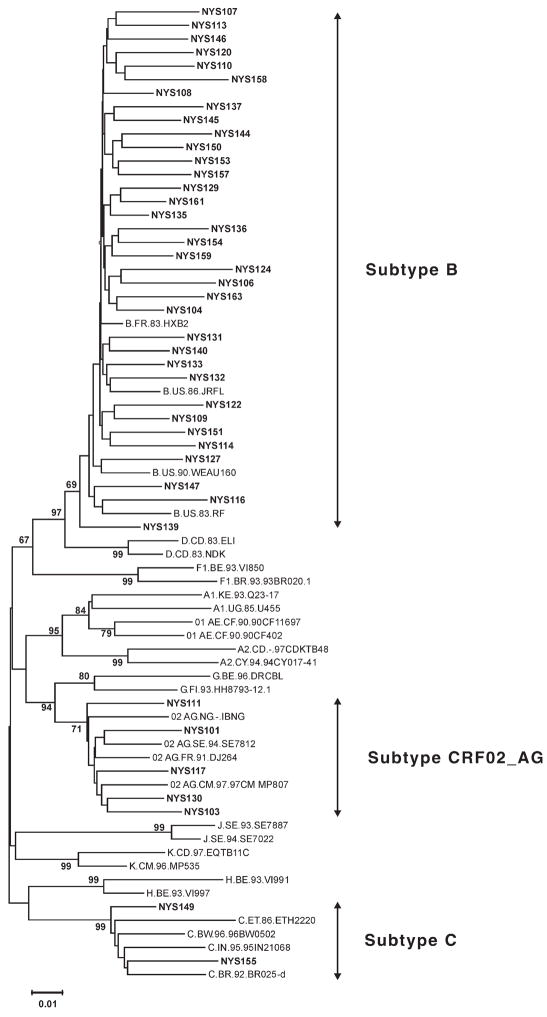
Phylogenetic analysis of pol region sequences suggested that 7 (16.7%; CI: 7.5% to 32.0%) infants were infected with a non–subtype B strain of HIV (Fig. 1). Two (4.8%; CI: 0.8% to 17.4%) infants were infected with putative subtype C virus, and 5 (11.9%; CI: 4.5% to 26.4%) were infected with a circulating recombinant form (CRF02). Six of the 7 infants infected with a non–subtype B strain resided in New York City, and one resided elsewhere in New York State at the time of delivery. None of the infants infected with non–subtype B strains of HIV had any major drug-resistance mutations. Four of the 7 (57.1%) infants infected with a non–subtype B strain were exposed to ARVs in the antepartum and/or intrapartum periods, and 3 (42.9%) had no ARV exposure in either period. Each non–subtype B strain carried one or more common, subtype-specific polymorphisms classified as minor protease-resistance mutations, including the protease-specific polymorphisms K20I and M36I in the CRF02 strains, and M36I, L63P, and I93L in the subtype C strains.
FIGURE 1.
Phylogenetic tree of pol sequences from HIV-infected infants. A phylogenetic tree was constructed by the neighbor-joining method, using the Kimura 2-parameter model. Bootstrap values of key branch nodes are indicated for 1000 data sets. Subtype prediction based on this analysis is shown to the right. Sequences from New York State infants diagnosed in 2001 and 2002 are indicated by boldface type.
DISCUSSION
Although the rate of MTCT has decreased over time, the proportion of infected infants who acquire drug-resistant virus seems to be rising. The prevalence of drug-resistance mutations increased 1.6-fold over the prevalence of 12.1% observed among infants diagnosed in 1998–1999.11 There was little or no change in the prevalence of resistance mutations to protease inhibitors and NRTIs, but the prevalence of NNRTI-resistance mutations rose by 3.6-fold in comparison with the 1998–1999 study. Therefore, the overall rise in drug-resistance mutations seems predominantly to be due to an increase in the prevalence of NNRTI-resistance mutations. These findings from the perinatally infected pediatric population of New York State are consistent with data from the adult ARV-naive population, in which increases in the prevalence of NNRTI resistance ranging from 2.0- to 4.3-fold have been reported over similar periods.5,9,10
There were several important limitations to the study that should be considered. First, we did not have access to maternal specimens, previous genotype results, or maternal ARV history before pregnancy, which would have allowed additional analysis of possible risk factors associated with transmission of drug-resistant virus. Second, we did not have data on the country of origin of the mother, which would have provided additional insight into the analysis of non–subtype B strains. Third, in the current study, we used a different genotyping assay from that used in the 1998–1999 study, and therefore, the observed increase in drug resistance could be attributed to differences in assay sensitivity. However, we think this is unlikely given that both studies used proviral DNA and given that the amplification success rates of 80.5% for the 1998–1999 cohort and 89.3% for the 2001–2002 cohort were comparable.
