Abstract
To investigate the viral features of long-term nonprogressive HIV-1 infection and the selection of viral genomes, we studied serial complete HIV-1 sequences obtained from a mother–child pair, both long-term nonprogressors. Analysis of four genomic sequences demonstrated that all viral genes were intact, lacking major deletions or premature stop codons to easily explain the slow disease progression. These data suggest that viral attenuation, if present, was caused by subtle sequence variations or virus–host interactions. Serial sequences from an HIV-1-infected mother–child pair afforded us the opportunity to examine the immune selection of HIV-1 sequences years after transmission between individuals. We demonstrated that the daughter's strains were most likely subjected to immunoselection or immunoediting according to the presence of novel MHC class I alleles that differed between mother and daughter. An analysis of nef-specific cytotoxic T-lymphocyte responses in the child, whose HIV-1 nef sequence differed from the maternal nef, supported this interpretation. This study highlights the potential of full genome analysis in the investigation of pathogenesis and immune selection during HIV-1 evolution.
Genetic analysis of epidemiologically linked HIV-1 strains from close contacts, including mother and child pairs, represents a valuable tool for a better understanding of viral biology, epidemiology, transmission, and disease progression. Long-term nonprogressive HIV-1 infection offers an opportunity to examine the correlates of protection from disease progression. Mother-to-child transmission (MTCT) makes it possible to study both transmission and pathogenesis in a setting where both individuals share half of their genetic background.
The genetic differences between HIV-1 strains from a mother–child pair can be attributed in some cases to selective transmission as well as to divergent viral evolution after transmission. One study observed that minority viral variants in the mother may be transmitted to the child, establishing a new infection with a more homogeneous viral population than that in the mother. The genetic distance between HIV-1 strains derived from mother and child in these pairs is greater than the distance between sequential isolates from the same individual.1 Other studies have shown that the selection of viral strains may vary, with transmission of multiple variants or no evidence of selection of minor strains.2,3
A small but significant fraction of HIV-1-infected individuals does not progress to AIDS. Instead, it remains asymptomatic for greater than 10 years, with normal numbers of CD4+ T-lymphocytes without antiretroviral therapy (ART); these individuals are called long-term nonprogressors (LTNP). Nonprogression or slow progression of the disease can be attributed to viral, immunologic, and host characteristics and their interrelations.4–6 A number of studies including investigation of epidemiologically linked cohorts revealed that infection with attenuated viruses may result in persistent infection without disease progression. These strains can harbor large deletions or modifications throughout the genome. There is also evidence, however, that the pathogenicity and biological properties of the particular viral strain can be influenced by small, even point mutations or other gene variations in critical sites of the genome.5
The pace of disease progression varies in infected children. Ganeshan et al.7 reported that greater viral genetic distances were observed in viral strains from children with slow disease progression compared to those with rapid progression; these greater distances correlated with a higher accumulation rate of nonsynonymous base substitutions per potential mutational site rather than with a difference in replication kinetics. These results suggest that a stronger host selection pressure may be at play in the slow progressors.
Disease progression is dependent on the effectiveness of the cellular immune control of HIV-1 and heterozygosity of HLA class I alleles has been associated with delayed progression.4 A study of the role of the immune response in MTCT demonstrates that transmission of viral strains possessing amino acid substitutions within targeted cytotoxic T-lymphocyte (CTL) epitopes plays an important role in vertical transmission of the virus.8 Distinct immunoselective pressure driven by cellular immune responses can also contribute to the diversification of the viral strains in mother and child during the course of infection. The immunoediting of the virus under the selection pressure of the host immune system also results in the development of escape variants capable of evading the host HLA-restricted immune response through mutations in CTL epitopes.8,9 It was recently reported that a viral variant bearing crucial escape mutations was transmitted to a host lacking the HLA alleles driving the selection, whereupon reversion of the escape mutations in the new host occurred.10 MTCT of HIV-1 is particularly interesting in part because the mother and child share half of the MHC class I alleles between them. Theoretically, in vertical transmission, both escape and reversion can take place in the evolution of the viral strains.
A strong cell-mediated immune response can suppress the viral load and prevent or slow disease progression. The host immunogenetic background, including mutations in chemokine receptor genes for CCR5 and CCR2, influences the pace of HIV-1 disease progression. Humoral immune responses, including neutralizing antibodies against HIV-1 and other antibodies directed against CCR5, have been associated with LTNP infection.11 Taken together, there are multiple possible determinants of slow or no disease progression and analysis of each individual case can increase our understaning of viral pathogenicity and virus–host interactions.
Most studies of MTCT and LTNP to date have analyzed selected viral genes or gene fragments, with little attention devoted to complete genomic sequences. Investigation of full-length viral sequences provides the opportunity to perform complex genetic analysis of the parental and transmitted HIV-1 strains. To investigate the viral features of long-term nonprogressive HIV-1 infection and the selection of viral genomes, we studied serial complete HIV-1 sequences obtained from a mother–child pair, both long-term nonprogressors. Serial sequences from this pair provided the opportunity to examine the immune selection of HIV-1 sequences years after transmission from mother to child.
We analyzed HIV-1 strains from a mother and daughter of Hispanic origin who were followed at Stony Brook University Hospital in Stony Brook, NY. The investigation was approved by the institutional review board at each site and the mother signed informed consent for herself and her daughter. The mother's risk for infection was intravenous drug use (IDU), and she transmitted HIV-1 to her daughter perinatally in 1984, possibly shortly after her primary HIV-1 infection. Both mother and daughter remained well, except for asthma in the mother, and both maintained normal CD4+ T cell numbers for more than 10 years without antiretroviral therapy; they were therefore classified as LTNP (Table 1). CD4+ T-lymphocyte subsets were determined by using standard flow cytometry, and plasma viral loads were determined by using the NASBA procedure (BioMerieux). Interestingly, the viral load was consistently similar in both individuals and relatively high (approximately 35,000 copies/ml of plasma) compared to typical patients with slow or nonprogressive disease (Table 1). The mother maintained a normal CD4+ cell count (CD4 >500/μl) until 1996 without antiviral therapy. In December 1996, when her CD4+ cell count fell to 396/μl, she started ART with AZT, 3TC, and ritonavir. In 1998 she suffered a severe asthma attack and died.
Table 1.
Clinical Features of Mother and Daughter
| Date | CD4+ T cells | Viral load | Clinical status |
|---|---|---|---|
| Mother | |||
| 1984 | Asymptomatic; gave birth to girl HIV-infected perinatally | ||
| 1992a | 650 | 41,400 | Asymptomatic |
| 1993 | 680 | 35,000 | Asymptomatic |
| 1995 | 630 | Occasional asthma | |
| 1996 | 396 | 32,000 | Occasional asthma; starts ART (ritonavir, AZT, 3TC) |
| 2/97 | 984 | 5,000 | Asymptomatic |
| 1998 | Died from asthma attack | ||
| Daughter | |||
| 1984 | Born, HIV-infected perinatally | ||
| 1993 | 1090 | 35,000 | Asymptomatic |
| 1994 | 860 | 29,000 | Asymptomatic |
| 9/95 | 660 | Subtle neurologic deterioration, poor performance in school. | |
| 11/95 | 3,400 | Started ART (AZT, 3TC, ddI) | |
| 4/97 | 13,000 | Well, improvement in academic performance |
This woman was investigated as part of a mother-to-child transmission study during a subsequent pregnancy in 1992. She did not transmit HIV-1 to her second child and is identified as Mother 11 in a previous publication.21
The daughter, infected vertically in 1984, remained asymptomatic until 1995, when she began to experience subtle neurologic changes, with loss of intellectual ability and worsening academic performance in school. At the same time, her CD4+ T cell count fell from 860/μl in 1994 to 660/μl in 1995. Based on these changes, she was started on ART with AZT, 3TC, and ddI. She responded clinically, with clear improvement in her academic performance. Currently, at 22 years of age, she is in very good health, with excellent control of viral replication. She is married and gave birth to a child in 2004 who is HIV-1 negative.
Plasma samples were obtained from the mother in 1993 [Sample M1, asymptomatic stage, viral load (VL) = 35,000 copies/ml], and 1996, before ART began (Sample M2, VL = 32,000 copies/ml). The daughter's samples were obtained in 1993 (Sample D1, VL = 35,000 copies/ml), and in 1995, approximately 1 month after initiating ART (Sample D2, VL = 3400 copies/ml). Both mother and daughter were homozygous for the wild-type CCR5 gene. The HLA haplotype of the mother was A19, A1, B44, B35 (B*3501), Cw16, and Cw7. The child shared the A19, B44, and Cw16 alleles with the mother and her unique alleles were A2 (A*0201), B14, and Cw8.