Although the overall prevalence of NRTI-resistance mutations seems to have remained stable over the period of 1998 to 2002, the mutation patterns associated with NRTI resistance have changed over time. The absence of TAMs in the current analysis is in contrast to previous results in which 5 of 11 infants with resistance mutations had one or more TAMs.11 This result was unexpected given that (1) zidovudine is a commonly used ARV, especially in the perinatal setting; (2) TAMs are frequently detected in the general ARV-treated population;3 and (3) the prevalence of NRTI-resistance mutations, including TAMs, has increased in the adult ARV-naive population during this period.5,9,10 It is not clear whether this apparent shift in mutation patterns in our pediatric population reflects a corresponding change in HIV-infected women of childbearing age or whether it is an artifact of the relatively small number of infants analyzed.
Consistent with previous findings, perinatal ARV exposure was not associated with the occurrence of drug-resistance mutations.11 There are several plausible explanations to account for the occurrence of drug-resistant HIV in infants despite the absence of selective drug pressure in the perinatal period. For example, resistance mutations could have emerged in the mother during ARV treatment that occurred before pregnancy, with drug-resistant variants persisting at high levels after treatment was discontinued. Although a rapid shift from resistant to wild-type virus is observed frequently after removal of selective drug pressure, drug-resistant variants have been shown to persist at detectable levels for several months after discontinuation of treatment.17,18 Furthermore, persistence of drug-resistant HIV in the absence of selective drug pressure has been observed in the genital tract.19 Alternatively, the mother could have become infected with a drug-resistant strain of HIV, either before or during pregnancy, and could have transmitted this strain to the infant while remaining ARV naive. In a study of HIV-infected pregnant women in St. Louis, MO, 3 of 18 (17%) ARV-naive women were found to have NNRTI-resistant virus.20 MTCT of HIV during primary infection of the mother has been observed,21 and long-term persistence of drug-resistant variants acquired in primary infection has been well documented.22–26 Finally, it is possible that the mother was exposed to ARV drugs during the pregnancy, but her treatment was not documented.
The prevalence rate of 16.7% for non–subtype B HIV strains in this infant cohort is considerably higher than prevalence estimates reported for other US cohorts, with estimates ranging from 1.2% to 5.4% in the adult population.27–29 We have estimated the prevalence of non–subtype B strains to be 4.4% in our previously described infant cohort,11 suggesting that subtype diversity among the perinatally infected population of New York State may be increasing. This is particularly important given that false-negative HIV DNA PCR test results have been reported in the United States for infants infected with non–subtype B strains of HIV.30,31 It is not clear whether the apparent rise in non–subtype B infections reflects increasing subtype diversity in the population of HIV-infected women giving birth in New York State or whether women infected with non–subtype B strains are more likely to become pregnant or deliver than women infected with subtype B strains.
The current guidelines for the use of antiretroviral agents in pediatric HIV infection recommend that resistance testing be considered in newly diagnosed infants younger than 12 months before initiation of therapy and emphasize the importance of monitoring the frequency of drug-resistant HIV in newly infected infants.32 The guidelines also indicate that in cases where perinatal exposure to a non–subtype B strain is suspected, a negative diagnostic test should be interpreted with caution, and the use of tests shown to be sensitive to non–B strain detection is recommended.32 The results of our study indicate that drug-resistant and non–subtype B HIV strains seem to be increasing in the perinatally infected population. Data on the clinical benefit of resistance testing upon diagnosis of HIV infection in infants are currently lacking. However, based on the results of this study, drug-resistance testing of all infants upon confirmation of HIV infection is warranted. Furthermore, infection with a non–subtype B HIV strain should be considered in interpretation of diagnostic test results and in developing disease monitoring and treatment strategies for the pediatric population.
Acknowledgments
Supported by the Emerging Infection and HIV Training and Research Program grants 1D43TW007384-01 and 2D43TW000233-11 from the NIH Fogarty International Center.