Virion-derived RNA was extracted from plasma specimens by using the RNAgen kit (Promega). Reverse transcription was performed by using a PE Biosystems RT-PCR kit and long cDNA fragments were amplified by nested polymerase chain reaction (PCR) using rTh polymerase (GeneAmp XL PCR Kit, PE Biosystems, Foster City, CA). Nearly full-length HIV-1 genomes (nt 601–9539 according to HXB2) were obtained from several overlapping fragments by reverse transcription and long PCR amplification of genomes; the procedure was performed as described previously, with slight modifications.12 The four overlapping, amplified sets of fragments derived from samples M1, M2, and D1 encompassed nearly full genome sequences and corresponded to HIV-1 nt positions 601–2334, 2043–5220, 4956–9173, and 9060–9540 (reference strain HXB2). In the D2 strain, the 4956–9173 fragment could not be amplified as a single sequence and was composed of two fragments, 4956–6991 and 6983–9173. Both strands of DNA templates were directly sequenced by using an automated fluorescent sequencer (Applied Biosystems) at the Wadsworth Center Molecular Genetics Core facility. The sequences of the LTRs that could not be obtained from the ends of the RNA genome were derived from proviral long terminal repeat (LTR) fragments by PCR amplification and direct sequencing. DNA was extracted from peripheral blood lymphocytes sampled at approximately the same time points as were the plasma samples by using the QIAMP DNA minikit (Qiagene). The DNA fragments spanning the LTR and gag regions (nt 9428–1064 and 601–2334) were amplified and subjected to direct sequencing as described above. The viral genomic sequences were completed by adding these proviral LTR DNA sequences (nt 456–600 and 9540–9719). In some cases, the heterogeneity of the PCR products representing the LTRs was assessed through cloning by using the TOPO TA kit (Invitrogen, Carlsbad, CA). Plasmid DNA was isolated with the QIAprep 8 Turbo Miniprep Protocol (QIAgen) and multiple clones were sequenced. Similarly, selected gag fragments obtained by using reverse transcriptaes (RT)-PCR were cloned and sequenced.
The sequences were aligned with each other and with reference subtype B sequences (available at the Los Alamos National Laboratory HIV database; http://hiv-web.lanl.gov) by using the CLUSTAL W program and then corrected manually. The sequences were also compared to the subtype B consensus sequence and the differences were subjected to further analysis. The sequence identities between the isolates were calculated by using the Genedoc program package. Synonymous and nonsynonymous substitution rates were calculated with a SNAP (Synonymous/Nonsynonymous Analysis Program) based on the method of Nei and Gojobori (http://hiv-web.lanl.gov). HIV-1 coreceptor usage predictions were based on evaluation of the V3 loop env sequences.13 The phylogenetic trees were constructed by using the Treecon phylogenetic software package;14 pairwise genetic distances were calculated by the Kimura two-parameter method, and the tree topologies were inferred by using the neighbor-joining method.
The putative CTL epitopes related to the MHC class I haplotypes of the mother and daughter were identified from the genomic sequences by using the previously characterized epitopes deposited in the Los Alamos HIV immunology database. Sequence variations in particular strains were compared to each other and the differences were recorded. Modifications suggesting disruption of the epitope were classified as “mutations,” whereas changes restoring the epitope were classified as reverse mutations. Epitopes in which no changes were recorded were not taken into account.
The ELISPOT assay was performed as previously reported.15 Briefly, 96-well Millipore plates (Millipore Corp, Bedford, UK) were coated with 1-DIK monoclonal antibody (Mab) at 15 μg/ml (Mabtech, Nacka, Sweden). Peripheral blood mononuclear cells (PBMC) from HIV-1-infected subjects were added to the plates in duplicate at 1 × 105/well and stimulated with either phytohemagglutinin (PHA) at 20 μg/ml (positive control, Murex Biotech, Dartford, UK), R10 media alone (peptide control), or 2 μg/ml peptides. Following incubation, anti-interferon (IFN)-γ-Mab 7-B6-1 at 1 μg/ml (Mabtech) was used as a secondary antibody. Plates were developed with an alkaline phosphatase-conjugate substrate kit (Bio-Rad Laboratories, Hercules, CA) and the spots were counted with the AID ELISPOT reader and software (Autoimmun Diagnostika, Strasbourg). Spot forming units (SFU) were defined as the average number of spots in duplicate wells, and HIV-1-specific SFU (HIVSFU) was defined as SFU minus the average number of spot forming cells in the negative control wells.
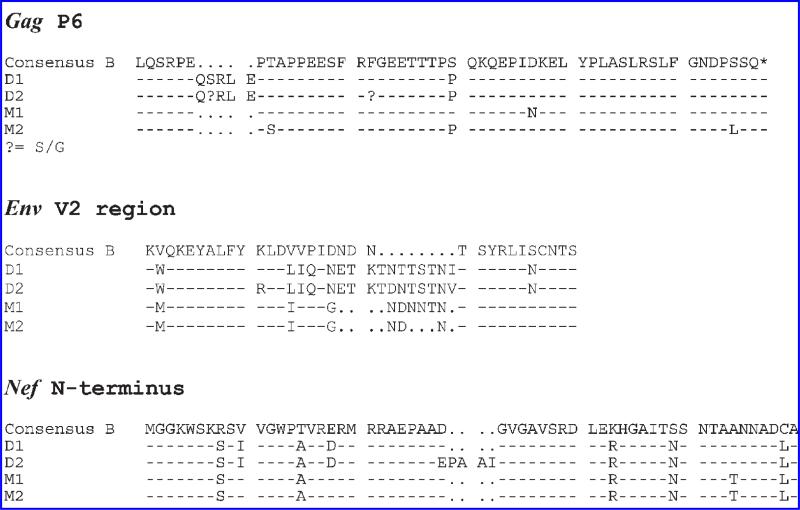
Four full-genome RNA sequences (two from the mother, M1 and M2, and two from the daughter, D1 and D2) were obtained and characterized [GenBank accession numbers: Daughter 1993-D1 (code WCD32P0793): DQ487188; Daughter 1995-D2 (code WCD32P1195): DQ487189; Mother 1993-M1 (code WCM32P0793): DQ487190; Mother 1996-M2 (code WCM32P0896): DQ487191]. The analysis of the four sequences indicated that all viral genes were intact, with no major deletions or premature stop codons that could have easily explained the slow progress of the disease, although the contribution of minor sequence variations (point mutations, insertions or deletions) could not be excluded. Several deletions and insertions were observed between mother and child strains, e.g., in gag p6 protein region (Fig. 1), and also in vif, vpu. The frequency of the p6 insertion was determined by analysis of the proviral sequences of the gag gene region. The insertion was present in 0 out of 20 clones from the mother and 12 out of 12 clones from the child. Thus, this insertion probably evolved during the course of the infection in the child or possibly circulated in the mother as a minor form.
FIG. 1.
Amino acid sequences of the Gag p6, Env V2, and Nef regions featuring length polymorphism between the isolates from the mother and the child. Consensus subtype B sequences were included in the alignments. Dashes represent identical sequences; dots represent deletions.
Comparison of two strains from the mother from 1993 and 1995 revealed insertions and deletions only in the highly variable domains of the env gene. The genomic comparison of two strains from the child showed the length polymorphism in env and nef (Fig. 1). V2 extensions were observed in the samples derived from the child and the V2 domains were longer as compared to those derived from the mother. Moreover, multiple N-glycosylation sites were determined within these extensions. Analysis of the V3 loop of the gp120 proteins predicted CCR5 usage as a major coreceptor in all isolates, and the GPGR motif in the principal neutralization domain was conserved in all isolates.
Phylogenetic analyses of these four sequences, along with genomic subtype B reference strains, demonstrated the linkage between mother and child isolates; furthermore, the bootstrap values indicated that the sequences from the mother and daughter clustered separately. The genetic distance (percentage of identity) between the strains derived from the mother and the child at the same time point after 11 years of separate evolution was 94% throughout the genome, ranging from 87% in vpu gene, and 88% in the env, to 97% in the LTR. The genetic distance between two strains derived from the mother was 97% (ranging from 95% in env to 100% in rev and vpu). The genetic distance between two strains from the child was 97% (ranging from 95% in env and nef to 99% in rev and tat). At the amino acid level, the percentage of identities in particular genes between two strains derived from the mother ranged from 93% (env) to 100%; strains from the child ranged from 92% to 99%; and the identities between strains from the mother and child ranged from 80% to 96%. The ratio between synonymous substitution per potential synonymous site (ds) and nonsynonymous substitution per potential nonsynonymous site (dn) was on average greater when strains from the same individual were compared, and the values differed for particular genes. In general, the average ds/dn from all genes was higher in the genomes from the same individuals, 3018 ± 1605 between two strains from the mother, 3588 ± 2832 from the child, and 2263 ± 1048 between the second strains from the mother and the child. As expected, the lowest ds/dn values were observed in the most variable env gene.