The authors acknowledge the assistance of Renee Hallack, Joseph Smith, and Kimdar Kemal for technical assistance and valuable comments on the article. We thank the Molecular Genetics Core Facility at the Wadsworth Center for performing DNA sequencing.
References
- 1.Perinatal HIV Guidelines Working Group. Public Health Service Task Force recommendations for the use of antiretroviral drugs in pregnant HIV-1 infected women for maternal health and interventions to reduce perinatal HIV-1 transmission in the United States. 2005 February 25; Available at: http://aidsinfo.nih.gov/guidelines/perinatal/PER_022405.pdf.
- 2.Mofenson LM. Successes and challenges in the perinatal HIV-1 epidemic in the United States as illustrated by the HIV-1 serosurvey of childbearing women. Arch Pediatr Adolesc Med. 2004;158:422–425. doi: 10.1001/archpedi.158.5.422. [DOI] [PubMed] [Google Scholar]
- 3.Richman DD, Morton SC, Wrin T, et al. The prevalence of antiretroviral drug resistance in the United States. AIDS. 2004;18:1393–1401. doi: 10.1097/01.aids.0000131310.52526.c7. [DOI] [PubMed] [Google Scholar]
- 4.Boden D, Hurley A, Zhang L, et al. HIV-1 drug resistance in newly infected individuals. JAMA. 1999;282:1135–1141. doi: 10.1001/jama.282.12.1135. [DOI] [PubMed] [Google Scholar]
- 5.Little SJ, Holte S, Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 6.Weinstock HS, Zaidi I, Heneine W, et al. The epidemiology of antiretroviral drug resistance among drug-naive HIV-1–infected persons in 10 US cities. J Infect Dis. 2004;189:2174–2180. doi: 10.1086/420789. [DOI] [PubMed] [Google Scholar]
- 7.Wensing AM, van de Vijver DA, Angarano G, et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J Infect Dis. 2005;192:958–966. doi: 10.1086/432916. [DOI] [PubMed] [Google Scholar]
- 8.Yerly S, Kaiser L, Race E, et al. Transmission of antiretroviral-drug–resistant HIV-1 variants. Lancet. 1999;354:729–733. doi: 10.1016/S0140-6736(98)12262-6. [DOI] [PubMed] [Google Scholar]
- 9.Grant RM, Hecht FM, Warmerdam M, et al. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288:181–188. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 10.Simon V, Vanderhoeven J, Hurley A, et al. Evolving patterns of HIV-1 resistance to antiretroviral agents in newly infected individuals. AIDS. 2002;16:1511–1519. doi: 10.1097/00002030-200207260-00008. [DOI] [PubMed] [Google Scholar]
- 11.Parker MM, Wade N, Lloyd RM, Jr, et al. Prevalence of genotypic drug resistance among a cohort of HIV-infected newborns. J Acquir Immune Defic Syndr. 2003;32:292–297. doi: 10.1097/00126334-200303010-00008. [DOI] [PubMed] [Google Scholar]
- 12.Charbonneau TT, Wade NA, Weiner L, et al. Vertical transmission of HIV in New York State: a basis for statewide testing of newborns. AIDS Patient Care STDS. 1997;11:227–236. doi: 10.1089/apc.1997.11.227. [DOI] [PubMed] [Google Scholar]
- 13.Johnson VA, Brun-Vezinet F, Clotet B, et al. Update of the drug resistance mutations in HIV-1: 2005. Top HIV Med. 2005;13:51–57. [PubMed] [Google Scholar]
- 14.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 16.Kemp SD, Shi C, Bloor S, et al. A novel polymorphism at codon 333 of human immunodeficiency virus type 1 reverse transcriptase can facilitate dual resistance to zidovudine and L-2′,3′-dideoxy-3′-thiacytidine. J Virol. 1998;72:5093–5098. doi: 10.1128/jvi.72.6.5093-5098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quiros-Roldan E, Airoldi M, Moretti F, et al. Genotype resistance profiles in patients failing an NNRTI-containing regimen, and modifications after stopping NNRTI therapy. J Clin Lab Anal. 2002;16:76–78. doi: 10.1002/jcla.10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller V, Sabin C, Hertogs K, et al. Virological and immunological effects of treatment interruptions in HIV-1 infected patients with treatment failure. AIDS. 2000;14:2857–2867. doi: 10.1097/00002030-200012220-00007. [DOI] [PubMed] [Google Scholar]