Serial sequences from this HIV-1-infected mother–child pair afforded us the opportunity to examine the immune selection of HIV-1 sequences years after transmission between individuals. The sequences of characterized epitopes deposited in the Los Alamos HIV database were identified in all four genomes by using the individual's HLA type and viral sequences. The numbers of epitopes presented in the context of particular MHC class I molecules specific either for the mother, the child, or for both of them were analyzed in relation to the clade B consensus reference sequences. The analysis was focused on the possible development of escape mutations. Therefore, differences in the sequences of the putative CTL epitopes in the strains from the mother and the child were recorded, while epitopes that were either conserved or mutated in both mother and child samples were not taken into account. Indeed, no significant differences in the frequency of development of escape or reverse mutations were noted in the epitopes related either to the shared HLA alleles or alleles unique for the mother. On the other hand, the analysis of 87 epitopes restricted to HLA alleles of the child revealed 10 new mutations in the viral D2 strain (two of them were not present in the D1 strain) as compared to the sequences from the mother, while only one mutation unique for the strain (M2) from the mother was recorded. The sequences of these epitopes are presented in Table 2.
Table 2.
CTL Epitopes Presented in the Context of MHC Class I Allelesa
| Gene | HLA restriction | Epitope sequence | D2 isolate | M2 isolate |
|---|---|---|---|---|
| vpr | A*0201 | AIIRILQQL | AILRILQQL | AIIRILQQL |
| gag | A2 | HAGPIAPGQMREPRG | QAGPIAPGQMRDPRG | HAGPIAPGQMREPRG |
| pol | A2 | ALVEICTEM | ALIEICTEM | ALVEICTEM |
| LWVTVYYGVPVWKEATTTL | WWVTVYYGVPVWKEATNTL | LWVTVYYGVPVWKEATTTL | ||
| env | A2 | FCA | FCA | FCA |
| pol | A2, A*0201 | VIYQYMDDL | CIYQYMDDL | VIYQYMDDL |
| env | A2, A*0201 | LLNATAIAV | LLNAIAIAV | LLNATAIAV |
| env | A2, A2.1 | KLWVTVYYGV | NWWVTVYYGV | KLWVTVYYGV |
| env | B14 | VERYLKDQQL | VERYLRDQQL | VERYLKDQQL |
| env | B14, B*1402 | ERYLKDQQL | ERYLRDQQL | ERYLKDQQL |
| env | Cw8 | CTNVSTVQC | CQNVSTVQC | CTNVSTVQC |
| env | A2.1 | WLWYIKIFI | WLWYIKIFI | WLWYIRIFI |
The table lists sequences of CTL epitopes that were conserved in the mother's viral strain (M2) and mutated in the child's isolate (D2), or were conserved in the child's isolate (D2) and mutated in the mother's isolate (M2). Amino acid residues in the child's and mother's viral peptide sequences distinct from the characterized epitope reference sequences are in bold. The number of mutations observed in the child's HIV-l isolate (D2), but not in the mother's M2 isolate, was significantly higher than the number of mutations unique for the mother in the M2 isolate (10 vs. 1 out of 87, χ2 < 0.01 determined by the Fisher's test).
To functionally analyze the association between the immune responses, T cell escape, and virus mutations in the child, we selected 15-mers overlapping peptides from the clade B nef consensus sequence1 which correspond to the sequences that contained mutations in the child. We then determined the CTL responses of PBMCs from the child against those peptides by ELISPOT assay (Table 3). Four strong positive responses were seen, suggesting those mutations observed in virus from the child but not in the virus from the mother were likely due to the CTL pressure. The functional study of the immune response against nef revealed a strong association between T cell responses and escape variants. Since the child was on treatment it is hard to associate the T cell responses with the clinical outcome. But broad nef responses to several epitopes with wild-type sequences might contribute to delayed progression at an early stage (i.e., before the virus mutated).
Table 3.
ELISPOT Responses to HIV-1 Epitopes Derived from the Nef Protein
| Peptide ID | Peptide sequence | Peptide sequence D2 isolate | Spotsa |
|---|---|---|---|
| NEG | 0 | ||
| NEG | 0 | ||
| nef-5139 | MGGKWSKRSVVGWPT | MGGKWSKSSIVGWPA | 0 |
| nef-5140 | WSKRSVVGWPTVRER | WSKSSIVGWPAVRDR | 0 |
| nef-5141 | SVVGWPTVRERMRRA | SIVGWPAVRDRMRRA | 270 |
| nef-5142 | WPTVRERMRRAEPAA | WPAVRDRMRRAEPAA | 350 |
| nef-5156 | PVRPQVPLRPMTYKA | PVRPQVPLRPMTFKA | 410 |
| nef-5157 | QVPLRPMTYKAAVDL | QVPLRPMTFKAAVDL | 10 |
| nef-5158 | RPMTYKAAVDLSHFL | RPMTFKAAVDLSHFL | 110 |
| nef-5160 | VDLSHFLKEKGGLEG | VDLSHFLKEMGGLDG | 0 |
| nef-5161 | HFLKEKGGLEGLIYS | HFLKEMGGLDGLIYS | 0 |
| nef-5162 | EKGGLEGLIYSQKRQ | EMGGLDGLIYSQRRQ | 0 |
| nef-5169 | FPDWQNYTPGPGIRY | FPDWQNYTPGPGTRF | 0 |
| nef-5170 | QNYTPGPGIRYPLTF | QNYTPGPGTRFPLTF | 0 |
| nef-5171 | PGPGIRYPLTFGWCF | PGPGTRFPLTFGWCF | 450 |
| nef-5177 | VEEANEGENNSLLHP | VEEANEGETNNLLSP | 0 |
| nef-5178 | NEGENNSLLHPMSLH | NEGETNNLLSPMSLH | 0 |
| nef-5179 | NNSLLHPMSLHGMDD | TNNLLSPMSLHGMED | 0 |
| nef-5180 | LHPMSLHGMDDPERE | LSPMSLHGMEDPEKE | 0 |
| nef-5181 | SLHGMDDPEREVLEW | SLHGMEDPEKEVLVW | 0 |
| nef-5184 | LEWKFDSRLAFHHMA | LEWKFDSRLAFHHIA | 0 |
| nef-5185 | FDSRLAFHHMARELH | FDSRLAFHHIAKELH | 0 |
| nef-5186 | LAFHHMARELHPEYY | LAFHHIAKELHPEYY | 0 |
Spot forming units/million PBMCs.
The CTL responses of the childís PBMCs against selected overlapping peptides from Clade B nef consensus sequence peptides1 were determined by ELISPOT assay. Sequences of the synthetic peptides used in the assay (peptide sequence) and the sequences derived from the child's second isolate are presented. Amino acid residues in the child's viral peptide seqences distinct from the consensus sequences are in bold.
The data presented in this study characterize the evolution of the vertically transmitted HIV-1 strain. Although more full-genome sequencing of multiple mother–child pairs would be helpful, we have clearly documented the genetic distance throughout the genome between strains derived from the mother and the child after 8 years of separate evolution. On the other hand, phylogenetic analysis revealed the epidemiologic linkage and the genetic relationship between the strains compared to a panel of reference subtype B sequences. The fact that the ratio between synonymous substitution per potential synonymous site (ds) and nonsynonymous substitution per potential nonsynonymous site (dn) was on average larger when strains from the same individual were compared indicates that the strains from the mother and the child diverged due to the distinct selection pressure.
Previous studies revealed the immunoediting of the viral antigens under selective pressure after vertical transmission of the virus. Those findings were obtained by the analysis of particular genes or their fragments harboring only selected epitopes. In this study we have analyzed and compared to each other complete HIV-1 genomes and identified a higher number of mutations in the transmitted strains in epitopes presented in the context of MHC class I alleles that were unique for the child than in the maternal viral sequences. This examination was based on the CTL epitopes that have been identified and are in the Los Alamos HIV Database. The results of this analysis suggest the development or selection of the virus variants in the child as a consequence of the different selection pressure, as one-half of MHC class I alleles are not shared. This resulted in selection of viral variants harboring variations in the CTL epitopes presented in the context of MHC class I molecules from the child.
The data presented in this study indicate that slow disease progression is not always related to infection with an apparently defective virus since all viral genes were intact with no large deletions or premature stop codons. The replicative competence of the HIV-1 strain is suggested by the viral load (29,000–40,000 range) in the course of infection. This medium viral load also suggests that the infection was not effectively controlled by the immune system. We have focused our analysis on small and unusual sequence variations that could contribute to the very slow disease progression. As expected in LTNP infection, the analysis of the env gene V3 region predicted CCR5 coreceptor usage. The tetrameric tip GPGR was conserved in the V3 principal neutralization domain. Extended V2 loops and multiple N-glycosylation sites, present in the isolates, can be related to the slow progression of the disease as has been described.16,17 Moreover, it has been shown that these extensions correlate with the usage of the CCR5 coreceptor.18 Interestingly, the shortest V2 loop was determined in the second isolate from the mother, obtained when her CD4+ cell count declined.