- 19.Newstein M, Losikoff P, Caliendo A, et al. Prevalence and persistence of nonnucleoside reverse transcriptase inhibitor mutations in the female genital tract. J Acquir Immune Defic Syndr. 2005;38:364–366. [PubMed] [Google Scholar]
- 20.Juethner SN, Williamson C, Ristig MB, et al. Nonnucleoside reverse transcriptase inhibitor resistance among antiretroviral-naive HIV-positive pregnant women. J Acquir Immune Defic Syndr. 2003;32:153–156. doi: 10.1097/00126334-200302010-00006. [DOI] [PubMed] [Google Scholar]
- 21.Van Tine BA, Shaw GM, Aldrovandi G. Mother-to-infant transmission of the human immunodeficiency virus during primary infection. N Engl J Med. 1999;341:1548. doi: 10.1056/NEJM199911113412014. [DOI] [PubMed] [Google Scholar]
- 22.Brenner BG, Routy JP, Petrella M, et al. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. J Virol. 2002;76:1753–1761. doi: 10.1128/JVI.76.4.1753-1761.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briones C, Perez-Olmeda M, Rodriguez C, et al. Primary genotypic and phenotypic HIV-1 drug resistance in recent seroconverters in Madrid. J Acquir Immune Defic Syndr. 2001;26:145–150. doi: 10.1097/00042560-200102010-00006. [DOI] [PubMed] [Google Scholar]
- 24.Gandhi RT, Wurcel A, Rosenberg ES, et al. Progressive reversion of human immunodeficiency virus type 1 resistance mutations in vivo after transmission of a multiply drug-resistant virus. Clin Infect Dis. 2003;37:1693–1698. doi: 10.1086/379773. [DOI] [PubMed] [Google Scholar]
- 25.Little SJ, Dawson K, Hellmann NS, et al. Persistence of transmitted drug-resistant virus among subjects with primary HIV infection deferring antiretroviral therapy. Antivir Ther. 2003:8. [Google Scholar]
- 26.Smith MS, Koerber KL, Pagano JS. Long-term persistence of zidovudine resistance mutations in plasma isolates of human immunodeficiency virus type 1 of dideoxyinosine-treated patients removed from zidovudine therapy. J Infect Dis. 1994;169:184–188. doi: 10.1093/infdis/169.1.184. [DOI] [PubMed] [Google Scholar]
- 27.Weidle PJ, Ganea CE, Irwin KL, et al. Presence of human immunodeficiency virus (HIV) type 1, group M, non-B subtypes, Bronx, New York: a sentinel site for monitoring HIV genetic diversity in the United States. J Infect Dis. 2000;181:470–475. doi: 10.1086/315253. [DOI] [PubMed] [Google Scholar]
- 28.Brodine SK, Starkey MJ, Shaffer RA, et al. Diverse HIV-1 subtypes and clinical, laboratory and behavioral factors in a recently infected US military cohort. AIDS. 2003;17:2521–2527. doi: 10.1097/00002030-200311210-00016. [DOI] [PubMed] [Google Scholar]
- 29.Delwart EL, Orton S, Parekh B, et al. Two percent of HIV-positive US blood donors are infected with non–subtype B strains. AIDS Res Hum Retroviruses. 2003;19:1065–1070. doi: 10.1089/088922203771881149. [DOI] [PubMed] [Google Scholar]
- 30.Kline NE, Schwarzwald H, Kline MW. False negative DNA polymerase chain reaction in an infant with subtype C human immunodeficiency virus 1 infection. Pediatr Infect Dis J. 2002;21:885–886. doi: 10.1097/00006454-200209000-00023. [DOI] [PubMed] [Google Scholar]
- 31.Obaro SK, Losikoff P, Harwell J, et al. Failure of serial human immunodeficiency virus type 1 DNA polymerase chain reactions to identify human immunodeficiency virus type 1 clade A/G. Pediatr Infect Dis J. 2005;24:183–184. doi: 10.1097/01.inf.0000151040.57772.40. [DOI] [PubMed] [Google Scholar]
- 32.Working Group on Antiretroviral Therapy and Medical Management of HIV-Infected Children. Guidelines for the use of antiretroviral agents in pediatric HIV infection. 2005 November 3; Available at: http://aidsinfo.nih.gov/guidelines/pediatric/PED_11032005.pdf.