Five amino acid insertions into the gag gene region coding the p6 peptide to the site adjacent to the PTAPP motif have been found to be important for interactions with the Tsg101 cellular protein;19 five amino acid insertions into that gag region were found in the child's strains but not in the mother's. The biological significance of the insertion to the adjacent site is not clear, but a recent study has shown that the frequent presence of p6 duplication was not associated with differences in transmission, viral load, or disease progression in infants. Interestingly, the R77Q variation in the Vpr protein was present in all isolates. This sequence variation was associated with non-progressive infection; it may contribute to slow disease progression because it dramatically reduces cellular apoptosis.20 More sequence variations were found also in other regulatory genes (variation in the splicing site in the tat gene, in the mother only) but their biological significance remains to be evaluated.
In this study of HIV-1 strains derived from a mother–child pair of LTNPs the viral strains were not obviously attenuated and not effectively suppressed by a CTL response. The most plausible explanation for the slow progress of the disease in both individuals is a combination of factors involving genetic variations in the virus genomes that could influence viral fitness, and the interaction of the viral sequences with the immune response and genetic background of the mother and child. Taken together, this study also highlights the potential of full genome analysis of virion RNA in the investigation of virus–host interrelations and immune selection during HIV-1 evolution.
ACKNOWLEDGMENTS
We thank the study subjects, M. Shudt, and the Wadsworth Center Molecular Genetics Core Laboratory for DNA sequence analysis, S. Philpott (Wadsworth Center) for CCR5 usage prediction, and J. Vorlicek, Institute of Physiology, Prague, for statistical analysis. This study was funded by NIH Research Grant D43 TW000233 provided by the Fogarty International Center, the National Institute on Drug Abuse, and the National Heart Lung and Blood Institute, by RO1-AI-42555 from the U.S. National Institute of Allergy and Infectious Diseases and by Grant AVOZ50520514 from the Academy of Sciences of the Czech Republic. T.D. and S.R.J. are funded by Medical Research Council, UK. Y.H. Zhang is funded by Drs. R.C. and E.Y. Lee Charitable Foundation, Hong Kong.
Footnotes
This article has been cited by:
1. Terese L. Katzenstein, Ann Berith Petersen, Louise Bruun Jørgensen, Tine Johanne Strand, Tina Vasehus Madsen, Jan Gerstoft. 2008. Seventeen-Year-Old Mother-to-Child HIV Type 1 Transmission Identified by Phylogeny and Signature PatternsSeventeen-Year-Old Mother-to-Child HIV Type 1 Transmission Identified by Phylogeny and Signature Patterns. AIDS Research and Human Retroviruses 24:8, 1117-1120. [Abstract] [PDF] [PDF Plus]
REFERENCES
- 1.Leitner T, Foley B, Hahn B, et al., editors. HIV Sequence Compendium 2005. Los Alamos National Laboratory; Los Alamos, NM: 2005. [Google Scholar]
- 2.Dickover RE, Garratty EM, Plaeger S, Bryson YJ. Perinatal transmission of major, minor, and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J Virol. 2001;75:2194–2203. doi: 10.1128/JVI.75.5.2194-2203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhofstede C, Demecheleer E, De Cabooter N, Gaillard P, Mwanyumba F, Claeys P, Chohan V, Mandaliya K, Temmerman M, Plum J. Diversity of the human immunodeficiency virus type 1 (HIV-1) env sequence after vertical transmission in mother. J Virol. 2003;77:3050–3057. doi: 10.1128/JVI.77.5.3050-3057.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchbinder S, Vittinghoff E. HIV-infected long-term nonprogressors: Epidemiology, mechanisms of delayed progression, and clinical and research implications. Microbes Infect. 1999;1:1113–1120. doi: 10.1016/s1286-4579(99)00204-x. [DOI] [PubMed] [Google Scholar]
- 5.Alexander L, Weiskopf E, Greenough TC, Gaddis NC, Auerbach MR, Malim MH, O'Brien SJ, Walker BD, Sullivan JL, Desrosiers RC. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J Virol. 2000;74:4361–4376. doi: 10.1128/jvi.74.9.4361-4376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang G, Burger H, Chappey C, Rowland-Jones S, Visosky A, Chen C-H, Moran T, Townsend L, Murray M, Weiser B. Analysis of transition from long-term nonprogressive to progressive infection identifies sequences that may attenuate HIV type 1. AIDS Res Hum Retroviruses. 2001;17:1395–1404. doi: 10.1089/088922201753197060. [DOI] [PubMed] [Google Scholar]
- 7.Ganeshan S, Dickover RE, Korber BT, Bryson YJ, Wolinsky SM. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goulder PJ, Brander C, Tang Y, Tremblay C, Colbert RA, Addo MM, Rosenberg ES, Nguyen T, Allen R, Trocha A, Altfeld M, He S, Bunce M, Funkhouser R, Pelton SI, Burchett SK, McIntosh K, Korber BT, Walker BD. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 9.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 10.Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 11.Pastori C, Weiser B, Barassi C, Uberti-Foppa C, Ghezzi S, Longhi R, Calori G, Burger H, Kemal K, Poli G, Lazzarin A, Lopalco L. Long lasting CCR5 internalization by antibodies in a subset of long term non progressors: A possible protective effect against disease progression. Blood. 2006;107:4825–4833. doi: 10.1182/blood-2005-06-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang G, Zhu G, Burger H, Keithly JS, Weiser B. Minimizing DNA recombination during long RT-PCR. J Virol Methods. 1998;76:139–148. doi: 10.1016/s0166-0934(98)00133-5. [DOI] [PubMed] [Google Scholar]
- 13.Philpott S, Weiser B, Anastos K, Kitchen CM, Robison E, Meyer WA, 3rd, Sacks HS, Mathur-Wagh U, Brunner C, Burger H. Preferential suppression of CXCR4-specific strains of HIV-1 by antiviral therapy. J Clin Invest. 2001;107:431–438. doi: 10.1172/JCI11526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van de Peer Y, De Wachter R. TREECON for Windows: A software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 15.Kaul R, Dong T, Plummer FA, Kimani J, Rostron T, Kiama P, Njagi E, Irungu E, Farah B, Oyugi J, Chakraborty R, MacDonald KS, Bwayo JJ, McMichael A, Rowland-Jones SL. CD8(+) lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J Clin Invest. 2001;107:1303–1310. doi: 10.1172/JCI12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shioda T, Oka S, Xin X, Liu H, Harukuni R, Kurotani A, Fukushima M, Hasan MK, Shiino T, Takebe Y, Iwamoto A, Nagai Y. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: Association of V2 extension with slow disease progression. J Virol. 1997;71:4871–4881. doi: 10.1128/jvi.71.7.4871-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang B, Spira TJ, Owen S, Lal RB, Saksena NK. HIV-1 strains from a cohort of American subjects reveal the presence of a V2 region extension unique to slow progressors and non-progressors. AIDS. 2000;14:213–223. doi: 10.1097/00002030-200002180-00002. [DOI] [PubMed] [Google Scholar]
- 18.Masciotra S, Owen SM, Rudolph D, Yang C, Wang B, Saksena N, Spira T, Dhawan S, Lal RB. Temporal relationship between V1V2 variation, macrophage replication, and coreceptor adaptation during HIV-1 disease progression. AIDS. 2002;16:1887–1898. doi: 10.1097/00002030-200209270-00005. [DOI] [PubMed] [Google Scholar]
- 19.Colgrove RC, Millet A, Bauer GR, Pitt J, Welles SL. Gag-p6 Tsg101 binding site duplications in maternal-infant HIV infection. AIDS Res Hum Retroviruses. 2005;21:191–199. doi: 10.1089/aid.2005.21.191. [DOI] [PubMed] [Google Scholar]
- 20.Lum JJ, Cohen OJ, Nie Z, Weaver JG, Gomez TS, Yao XJ, Lynch D, Pilon AA, Hawley N, Kim JE, Chen Z, Montpetit M, Sanchez-Dardon J, Cohen EA, Badley AD. Vpr R77Q is associated with long-term nonprogressive HIV infection and impaired induction of apoptosis. J Clin Invest. 2003;111:1547–1154. doi: 10.1172/JCI16233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang G, Burger H, Grimson R, Tropper P, Nachman S, Moore R, Reyelt C, Hutcheon N, Baker D, Weiser B. Maternal plasma HIV-1 RNA level: A determinant and projected threshold for mother-to-child transmission. Proc Natl Acad Sci USA. 1995;92:12100–12104. doi: 10.1073/pnas.92.26.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]